Abstract
Many important virulence genes of pathogenic bacteria are preferentially expressed in vivo. We used the recently developed in vivo-induced antigen technology (IVIAT) to identify Vibrio vulnificus genes induced in vivo. An expression library of V. vulnificus was screened by colony blot analysis by using pooled convalescent-phase serum that had been thoroughly adsorbed with in vitro-expressed V. vulnificus whole cells and lysates. Twelve clones were selected, and the sequences of the insert DNAs were analyzed. The DNA sequences showed homologies with genes encoding proteins of diverse functions: these functions included chemotaxis (a methyl-accepting chemotaxis protein), signaling (a GGDEF-containing protein and a putative serine/threonine kinase), biosynthesis and metabolism (PyrH, PurH, and IlvC), secretion (TatB and plasmid Achromobacter secretion [PAS] factor), transcriptional activation (IlvY and HlyU), and the activity of a putative lipoprotein (YaeC). In addition, one identified open reading frame encoded a hypothetical protein. Isogenic mutants of the 12 in vivo-expressed (ive) genes were constructed and tested for cytotoxicity. Cytotoxic activity of the mutant strains, as measured by lactate dehydrogenase release from HeLa cells, was nearly abolished in pyrH, purH, and hlyU mutants. The intraperitoneal 50% lethal dose in mice increased by ca. 10- to 50-fold in these three mutants. PyrH and PurH seem to be essential for in vivo growth. HlyU appears to be one of the master regulators of in vivo virulence expression. The successful identification of ive genes responsible for the in vivo bacterial virulence, as done in the present study, demonstrates the usefulness of IVIAT for the detection of new virulence genes.
The expression of virulence determinants in bacteria is known to be regulated by various environmental and host factors (38). During host-parasite interactions, many novel genes that not expressed during in vitro growth have been demonstrated to be coordinately regulated or stimulated by host factors encountered in vivo (20). The usefulness of the information concerning virulence expression gained from in vitro studies is therefore incomplete in relation to in vivo bacterial pathogenesis.
Vibrio vulnificus, an opportunistic pathogen, experiences a dramatic environmental change during its infection process. V. vulnificus is an estuarine bacterium that preferentially affects individuals who are heavy drinkers of alcohol and patients with underlying hepatic diseases and other immunocompromised conditions. The pathogen frequently causes fatal septicemia with a rapid progress, resulting in a mortality rate of more than 50% within a few days. The putative virulence factors of V. vulnificus reported so far include a hemolysin (15), a protease (29), phospholipase A2 (55), siderophores (53), and capsular polysaccharides (61a). We reported that the ToxRS system of V. vulnificus, a transmembrane signal-transducing transcription activator, regulated the expression of the hemolysin gene vvhA (32). The ToxRS system was reported to play an important role in regulating in vivo virulence gene expression during V. cholerae infection in a mouse model (33). However, whether the V. vulnificus ToxRS system plays an important role in regulating in vivo virulence gene expression during infection needs further study. V. vulnificus, while infecting the susceptible hosts, passes through gastric acidity, experiences an abrupt pH increase in the duodenum, receives bile secretion, invades into intestinal mucosa, and eventually enters the bloodstream where the pathogen multiplies. During this complicated infection process, V. vulnificus should be able to sense changes in the environmental parameters in the host milieu. The changing signals are likely relayed to specific genes by cognate signal transduction systems, resulting in the expression of specific virulence factors (33). Virulence factors required for in vivo survival and growth of V. vulnificus are expected to be produced at the right place and time in a tightly regulated fashion, as reported for other pathogens (21, 33, 37). To date, reports concerning the in vivo expression of V. vulnificus virulence factors are very limited. The in vivo-expressed (ive) genes may serve new targets for antimicrobial therapy or vaccine development.
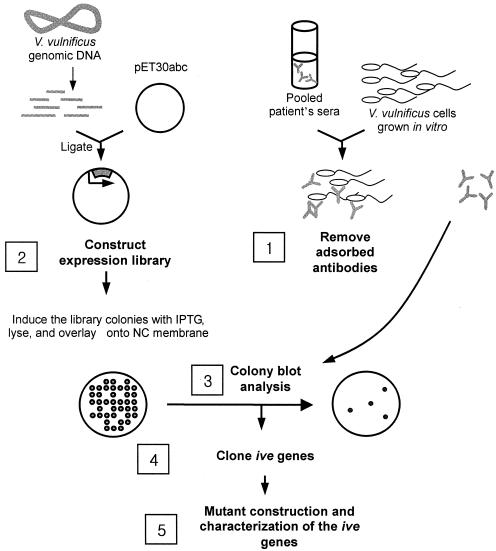
In the present study, we used in vivo-induced antigen technology (IVIAT), a novel method designed to screen microbial genes expressed specifically during human infections, to identify V. vulnificus genes expressed preferentially in vivo. IVIAT identifies genes expressed during an actual human infection rather than in an animal model (17). We used pooled convalescent-phase human serum to probe genes specifically expressed in vivo. An experimental scheme of IVIAT used in the present study is presented in Fig. 1. This new technology allowed us to identify several novel genes that likely play important roles in the survival and replication of V. vulnificus in humans.
FIG. 1.
