Abstract
We identified and characterized a novel staphylococcal enterotoxin-like putative toxin, which is named SER. Nucleotide sequencing analysis of the ser gene revealed that ser was most closely related to the seg gene. The ser gene product, SER, was successfully expressed as a recombinant protein in an Escherichia coli expression system, and recombinant SER (rSER) showed significant T-cell stimulation activity. The SER production in ser-harboring Staphylococcus aureus strains was confirmed by Western blot analysis using anti-rSER antibody. Moreover, ser was seen to be encoded by at least two types of plasmids. In particular, one kind of plasmid encoding the ser gene has been known as a sed- and sej-carrying pIB485-related plasmid.
Staphylococcus aureus is an important pathogen in humans and animals because this bacterium produces a wide variety of exotoxins, including staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin 1 (TSST-1) (8, 15). SEs are emetic toxins and are causative agents in staphylococcal food poisoning in humans, although their mechanisms of emetic activity are not fully understood (5). Also, SEs and TSST-1 are superantigens (SAgs), which have the ability to stimulate large populations of T cells that have a particular Vβ element of the T-cell receptor (TCR). This gives rise to symptoms that are characteristic of toxic shock syndrome in humans. Five major serological types, SEA through SEE, have been characterized (5). However, in recent years, many new types of SE (i.e., SEG, SEH, SEI, SEJ, SEK, SEL, SEM, SEN, SEO, SEP, and SEQ) have been reported (10, 12, 13, 17, 22, 23, 24, 27, 30, 31). In addition, the determination of the complete genome sequences of several S. aureus strains has revealed that they maintain many SAg-related genes (staphylococcal exotoxin-like genes set1 to set26) in their genomes (2, 13, 29). Staphylococcal SAgs constitute quite a large family of structurally related proteins.
On the other hand, it has been known that these SAg genes are associated with mobile genetic elements such as pathogenicity islands, prophages, and plasmids. SEB, SEC, SEG, SEI, SEM, SEN, SEO, SEK, SEL, SEQ, and TSST-1 are encoded by pathogenicity islands (2, 12, 13, 14, 30). SEA, SEE, and SEP are encoded by prophages (6, 7, 13), whereas SED and SEJ are encoded by a plasmid known as pIB485 (3, 31). These facts imply that these SAg genes transfer between staphylococcal strains by horizontal transfer. There is a possibility that these mobile genetic elements play an important role in the evolution of S. aureus as a pathogen. To clarify the pathogenicity or virulence of S. aureus, it is important to bring to light the full extent of the diversity of staphylococcal SAgs. Here we describe a new staphylococcal superantigen-like putative toxin that is phylogenetically related to SEs, named SER.
In September of 1997, an outbreak of food poisoning occurred at a lunch box shop in the Fukuoka prefecture of Japan. Ten persons ate food prepared by the shop, and four of them developed nausea, vomiting, and diarrhea within 1.5 to 5 h. Many S. aureus isolates were obtained from patient feces and swabs of the lunch boxes (8.0 × 106 isolates/lunch box swab). Other food-borne pathogens were not isolated. Four S. aureus isolates (Fukuoka 5, Fukuoka 6, Fukuoka 7, and Fukuoka 8) were subjected to PCR analysis for detection of SE genes according to the method of Omoe et al. (21). However, no isolates harbored sea, seb, sec, sed, see, seg, seh, or sei genes. We hypothesized that this outbreak was caused by an unidentified SE and so started a genetic analysis of these staphylococcal isolates. We initially tested whether or not these isolates harbored known enterotoxin-like nucleotide sequences. The bacterial strains and plasmids that were used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | SE genotype or characteristic | Source or reference |
|---|---|---|
| S. aureus | ||
| Isolates from SE-unidentified food poisoning outbreak | ||
| Fukuoka 5 | sej ser, carrying pF5 | Isolated from swab of lunch box |
| Fukuoka 6 | sej ser, carrying pF6 | Isolated from patient feces |
| Fukuoka 7 | sej ser, carrying pF7 | Isolated from patient feces |
| Fukuoka 8 | —a | Isolated from feces obtained from worker at lunch box shop |
| sed-harboring strain | ||
| 1151-7NG | sed sej ser, carrying p1151 | M. S. Bergdoll |
| 196E | sea sed sej ser, carrying p196 | M. S. Bergdoll |
| FRI361 | sec2 sed seg sei sej ser, carrying p361 | M. S. Bergdoll |
| 11727 | sea sed sej ser, carrying p727 | Food poisoning |
| 11740 | sea sed sej ser, carrying p740 | Food poisoning |
| IVM7 | sed seg sei sej ser, carrying pI7 | Healthy human |
| IVM16 | sed seg sei sej ser, carrying pI16 | Healthy human |
| IVM46 | sed seg sei sej ser, carrying pI46 | Healthy human |
| seg reference strain | ||
| Fukuoka 1 | seg sei | 21 |
| Plasmids | ||
| pF5 | sej ser | This work |
| pF6 | pF5-like plasmid, sej ser | This work |
| pF7 | pF5-like plasmid, sej ser | This work |
| p1151 | pIB485-like plasmid, sed sej ser | This work |
| p196 | pIB485-like plasmid, sed sej ser | This work |
| p361 | pIB485-like plasmid, sed sej ser | This work |
| p727 | pIB485-like plasmid, sed sej ser | This work |
| p740 | pIB485-like plasmid, sed sej ser | This work |
| pI7 | pIB485-like plasmid, sed sej ser | This work |
| pI16 | pIB485-like plasmid, sed sej ser | This work |
| pI46 | pIB485-like plasmid, sed sej ser | This work |
| E. coli | ||
| Strains | ||
| DH5α | Toyobo | |
| KRX1 | DH5α carrying pKRX1, Aprser | This work |
| Plasmids | ||
| pBluescriptSK− | Apr | Stratagene |
| pGEM3Zf(+) | Apr | Promega |
| pGEX-6P-1 | Apr, GST fusion expression vector | Pharmacia |
| pKOG4 | Apr, pGEM3Zf(+) with seg | 21 |
| pKOH1 | Apr, pGEM3Zf(+) with seh | 21 |
| pKOI6 | Apr, pGEM3Zf(+) with sei | 21 |
| pKO311 | Apr, pBluescript SK− with sej and ser | This work |
| pKOJ1 | Apr, pGEM3Zf(+) with sej | This work |
| pKOR1 | Apr, pGEM3Zf(+) with ser | This work |
| pKRX1 | Apr, pGEX-6P-1 with ser | This work |
—, negative for sea, seb, sec, sed, see, seg, seh, sei, sej, and ser.
Identification of a seg-like sequence in S. aureus strain Fukuoka 5.
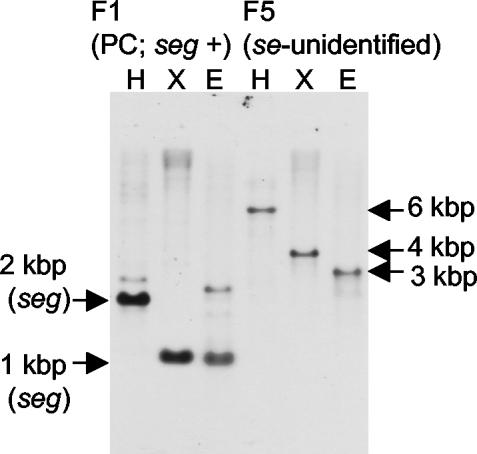
HindIII-, XbaI-, and EcoRI-digested total DNA (2 μg) of S. aureus strain Fukuoka 5 (food poisoning outbreak-related strain, SE unidentified) was subjected to Southern blot analysis (25) using digoxigenin (DIG)-labeled seg, seh, and sei probes digested from pKOG4, pKOH1, and pKOI6, respectively (21). The membranes were washed, and the signals were detected by chemiluminescence according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). The Fukuoka 5 strain contained a nucleotide sequence that hybridized to the seg probe, a 6.0-kbp HindIII fragment, a 4.0-kbp XbaI fragment, and a 3.0-kbp EcoRI fragment, under low-stringency conditions (37°C) (Fig. 1). Under high-stringency conditions (65°C), the seg probe hybridized to only homologous nucleotide sequence (seg) in the Fukuoka 1 strain (positive control; seg and sei positive) total DNA, and no signal was observed in the Fukuoka 5 strain. The seh and sei probes did not hybridize to Fukuoka 5 total DNA under low-stringency conditions (data not shown).
FIG. 1.