IVIAT. Convalescent-phase sera from patients who survived V. vulnificus septicemia were pooled and adsorbed exhaustively with a virulent clinical isolate (CMCP98K) grown in vitro (step 1). An expression library of V. vulnificus CMCP98K was generated in E. coli BL21(DE3) (step 2), and the clones were probed with the adsorbed serum (step 3). Reactive clones, which were producing V. vulnificus antigens that are expressed during septicemic infection but not during in vitro cultivation, were purified, and their cloned DNA was sequenced. In vivo-expressed (ive) genes were identified in full length in the GenBank bacterial genome database and cloned by PCR (step 4). The pathogenic roles of the ive gene products were investigated by testing the phenotypes of the null mutants of each gene (step 5).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are listed in Table 1. V. vulnificus CMCP6 and CMCP98K are clinical isolates from the Chonnam National University Hospital. V. vulnificus MO6-24/O is also a clinical isolate and was provided by J. Glenn Morris, Jr., of the University of Maryland. The strains were identified by conventional biochemical tests and PCR analysis with V. vulnificus-specific primers (31). E. coli strains were grown in Luria-Bertani (LB) or brain heart infusion (BHI) medium, and V. vulnificus strains were grown in 2.5% NaCl heart infusion (HI) medium. Antibiotics were used as follows: ampicillin (AMP) at 100 μg/ml, kanamycin (KAN) at 50 μg/ml, chloramphenicol (CHL) at 30 μg/ml, and tetracycline (TET) at 12.5 μg/ml for E. coli and AMP at 20 μg/ml, CHL at 2 μg/ml, and TET at 2 μg/ml for V. vulnificus.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| V. vulnificus | ||
| MO6-24/O | Clinical isolate | 47 |
| CMCP6 | Clinical isolate | Chonnam National University Hospital |
| CMCP98K | Clinical isolate | Chonnam National University Hospital |
| CMM1701 | CMCP6 with the insertion mutation in Vv-ive-1 gene; Cmr | This study |
| CMM1702 | CMCP6 with the insertion mutation in Vv-ive-2 gene; Cmr | This study |
| CMM1703 | CMCP6 with the insertion mutation in Vv-ive-3 gene; Cmr | This study |
| CMM1704 | MO6-24/O with the insertion mutation in Vv-ive-4 gene; Cmr | This study |
| CMM1705 | CMCP6 with the insertion mutation in Vv-ive-5 gene; Cmr | This study |
| CMM1706 | CMCP6 with the insertion mutation in Vv-ive-6 gene; Cmr | This study |
| CMM1707 | CMCP6 with the deletion mutation in Vv-ive-7 gene | This study |
| CMM1708 | MO6-24/O with the deletion mutation in Vv-ive-8 gene | This study |
| CMM1709 | MO6-24/O with the insertion mutation in Vv-ive-9 gene; Cmr | This study |
| CMM1710 | CMCP6 with the deletion mutation in Vv-ive-10 gene | This study |
| CMM1711 | CMCP6 with the insertion mutation in Vv-ive-11 gene; Cmr | This study |
| CMM1712 | CMCP6 with the insertion mutation in Vv-ive-12 gene; Cmr | This study |
| CMM1715 | CMM1710 containing pCMM1701; Ampr Tcr | This study |
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Laboratory collection |
| SY327 λpir | Δ(lac pro) argE(Am) rif nalA recA56 λpir lysogen | 39 |
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tcr::Mu λpirR6K lysogen; Kmr | 39 |
| Plasmids | ||
| pET30a,b,c | Inducible-expression vectors carrying N-terminal His · Tag/thrombin/S · Tag/enterokinase configuration, providing three different reading frames at the multicloning site; Kmr | Novagen |
| pDM4 | A suicide vector with ori R6K sacB, Cmr | 40 |
| pLAFR3 | IncP cosmid vector; Tcr | 54 |
| pLAFR3II | pLAFR3 with bla inserted at the cos site | This study |
| pRK2013 | IncP Kmr Tra Rk2+repRK2 repE1 | 9 |
| pCMM1701 | pLAFR3I1 with hlyU ORF | This study |
| pUTKm1 | Tn5-based insertion delivery plasmid; Ampr | 23 |
Cmr, CHL resistance; Tcr, TET-resistance; Kmr, KAN resistance; Ampr, AMP resistance.
Construction of an inducible expression genomic DNA library.
An expression library of V. vulnificus CMCP98K was generated as follows. V. vulnificus genomic DNA was purified by a conventional sodium dodecyl sulfate (SDS)-protease method described elsewhere (31), partially digested with Sau3AI, and separated by electrophoresis with a low-melting-point agarose gel (Amersham). DNA fragments of ca. 0.5 to 1.0 kb were cut from the agarose gel and extracted by using the QIAEX II gel extraction kit (Qiagen) after treatment with β-agarase I (New England Biolabs). The purified DNA inserts were ligated into the pET30abc expression vectors (Novagen), which had been cut by BamHI and treated with shrimp alkaline phosphatase (Roche). The resulting ligation mixture was electroporated into electrocompetent DH5α cells (Gibco-BRL). The transformants were spread onto LB agar plates containing 50 μg of KAN/ml. After overnight incubation, colonies on the plates were scraped and frozen at −70°C until use. An aliquot of the resulting library was grown in LB broth, and plasmids were isolated. The isolated plasmid library DNA was subsequently electroporated into the expression host E. coli BL21(DE3) (Novagen).