Southern blot analysis of SE-unidentified S. aureus Fukuoka 5 isolate under the low-stringency condition. HindIII (H)-, XbaI (X)-, and EcoRI (E)-digested total DNA of the S. aureus Fukuoka 5 strain (F5, food poisoning outbreak-related strain, SE unidentified) and the Fukuoka 1 strain (F1, positive control [PC]; seg and sei positive) were subjected to Southern hybridization using a seg probe.
Cloning and sequencing analysis of the ser gene.
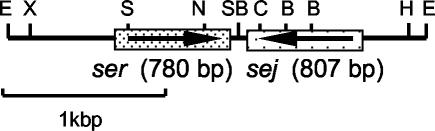
To clone the seg-like sequence in the Fukuoka 5 strain, a total DNA library was constructed using EcoRI-digested Fukuoka 5 DNA and lambda phage vector λZAPII (Stratagene, La Jolla, Calif.). The library was screened by plaque hybridization with a DIG-labeled seg probe under low-stringency conditions. About 104 clones were screened, and three positive clones were obtained. Positive clones were converted to plasmids by in vitro excision using a Rapid Excision Kit (Stratagene) according to the manufacturer's instructions. To verify the collected clones, these positive plasmids were labeled and subjected to hybridization to HindIII-, XbaI-, or EcoRI-digested Fukuoka 5 total DNA. One clone, pKO311, hybridized to a 6.0-kbp HindIII fragment, a 4.0-kbp XbaI fragment, and a 3.0-kbp EcoRI fragment of Fukuoka 5 total DNA. Thus, we concluded that pKO311 was a true positive clone. Nucleotide sequences of pKO311 were obtained for both strands by primer walking using an automatic DNA sequencer ABI 310 (Perkin-Elmer Applied Biosystems, Foster City, Calif.). pKO311 contained 2,751 bp of the insert. This insert contained two open reading frames (ORFs), and these ORFs could be transcribed in opposite directions. A search of BLAST at the DDBJ showed that one ORF was identical to a previously reported SE gene, sej. However, another ORF showed only a 65.9% homology with the seg gene. We concluded that this ORF is a new putative SE gene and designated it ser (Fig. 2). Figure 3 shows the nucleotide and deduced amino acid sequences for ser. The ser ORF encoded a polypeptide 259 amino acids in length. In the putative regulatory region of ser, we identified a potential promoter sequence and potential ribosomal binding site using the computer program Genetyx-Mac version 8.0 (Genetyx, Tokyo, Japan). The N-terminal signal peptide sequence of SER was predicted using the online signal peptide prediction software SignalP (http://www.cbs.dtu.dk/services/SignalP) (20). The mature form of SER was predicted to have a molecular weight of 27,049.
FIG. 2.
Diagram of cloned DNA containing SE genes. Subclone pKO311 contained 2,751 bp of insert. This insert contained two kinds of SE genes, sej and ser. The directions of transcription of the genes are shown by the arrows. The letters refer to sites cut by the restriction endonucleases: B, BglII; C, ClaI; E, EcoRI; H, HindIII; N, NdeI; S, StuI; SB, SnaBI; X, XbaI.
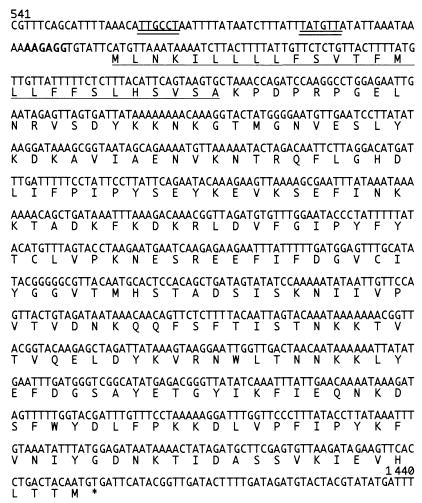
FIG. 3.
Nucleotide sequence of ser. Nucleotide sequence corresponding to positions 541 to 1440 of 2.8-kbp inset of pKO311 is shown. The putative ribosomal binding site sequence is in boldface. The putative promoter sequences are indicated by double underline. The deduced amino acid sequence with a putative signal peptide (underlined) and its stop codon (asterisk) are indicated below the nucleotide sequence.
Expression of recombinant SER in E. coli and T-cell-stimulating activity of recombinant SER.
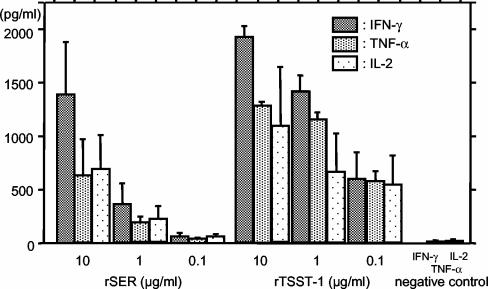
In order to construct the recombinant SER (rSER) expression plasmids, PCR primers were designed to amplify the gene fragment corresponding to the mature form of SER (SERS1, the sequence includes a 5′ BamHI site, 5′-CCCCGGATCCAAACCAGATCCAAGGCCTGGAG-3′; and SERAS2, the sequence includes a 5′ EcoRI site, 5′-CCCCGAATTCTCACATTGTAGTCAGGTGAACTT-3′).The ser gene was amplified by PCR using Pyrobest DNA polymerase (Takara, Kyoto, Japan), and the ser fragment was then subcloned into pGEM3Zf(+) (Promega, Madison, Wis.) and designated pKOR1. The nucleotide sequence of the ser gene in pKOR1 was verified by an ABI 310 automatic DNA sequencer (Perkin-Elmer Applied Biosystems). The ser fragment was then digested from pKOR1 with BamHI and EcoRI and was subcloned into the pGEX-6P-1 (Amersham Pharmacia Biotech, Piscataway, N.J.) glutathione S-transferase (GST) fusion expression vector. The resulting plasmids containing ser were named pKRX1. Expression, purification of GST-fused rSER, and the cleavage and removal of the GST tag from rSER were performed according to the method of Omoe et al. (21). The resulting mature rSER has five additional amino acid residues, GPLGS, at the N terminus. T-cell-stimulating activity of purified rSER was assessed by induction of gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin 2 (IL-2) production in murine splenocytes. Splenocytes isolated from specific-pathogen-free female C57BL/6 CrSIc mice (CLEA, Kanagawa, Japan) were stimulated by several concentrations of rSER or recombinant TSST-1 (rTSST-1). The cells were incubated at 37°C for 72 h in a humidified 5% CO2 atmosphere. Culture supernatants were harvested for use in IFN-γ, TNF-α, and IL-2 assays. Production levels of IFN-γ and TNF-α were determined by means of sandwich enzyme-linked immunoabsorbent assays (ELISA) (18, 19). The IL-2 concentration was determined by using the mouse IL-2 ELISA kit (Biosource International, Camarillo, Calif.) according to the manufacturer's instructions. rSER induced significant amounts of IFN-γ, TNF-α, and IL-2 in stimulated murine splenocyte culture supernatants (Fig. 4). This result suggests that SER would act as a superantigen and stimulate T cells via TCR Vβ and major histocompatibility complex class II binding. Further analyses of the molecular basis of T-cell stimulation activity of SER, such as the induction of selective expansion of T cells bearing particular TCR Vβ regions and the requirement for major histocompatibility complex class II molecules to stimulate T cells, is needed. In addition, the rSER expressed by our system induces cytokine production in murine splenocytes, although rSER has an additional five amino acid residues derived from the expression vector sequence at the N terminus. Many biologically active SEs have been successfully expressed in a similar manner previously (11, 21). It seems that the additional five amino acid residues do not have a significant effect on the T-cell-stimulating activity of SEs.
FIG. 4.
Production of IFN-γ, TNF-α, and IL-2 by murine splenocytes in response to stimulation with rSER. Mouse splenocytes isolated from C57BL/6 mice were stimulated by various concentrations of rSER and rTSST-1 (positive control). Production of IFN-γ, TNF-α, and IL-2 was measured by sandwich ELISA. Negative control cells were stimulated with phosphate-buffered saline rather than SAgs.
mRNA transcription of ser and sej in strain Fukuoka 5.