Elimination of antibodies reacting with in vitro-expressed V. vulnificus antigens from septicemic patients' sera.
Convalescent-phase sera were obtained from three patients who survived V. vulnificus septicemia. The three patients were 64- and 48-year-old males and a 54-year-old female. They all had underlying liver cirrhosis. Sera were taken 24, 29, and 12 days after the onset of symptoms from the three patients, respectively. All three cases were definitively diagnosed by isolating V. vulnificus from blood and necrotic tissues. The antibody titer of each serum was verified by enzyme-linked immunosorbent assay (ELISA), with V. vulnificus cell lysates as the immobilized antigens. Equal volumes (500 μl) of the three patients' sera were pooled and exhaustively absorbed with in vitro-expressed V. vulnificus antigens. The first adsorption of the pooled serum was conducted with 1011 CFU of V. vulnificus whole cells grown in 2.5% NaCl HI broth at 25°C. The serum was further adsorbed by V. vulnificus cell lysate immobilized onto a nitrocellulose membrane. A final adsorption step was carried out with heat-denatured V. vulnificus cell lysate immobilized on a nitrocellulose membrane. To make the bacterial lysate, 1011 CFU of V. vulnificus cells were sonicated on ice until complete disruption. For every adsorption step, the serum was incubated overnight at 4°C with gentle agitation on a rocking platform. The efficiency of each adsorption was monitored by ELISA. As the negative control, we used the serum from a 13-year-old girl who had never eaten raw shellfish because she does not like them. The serum was also thoroughly adsorbed with in vitro-expressed V. vulnificus whole cells, and the bacterial lysate by the same method used for the adsorption of the pooled convalescent-phase serum.
Library screening for V. vulnificus ive genes by using the colony immunoblot analysis.
The genomic expression library was plated onto BHI agar plates containing 50 μg of KAN/ml at the density of ca. 1,000 colonies per plate. The colonies were then replica plated onto BHI agar plates containing 50 μg of KAN/ml and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), followed by incubation for 5 h at 37°C to induce the expression of insert DNA fragments. The colonies were exposed to chloroform vapors to partially lyse the bacteria and the exposed proteins were adsorbed onto overlaid HyBond nitrocellulose membranes (Amersham). The membranes were carefully removed and saturated with a 5% skim milk solution in phosphate-buffered saline (PBS) containing 0.5% Tween 20. Colony Western blot analysis was performed with the adsorbed pooled serum as the primary antibody and peroxidase-conjugated rabbit anti-human immunoglobulin G (IgG) and IgM (Dako) as the secondary antibody. The positive signals were detected by using an enhanced chemiluminescence (ECL) kit and Hyperfilm ECL (Amersham). Reactive clones were collected and retested for reactivity by another round of colony immunoblot analysis. Each colony of the resulting reactive clones was grown overnight in 96-well culture plates and then replica plated onto BHI agar plates containing 50 μg of KAN/ml with or without IPTG. Colony immunoblot analysis was done as the first-round screening.
DNA sequencing.
DNA sequencing was done by the double-strand dideoxy-chain termination method by using an ABI Prism 377 automatic DNA sequencer (Perkin-Elmer Applied Biosystems) at the Korea Basic Science Institute in Kwangju, Korea. Sequence homologies to genes in the GenBank database were determined by using the BLAST algorithm of the National Center for Biotechnology Information at the National Library of Medicine.
Construction of V. vulnificus ive gene mutants by homologous recombination.
V. vulnificus ive isogenic mutant strains of the highly virulent V. vulnificus stain MO6-24/O or CMCP6 were constructed either by insertional inactivation method with a suicide vector or in-frame deletion by using allelic exchange method (39). For the insertional inactivation of Vv-ive-2, -3, -4, -5, -6, -9, -11, and -12 putative genes, truncated ive gene fragments were amplified by PCR and ligated into the suicide vector pDM4 (40). The primer sets used for the amplification of DNA fragments lacking 5′ and 3′ ends of each gene are listed in Table 2. Most of the PCR primers had overhangs recognized by XbaI, BglII, or BamHI to make the cloning into pDM4 easier. When the amplicon had a sequence recognized by a restriction enzyme near the end, the overhangs were not added to the primers. The ligated DNA was transformed into E. coli SY327 λpir. Plasmids amplified in E. coli SY327 λpir were isolated and transformed into E. coli SM10 λpir (39) and subsequently transferred to V. vulnificus MO6-24/O or CMCP6 by conjugation. CHL-resistant transconjugants that had the mobilized plasmid integrated into their chromosome by homologous recombination were selected on thiosulfate-citrate-bile-sucrose (TCBS) agar plates containing 2 μg of CHL/ml as described previously (40).
TABLE 2.