Total RNAs isolated from S. aureus Fukuoka 5 at 6 and 9 h after inoculation were subjected to Northern hybridization analysis according to the method of Sambrook et al. (25). To obtain the sej probe, PCR primers were designed to amplify the gene fragment corresponding to the mature form of SEJ (SEJS1, the sequence includes a 5′ BamHI site, 5′-CCCCGGATCCGATAGCAAAAATGAAAC-3′; SEJAS2, includes a 5′ EcoRI site, 5′-CCCCGAATTCCTAAACCAAAGGTAGACTTATTA-3′). The sej gene was amplified and subcloned into pGEM3Zf(+) (Promega). The resulting plasmid was designated pKOJ1. DIG-labeled antisense RNA probes were synthesized by using a DIG RNA labeling kit (Roche Diagnostics) employing pKOR1 (ser probe) and pKOJ1 (sej probe). Northern analysis using an ser antisense RNA probe confirmed 1.3- and 0.8-kbp transcripts. As well, an sej antisense probe hybridized 1.3- and 0.6-kbp transcripts (data not shown). The ser and sej mRNA might be transcribed from multiple promoters.
Preparation of polyclonal antibody and production of SER in S. aureus Fukuoka strains.
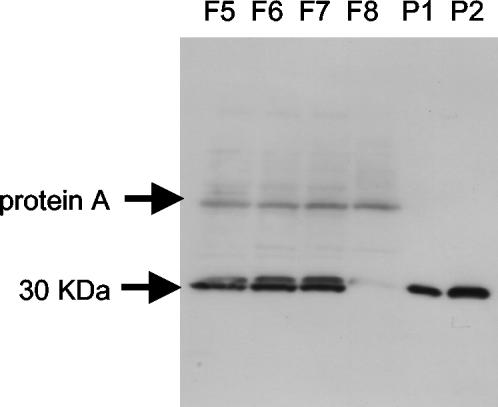
Anti-rSER serum was prepared by immunizing rabbits with purified rSER according to the method described by Shinagawa et al. (26). Titers of antiserum were monitored by means of ELISA. Monospecific rabbit anti-rSER antibody was affinity purified from hyperimmune serum by using an rSER-coupled Sepharose column. The anti-rSER antibody recognized only rSER, and no cross-reactivity with other SEs (SEA through SEE and SEG through SEI) was observed by Western blot analysis (data not shown). Using the anti-rSER antibody, we examined the SER productivity of S. aureus isolates from the food poisoning outbreak in Fukuoka. Culture supernatants of S. aureus strains were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes (Bio-Rad, Richmond, Calif.) according to the method described by Towbin et al. (28). The reactive signals were detected using an enhanced chemiluminescence system (Amersham Pharmacia Biotech) according to the manufacturer's instructions. These food poisoning-related isolates, strains Fukuoka 5, Fukuoka 6, and Fukuoka 7, produced and secreted significant amounts of SER into the culture supernatants (Fig. 5). Although the sizes of native and recombinant SER seem somewhat higher than the predicted molecular mass (27 kDa), it is not uncommon for SEs to show a higher molecular weight by sodium dodecyl sulfate-polyacrylamide gel electrophoresis than the amino acid sequence would suggest (22). One isolate, Fukuoka 8, did not produce SER. The lack of the ser gene in S. aureus strain Fukuoka 8 was confirmed by Southern hybridization (data not shown). The SE-unidentified food poisoning outbreak-related Fukuoka strains, which harbor ser and sej genes, were able to produce significant amounts of staphylococcal enterotoxin-like putative toxin, SER. There is a possibility that SER was the cause, at least in part, of this food poisoning outbreak, although we could not deny the possible involvement of another SE, SEJ. In this study, we have not assessed the emetic activity of SER by using monkeys. Monkeys have typically been the primary animal models used to assess the emetic activity of SEs (4), although the use of monkeys is severely restricted by their high cost and limited availability. Orwin et al. (23) recently reported that a novel SE-like SAg, named SEQ, lacked emetic activity in young pigtail monkeys. They suggested that it might be inappropriate to refer to SEQ as an enterotoxin, because SEs are defined by their capabilities to cause emesis after being orally administered to monkeys. Newly reported SEs, such as SEJ, SEK, SEL, SEM, SEN, SEO, and SEP, were so designated based on their sequence similarity with classical SEs, and they were not assessed as having emetic activity in monkeys. At present, we have only circumstantial evidence that SER is an emetic toxin. In the future, quantitative elucidation of the emetic activity of SERs and other novel SEs should be examined using monkeys in order to accurately elucidate the capability to cause food poisoning and to assess the risk related to newly reported SEs.
FIG. 5.
Confirmation of secretion of SER by S. aureus isolates by Western blot analysis. Using anti-rSER antibody, we examined SER productivity of S. aureus isolates from a food poisoning outbreak in Fukuoka. Culture supernatants from Fukuoka 5 (F5), Fukuoka 6 (F6), Fukuoka 7 (F7), and Fukuoka 8 (F8) were subjected to Western blot analysis. An approximately 30-kDa signal was detected by anti-rSER antibody. P1, 25 ng of rSER per well; P2, 50 ng of rSER per well.
ser and sej genes exist on two kinds of plasmids.
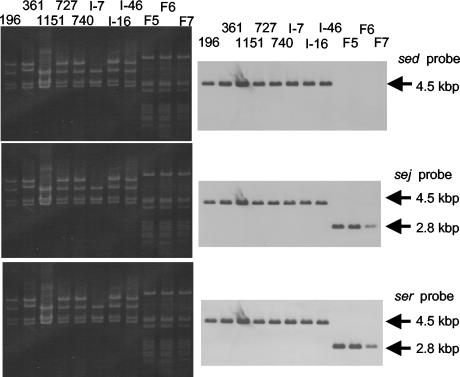
A search of FASTA showed that the 5′ flanking sequence of the ser gene has high homology with several staphylococcal plasmids, such as pN315B (GenBank, EMBL, and DDBJ accession number AP003139) or pMW2 (GenBank, EMBL, and DDBJ accession number AP004832). As well, a FASTA search using the sej flanking sequences of 2.8 kbp of the EcoRI fragment of pKO311 showed high homology with pIB485 (GenBank, EMBL, and DDBJ accession number AF053140), which contains the sed and sej genes (31). Thus, we purified plasmids from the Fukuoka 5, Fukuoka 6, and Fukuoka 7 strains and named them pF5, pF6, and pF7, respectively. Then, several pIB485-like plasmids were purified from S. aureus laboratory strains, food poisoning outbreak isolates, and healthy human isolates. These Fukuoka outbreak-related plasmids and pIB485-like plasmids were analyzed by Southern hybridization. Southern blot analysis with the ser probe showed that the ser gene exists on the pF5, pF6, and pF7 plasmids. As well, an sej probe revealed the same signals on the same membrane. We concluded that 2.8 kbp of the EcoRI fragment of pKO311 must be a part of the pF5 plasmid (Fig. 6). EcoRI-digested pIB485-like sed-harboring plasmids showed almost the same pattern as the original pIB485 plasmid. However, several types of length polymorphism were observed. All sed, ser, and sej probes hybridized to 4.5 kbp of the EcoRI fragment of pIB485-like plasmids, leading us to the conclusion that pIB485-like sed- and sej-harboring plasmids also contain the ser gene (Fig. 6). pIB485 was originally described as a sed-carrying plasmid by Bayles and Iandolo (3). Then, Zhang et al. reported the sej gene as a new SE gene which existed on a sed-carrying plasmid, pIB485 (31). We have also shown that sed and the sej-carrying pIB48-like plasmids also carry ser. At present, the evolutionary relationship between pF5 and pIB485 is unknown. In recent years, several studies have shown that most S. aureus strains harbor one or more SE genes (16, 21). The presence of multiple SE genes in S. aureus makes it less clear whether any single toxin is responsible for staphylococcal food poisoning. Previous studies have shown that SEA and SED are common causes of staphylococcal food poisoning (5). Because SED-producing S. aureus strains should harbor pIB485-like plasmids, the SED-producing S. aureus involved in food poisoning would produce the additional enterotoxin-related toxins, SER and SEJ. If SER and SEJ are proved to be emetic toxins, the food poisoning caused by SED-producing S. aureus may be assumed to be a complex phenotype of intoxication by multiple SEs.
FIG. 6.
ser and sej genes are carried by two kinds of staphylococcal plasmid. The pF5-like plasmids and pIB485-like plasmids were purified from S. aureus laboratory strains, food poisoning outbreak isolates, and healthy human isolates. These plasmids were digested by EcoRI and then subjected to Southern blot analysis. Lanes: 196E, p196E; 361, p361; 1151, p1151; 727, p727; 740, p740; I-7, pI7; I-16, pI16; I-46, pI46; F5, pF5; F6, pF6; and F7, pF7.