Primers used for the mutant construction
| Mutant strain | Primer and primer sequence used for mutation
|
Amplified or deleted (Δ) region/total ORF (bp) | |
|---|---|---|---|
| Name | Sequence (restriction enzyme)a | ||
| CMM1701 | Pive-1-1 | ggctccctatttttatttctcagc | |
| Pive-1-2 | aagcttcaccatatcttgatagatcgc (HindIII) | Δ481-1860/2,004 | |
| Pive-1-3 | aagcttaacgtcagcgaaagtgtcacc (HindIII) | ||
| Pive-1-4 | cgaaatcgtggtagctgttgacg | ||
| CMM1702 | Pive-2-1 | gaagatctcgatggcctaggtgtgttggg (BglII) | 12-631/900 |
| Pive-2-2 | gctctagacgccattgccaccaacacagc (XbaI) | ||
| CMM1703 | Pive-3-1 | gaagatcttctagtgattgtgaataccagc (BglII) | 456-993/1,161 |
| Pive-3-2 | gctctagaaacaactaagtagttattcacc (XbaI) | ||
| CMM1704 | Pive-4-1 | tggctcaggaagtaaaag | 120-618/726 |
| Pive-4-2 | gctctagatgtaaatgctgccaaatc (XbaI) | ||
| CMM1705 | Pive-5-1 | cgacatcggtggcccaaccatg | 378-1,170/1,593 |
| Pive-5-2 | gaagatctttgaaccagtaggccgccg (BamHI) | ||
| CMM1706 | Pive-6-1 | gctctagaccagcacgaaccattgcaac (XbaI) | 382-1,205/1,485 |
| Pive-6-2 | gaagatctcgctgatccctcaagctgatc (BglII) | ||
| CMM1707 | Pive-7-1 | ggcttcatttcatgaagccgc | |
| Pive-7-2 | aagctttccgaatagtctcacataggg (HindIII) | Δ34-384/393 | |
| Pive-7-3 | aagcttcaataccagttcccaaaaacc (HindIII) | ||
| Pive-7-4 | tggatgatgaatcaagttggtcc | ||
| CMM1708 | Pive-8-1 | atgcaatcgtaaactcacagc | |
| Pive-8-2 | aagcttggctttcatgtaatg (HindIII) | Δ10-165/231 | |
| Pive-8-3 | aagcttttagaaattcctgaacac (HindIII) | ||
| Pive-8-4 | gctcagggcttacactttctg | ||
| CMM1709 | Pive-9-1 | gctctagaactagccagcaactcttcac (XbaI) | 126-685/891 |
| Pive-9-2 | gaagatctacaatcgcttcatgacctga (BglII) | ||
| CMM1710 | Pive-10-1 | cgggatcctcgatatcgtcaactatag (BamHI) | |
| Pive-10-2 | cgggatccttttaagttcatgtgttgg (BamHI) | Δ13-291/297 | |
| Pive-10-3 | cgggatccgaataatgcttttgcgtg (BamHI) | ||
| Pive-10-4 | tacgtcggcactggcacctg | ||
| CMM1711 | Pive-11-1 | gaagatctgtaaaagttggcgttatggcg (BglII) | 97-710/810 |
| Pive-11-2 | gaagatcttgaacgttgtcttggcgagc (BglII) | ||
| CMM1712 | Pive-12-1 | gaagatctcaagaaaacaaccactctc (BglII) | 106-1,197/1,410 |
| Pive-12-2 | gaagatctatctaggttctctttggc (BglII) | ||
The restriction enzyme site is underlined.
Mutants of Vv-ive-1, -7, -8, and -10 genes were constructed by in-frame deletion. We designed two sets of primers amplifying the DNA fragments encompassing upstream or downstream of each gene (Table 2). One or both of each primer set had appropriate restriction overhang for cloning. Each amplicon was first cloned into TOPO PCR cloning vector (Invitrogen) and ligated each other. The DNA insert having in-frame deletion of each gene was recloned to pDM4 by using appropriate restriction enzymes, and the resulting plasmid was transformed into E. coli SM10 λpir. The plasmid in E. coli SM10 λpir was transferred to V. vulnificus MO6-24/O by conjugation, and the transconjugants were selected on TCBS agar plate containing CHL. The transconjugants were plated onto a 2.5% NaCl HI agar plate containing 10% sucrose to select clones that experienced the second homologous recombination events, forcing excision of the vector sequence and leaving only a mutated or wild-type (WT) allele of the genes. Each insertional or in-frame deletion mutation was confirmed by PCR of the chromosomal DNA from the respective mutant. The resulting mutant strains of the 12 putative ive genes are listed in the Table 1.
To complement the Vv-ive-10 (hlyU) mutant strain (CMM1710) with the WT allele in trans, we amplified a DNA fragment containing hlyU gene by PCR with a primer set (PhlyU-1, CGGGATCCTCGATATCGTCAACTATAG; PhlyU-4, TACGTCGGCACTGGCACCTG). The amplified DNA fragment was cloned into pLAFR3II plasmid, constructed by subcloning the bla gene from pUTKm1 (23) into BglII at the cos site of pLAFR3 (Table 1). The resulting plasmid (pCMM1701) was transferred into the hlyU mutant strain by the triparental mating with a conjugative helper plasmid, pRK2013 (9). The transconjugants were plated on TCBS agar containing AMP and TET, confirmed by PCR, and designated CMM1715.
Cytotoxicity assay of ive mutant strains.
Cytotoxic activity of bacteria was measured by lactate dehydrogenase (LDH) release from HeLa cell lines by using the CytoTox96 Non-Radioactive Cytotoxic Assay kit (Promega). One hundred thousand HeLa cells were plated into each well of 24-well cell culture plates and then incubated overnight. The next day, ive gene mutant strains suspended in fresh Dulbecco modified Eagle medium (DMEM) without fetal bovine serum were added to each well at a multiplicity of infection (MOI) of 100 and incubated for 90 min at 37°C in a 5% CO2 incubator. Released LDH in the supernatant was measured in accordance with the manufacturer's protocol.
LD50 determination.
V. vulnificus strains were cultured in 2.5% NaCl HI broth overnight at 37°C with agitation at 200 rpm. Subsequently, 1 ml of the overnight culture was inoculated into 100 ml of fresh 2.5% NaCl HI broth. The cultures were grown at 37°C and 200 rpm for 4.5 h, after which the cells were harvested by centrifugation and washed three times with PBS (pH 7.2). The cell pellet was resuspended and 10-fold serially diluted with PBS. Randomly bred specific-pathogen-free 8-week-old CD-1 mice were administrated with various doses of bacteria intraperitoneally. Five mice were used for each dose, and infected mice were observed for 48 h. The 50% lethal dose (LD50) of each mutant strain was calculated by the method of Reed and Muench (48).
Growth of the ive gene mutants in HeLa cell lysates.
HeLa cells were cultured as described above. Before tests, the culture was changed with fresh DMEM without fetal bovine serum and incubated for 90 min at 37°C in a 5% CO2 incubator. Then cells were scraped and sonicated to total lysis. Bacterial cells were inoculated into the cell lysate to the concentration of ca. 107 CFU/ml and incubated for 3 h. Viable cells were counted on 2.5% NaCl HI agar plates.
Nucleotide sequence accession number.
DNA sequences of the open reading frames (ORFs) identified in the present study have been deposited in the GenBank database as parts of a V. vulnificus whole genome sequence reported by our group under accession numbers AE016795 and AE016796.
RESULTS
Screening for ive genes of V. vulnificus by using IVIAT.
The convalescent-phase pooled serum was exhaustively adsorbed to remove antibodies reacting with in vitro-expressed proteins of V. vulnificus CMCP98K. The serum adsorption was conducted with in vitro-cultured V. vulnificus CMCP98K whole cells, lysed cell extracts, and heat-denatured cell extracts. Through the adsorption procedure, we intended to remove antibodies reacting with in vitro-expressed extracellular (or cell surface), cytoplasmic, and denatured fragment antigens. After the three rounds of adsorption, the adsorbed pooled convalescent-phase serum showed no significant reaction with in vitro-expressed V. vulnificus antigens by ELISA (data not shown). The resulting adsorbed convalescent-phase pooled serum was used to screen the pET30abc expression library. The adsorbed pooled serum contains antibodies reactive preferentially with in vivo-expressed antigens.
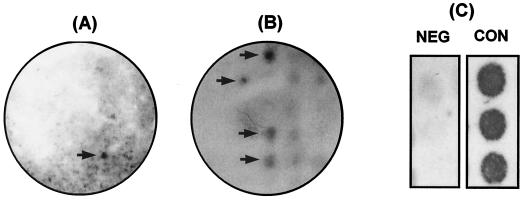
An expression library of V. vulnificus was generated by using the vector pET30abc in E. coli BL21(DE3). Clones from the expression library of V. vulnificus CMCP98K were probed with the adsorbed pooled serum by colony immunoblot analysis. Clones showing stronger signals on the IPTG-induced BHI agar plates than on the BHI agar plates without IPTG were selected. For the secondary screening, the clones showing positive signals at the primary screening were grown in 96-well culture plates and replica plated onto BHI agar plates with or without IPTG. Only the clones showing strong positivity at the secondary screening experiment were regarded to contain putative ive genes. At the tertiary screening step, we used the negative control serum, which was also thoroughly adsorbed with in vitro-expressed antigens, and made sure that the candidate ive gene products reacted specifically with the convalescent-phase serum. Candidate clones were grown in a 96-well culture plate and replica plated on BHI agar plates containing KAN and IPTG. Each clone was tested by the colony immunoblot analysis in duplicate or triplicate (Fig. 2). Insert DNAs in the positive clones were sequenced and analyzed by using the GenBank database for homology.
FIG. 2.
An example of the screening process for the confirmation that candidate gene products react specifically with the pooled convalescent-phase serum. Reactive clones on the primary screening plate (A) were picked up, transferred to a 96-well culture plate, and replica plated onto a IPTG-KAN-BHI agar plate for the secondary screening (B). Serum from a 13-year-old girl who had never eaten raw shellfish was adsorbed with in vitro-expressed antigens of V. vulnificus and used as a negative control. (C) Each clone was tested in duplicate or triplicate. NEG and CON, negative and pooled convalescent-phase sera, respectively. Arrows indicate positive clones.