The benefit for S. aureus to maintain multiple SAg-related genes, such as the SE gene, tst-1, and set, on genomes and plasmids is not fully understood. Ferens and Bohach (9) hypothesized that SAgs play an important role in modulating the host immune response and that they may contribute to the maintenance of a suitable environment for colonization of S. aureus on host mucosal membranes. As well, Arcus et al. (1) showed that SET3, i.e., staphylococcal exotoxin-like protein 3, did not exhibit any of the properties of an SAg. They also suggested that the SET family may have an entirely different function from SAgs, although SETs have a conserved SAg-like three-dimensional structure. It seems that these SAg and SAg-related proteins have multiple functions that contribute to their survival and localization in hosts and to the pathogenicity of S. aureus. SEs and TSST-1 are important toxins because they cause food poisoning and toxic shock syndrome. Moreover, it is also important to understand the functions of these SAgs and SAg-related toxins per se in order to clarify the nature of persistent infection of S. aureus and their pathogenicity.
Nucleotide sequence accession number. The nucleotide sequence of pK0311 was submitted to the GenBank, EMBL, and DDBJ databases and was assigned accession number AB075606.
Acknowledgments
This work was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (grant numbers 11760213, 12660281, and 14560259).
Editor: A. D. O'Brien
REFERENCES
- 1.Arcus, V. L., R. Langley, T. Proft, J. D. Fraser, and E. N. Baker. 2002. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 277:32274-32281. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W., and J. J. Iandolo. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171:4799-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergdoll, M. S. 1988. Monkey feeding test for staphylococcal enterotoxin. Methods Enzymol. 165:324-333. [DOI] [PubMed] [Google Scholar]
- 5.Bergdoll, M. S. 1989. Staphylococcus aureus, p. 463-523. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 6.Betley, M. J., and J. J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229:185-187. [DOI] [PubMed] [Google Scholar]
- 7.Couch, J. L., M. T. Soltis, and M. J. Betley. 1988. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J. Bacteriol. 170:2954-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferens, W. A., and G. A. Bohach. 2000. Persistence of Staphylococcus aureus on mucosal membranes: superantigens and internalization by host cells. J. Lab. Clin. Med. 135:225-230. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., S. R. Monday, T. J. Foster, G. A. Bohach, P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, D. L., K. Omoe, Y. Shimoda, A. Nakane, and K. Shinagawa. 2003. Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect. Immun. 71:567-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole-genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 15.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 16.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food. Prot. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 17.Munson, S. H., M. T. Tremaine, M. J. Betley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun. 66:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakane, A., A. Numata, M. Asano, M. Kohanawa, Y. Chen, and T. Minagawa. 1990. Evidence that endogenous gamma interferon is produced early in Listeria monocytogenes infection. Infect. Immun. 58:2386-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakane, A., A. Numata, and T. Minagawa. 1992. Endogenous tumor necrosis factor, interleukin-6, and gamma interferon levels during Listeria monocytogenes infection in mice. Infect. Immun. 60:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 21.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orwin, P. M., D. Y. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orwin, P. M., D. Y. Leung, T. J. Tripp, G. A. Bohach, C. A. Earhart, D. H. Ohlendorf, and P. M. Schlievert. 2002. Characterization of a novel staphylococcal enterotoxin-like superantigen, a member of the group V subfamily of pyrogenic toxins. Biochemistry 41:14033-14040. [DOI] [PubMed] [Google Scholar]
- 24.Ren, K., J. D. Bannan, V. Pancholi, A. L. Cheung, J. C. Robbins, V. A. Fischetti, and J. B. Zabriskie. 1994. Characterization and biological properties of a new staphylococcal exotoxin. J. Exp. Med. 180:1675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Shinagawa, K., M. Ishibashi, H. Yamamoto, N. Kunita, and K. Hisa. 1974. A consideration to immune doses of staphylococcal enterotoxin B to rabbits. Jpn. J. Med. Sci. Biol. 27:309-314. [DOI] [PubMed] [Google Scholar]
- 27.Su, Y. C., and A. C. Wong. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 61:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams, R. J., J. M. Ward, B. Henderson, S. Poole, B. P. O'Hara, M. Wilson, and S. P. Nair. 2000. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect. Immun. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, V. Kapur, and P. M. Schlievert. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3: implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227-233. [DOI] [PubMed] [Google Scholar]