Of ∼60,000 clones, 12 were confirmed to contain putative ive genes after the primary and secondary screening procedures. V. vulnificus ive gene candidates identified by the IVIAT are summarized in the Table 3. The 12 ive genes could be categorized into several classes: chemotaxis (a methyl-accepting chemotaxis protein [MCP]), signaling (a GGDEF-containing protein and a putative serine/threonine kinase), biosynthesis and metabolism (PyrH, PurH, and IlvC), secretion (TatB and plasmid Achromobacter secretion (PAS) factor), transcriptional activation (IlvY and HlyU), and a putative lipoprotein (YaeC). One ORF was predicted to encode a hypothetical protein.
TABLE 3.
In vivo-expressed gene candidates discovered by IVIAT in V. vulnificus
| Putative protein function and identified clone | Homologous protein in database (organism [% identity]) |
|---|---|
| Signaling | |
| Vv-Ive-1 | MCP VC0906 (V. cholerae [30]) |
| Vv-Ive-2 | GGDEF-containing protein STM3388 (S. enterica serovar Typhimurium LT2 [32]) |
| Vv-Ive-3 | Putative serine/threonine protein kinase STM2520 (S. enterica serovar Typhimurium LT2 [45]) |
| Biosynthesis and metabolism | |
| Vv-Ive-4 | Uridylate kinase (UMP kinase) PyrH VC2258 (V. cholerae [77]) |
| Vv-Ive-5 | AICAR transformylase/IMP cyclooxygenase PurH VC0276 (V. cholerae [92]) |
| Vv-Ive-6 | Putative acetohydroxyacid isomeroreductase IlvC VC0162 (V. cholerae [80]) |
| Secretion | |
| Vv-Ive-7 | Sec-independent protein translocase TatB VC0087 (V. cholerae [76]) |
| Vv-Ive-8 | PAS factor, a protein secretion factor (V. alginolyticus [85]) |
| Transcriptional activator | |
| Vv-Ive-9 | LysR-type transcriptional regulator IlvY VC0161 (V. cholerae [43]) |
| Vv-Ive-10 | Transcriptional activator HlyU VC0678 (V. cholerae [64]) |
| Known hypothetical protein | |
| Vv-Ive-11 | Putative lipoprotein gene YaeC VC0905 (V. cholerae [74]) |
| Vv-Ive-12 | Conserved hypothetical protein VC1606 (V. cholerae [56]) |
Construction of specific mutations in the putative ive genes.
Specific mutation in each of the 12 ive genes was constructed by insertion of a suicide plasmid in the chromosome or by in-frame deletion by using the allelic exchange method. Mutations of the ive genes were constructed with representative virulent strains MO6-24/O and CMCP6 (Table 1). The resulting mutants grew well on LB, HI, and BHI agar plates except for CMM1704, the putative pyrH mutant. The pyrH mutant strain showed smaller colonies on agar plates and retarded growth curve in HI broth (see Fig. 5).
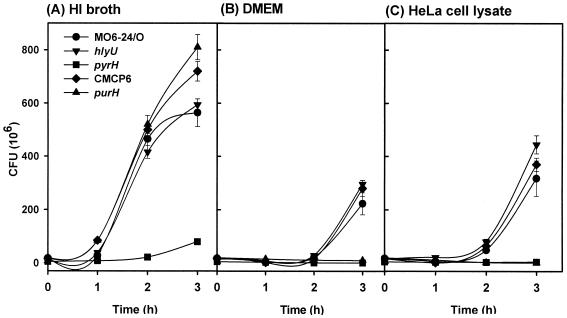
FIG. 5.
Growth of the ive gene mutants showing decreased virulence in HeLa cell lysates. The mutant strains of pyrH, hlyU, and purH homologues were inoculated into 2.5% NaCl HI (A), DMEM (B), and HeLa cell lysate (C) and then cultured for 3 h at 37°C in a 5% CO2 incubator. Viable cells were counted every 1 h on 2.5% NaCl HI agar plates. Growth of the mutants in HI broth, DMEM, and HeLa lysates was compared to that of appropriate isogenic WT strains MO6-24/O (pyrH and hlyU) or CMCP6 (purH). The error bar indicates the standard error of the mean from triplicate experiments.
Effect of the ive gene mutation on the cytotoxicity of V. vulnificus.
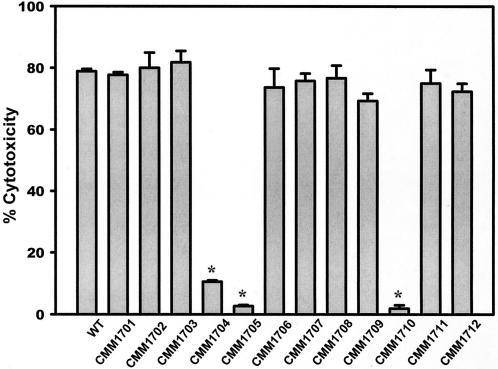
V. vulnificus cells are highly cytotoxic to cultured cell lines. The effects of the ive gene mutations on the cytotoxicity against HeLa cells were tested by using log-phase cultures. Mutants showing decreased LDH release when cultured with HeLa cells at an MOI of 100 for 90 min were screened. Among the mutants of the 12 ive genes, only the mutants of pyrH (CMM1704), purH (CMM1705), and hlyU (CMM1710) homologues showed significant decreases in cytotoxicity to HeLa cells (Fig. 3).
FIG. 3.
Effect of the ive gene mutation on the cytotoxicity of V. vulnificus. Bacterial cells were incubated with HeLa cells at an MOI of 100 for 90 min. The bars represent the mean percent release of LDH from four wells. WT stands for V. vulnificus WT MO6-24/O and CMCP6. The strains from CMM1701 to CMM1712 represent mutants of each ive genes identified in the present study (see Tables 1 and 3). Only the of pyrH, purH, and hlyU mutants showed a statistically significant decrease in cytotoxicity. An asterisk indicates statistical significance (P < 0.001) as determined by using the Student t test.
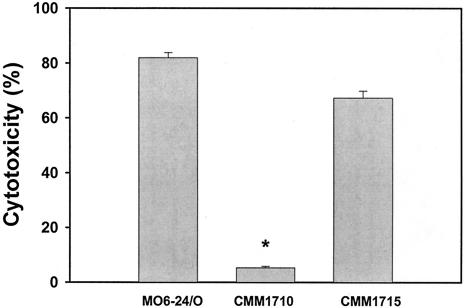
The CMM1710 strain, an hlyU in-frame deletion mutant, was not cytotoxic at all to HeLa cells even after 3 h of incubation, whereas the V. vulnificus WT strain killed almost 80% of the HeLa cells in 90 min. The cytotoxicity defect of CMM1710 could be complemented by a WT allele, along with its own promoter carried by the IncP plasmid pCMM1701 (Fig. 4).
FIG. 4.
Effect of the hlyU mutation on the cytotoxicity to HeLa cells. Log-phase bacterial cells were incubated with HeLa cells at an MOI 100 for 90 min. The bars represent the mean percent release of LDH from four wells. The cytotoxicity defect in the hlyU mutant (CMM1710) was almost completely complemented in the complemented strain (CMM1715). The asterisk indicates statistical significance (P < 0.001) as determined by using the Student t test.
Effect of the ive gene mutation on the lethality of V. vulnificus in mice.
To test the pathogenic significance of the three ive genes, mouse-killing activities of the three ive gene mutants showing significantly decreased cytotoxicity were compared to that of the isogenic WT strain. Intraperitoneal LD50s of the three mutants to specific-pathogen-free randomly bred CD-1 mice were significantly lower than that of the WT strain (Table 4). The LD50 to mice increased 53-, 14-, and 22-fold in CMM1710 (hlyU deletion mutant), CMM1704 (pyrH insertion mutant), and CMM1705 (purH insertion mutant), respectively. These data suggest that at least three of the ive genes identified in the present study are directly involved in the in vivo virulence of V. vulnificus.
TABLE 4.
Effect of the in vivo-expressed gene mutation on the LD50 of V. vulnificus
| Strain | Genotype | LD50 (CFU/mouse) | Fold increase in LD50 |
|---|---|---|---|
| MO6-24/O | V. vulnificus WT, a clinical isolate | 4.0 × 106 | |
| CMCP6 | V. vulnificus WT, a clinical isolate | 3.6 × 105 | |
| CMM1704 | MO6-24/O with the mutation in pyrH homolog (Vv-Ive-4) | 5.5 × 107 | 14 |
| CMM1710 | MO6-24/O with the mutation in hlyU homolog (Vv-Ive-10) | 2.1 × 108 | 53 |
| CMM1705 | CMCP6 with the mutation in purH homolog (Vv-Ive-5) | 8.0 × 106 | 22 |
Growth of the ive gene mutants showing decreased virulence in HeLa cell lysates.
The CMM1704 and CMM1705 strains, under microscope, appeared not to grow well when incubated with HeLa cells in DMEM in comparison with the WT and CMM1710 strains (data not shown). We speculated that the pyrH and purH mutants should have defects in utilizing host-derived factors for their growth. Growth of the mutants in HeLa cell lysates was compared to that of their isogenic WT strains. The WT strain did grow in DMEM, although their multiplication rate was much slower than in HI broth. HeLa cell lysates significantly stimulated their growth, suggesting that the strains could utilize factors in the lysate for growth. In HI broth, only the growth of CMM1704 was impeded (Fig. 5A), whereas no growth of CMM1704 and CMM1705 was observed in DMEM (Fig. 5B). Cell lysates could not stimulate the growth of the pyrH and purH mutants (Fig. 5C). On the other hand, CMM1710, the hlyU mutant, showed no growth difference from its isogenic mutant strain under any culture condition tested (Fig. 5). These results suggest that decreased virulence of the pyrH and purH mutants likely resulted from their in vivo growth defect. A functional defect in the in vivo virulence expression may be responsible for the decreased cytotoxicity and increased LD50 of the hlyU mutant.
DISCUSSION
Suitability of IVIAT in identifying ive genes of V. vulnificus.
There are currently several technologies designed to identify genes specifically expressed during infection. These include in vivo expression technology (IVET), recombinase-based in vivo expression technology (RIVET), signature-tagged mutagenesis (STM), differential fluorescence induction (DFI), and IVIAT (7, 8, 16, 17, 18, 20, 35, 59, 60). For the identification of V. vulnificus ive genes, technologies such as IVET, RIVET, and STM are not suitable since the harvesting efficiency of the pathogen from infected animals is very low. In infected animals, V. vulnificus disseminates systemically with no predilection for specific organs or tissues where V. vulnificus preferentially colonizes and multiplies. A sufficient number of V. vulnificus that are required for extensive screening of ive genes could not be retrieved from livers, spleens, hearts, or kidneys of infected mice (data not shown). Homogenization of whole body of infected mouse might be required to get sufficient number of in vivo-grown V. vulnificus. Moreover, the mouse model does not fully reproduce the human infection process, although mouse infection is the most widely used model for V. vulnificus virulence studies. For example, mice infected with the organism do not manifest skin and soft tissue necrosis, a hallmark of human infection. DFI also could not be used since encapsulated virulent V. vulnificus is resistant to phagocytosis (30).
In contrast, IVIAT avoids the use of animal or cell culture infection models by using serum from patients. The ive virulence factors identified by other technologies utilizing animal or cell culture models must be validated in the context of the actual human infection process. The pooled serum from convalescent-phase septicemic patients allows a direct identification of the widest array of antigens produced in the patients during different stages of infection (17). Since the major clinical manifestation of V. vulnificus is septicemia, convalescent-phase serum should contain high titers of antibodies against a wide range of ive antigens. We used a pooled convalescent-phase serum from three patients at different time points after the onset of symptoms to increase chances of identifying the broadest array of ive genes, including transiently expressed genes. In instances in which the main thrust of the work is directed toward obtaining the broadest possible understanding of the pathogenic mechanisms used by the microorganism, as was the case in the present study, it is more relevant to use a pool of sera rather than individual infected subject's serum. This is particularly important to represent antigens potentially produced at different stages of infection. If the goal was to find a candidate strategy for a diagnostic application, for instance, or a host immune response-independent diagnostic strategy, then it would be more relevant to use ive proteins that are produced throughout the infectious cycle, which would allow the use of serum from a single well-characterized patient.
In vivo-expressed genes associated with signaling.
Among the putative V. vulnificus ive genes identified, three were predicted to be associated with signal perception and transduction (Table 3). Living organisms monitor their environment and adjust their behavior, metabolism, and development in response to changes in physicochemical parameters. There are three known major classes of receptor proteins in prokaryotes, including (i) sensor histidine kinases of two-component signal transduction systems, (ii) chemotaxis transducers or MCPs, and (iii) diguanylate cyclases (42). Interestingly, one gene for each of these networks was identified in the present study.
The first clone, Vv-Ive-1 has a low level (∼30% at amino acid level) homology to a putative MCP gene of V. cholerae (19). Although Vv-Ive-1 showed low homology overall with known MCPs in GenBank, it had a characteristic MA (methyl-accepting chemotaxis-like) domain and a HAMP (histidine kinase, adenylyl cyclases, methyl-binding proteins, phosphatases) domain (data not shown), which are hallmarks of MCPs (13). Camilli and Mekalanos (7) reported that an MCP gene was induced during a V. cholerae infection in mice. When occupied by cognate chemoeffectors, MCP transduces signals to the flagellar motor to drive bacterial cells toward or away from the chemoeffectors (5). In Vibrio anguillarum, chemotactic motility is required for the invasion of fish, its natural host (46). The ive MCP of V. vulnificus identified in the present study might also play important roles in the invasion and progression of infection. Although the ive MCP mutant did not show any significant change in cytotoxicity and intraperitoneal LD50, intragastric LD50 of the mutant was eightfold higher than that of the isogenic WT strain (data not shown). The MCP might play an important role, whereas V. vulnificus invade humans through the oral route.
A putative diguanylate cyclase/phosphodiesterase gene was also identified. It showed homology to the GGDEF domain proteins (∼45% at amino acid level) that are reported to have diguanylate cyclase activity (2). According to the Pfam database, the GGDEF domain is present in more than 80 different proteins, most of which also contain signaling or two-component regulatory domains (3). GGDEF proteins of Acetobacter xylinum and Caulobacter crescentus were reported to play important roles in regulating important physiological functions of the organisms (1, 50). It is thus reasonable to suggest that the protein Vv-Ive-2 produced by the putative diguanylate/phosphodiesterase gene might play some role in the signaling pathway of V. vulnificus during the in vivo infection.
A putative serine/threonine kinase gene (Vv-ive-3) was also identified. This protein has 45% identity with a serine/threonine protein kinase of Salmonella enterica serovar Typhimurium LT2, the function of which is not yet identified. Bacterial two-component signal transduction systems are typically composed of a sensor protein kinase and a response regulator. A signal is sensed by the sensor kinase and relayed to the response regulator by a phosphotransfer mechanism. Since the 1970s, it has been widely accepted that histidine kinase is the only phosphorylating enzyme involved in signal transduction in prokaryotes. However, more recently, there have been sporadic reports on the presence of eukaryotic type serine/threonine or tyrosine kinases in bacteria (36, 41, 43, 58, 62). Additional studies of the pathophysiological functions of these putative bacterial serine/threonine protein kinases are needed. Such studies have been done in several similar kinases. For example, Streptomyces AfsK phosphorylates the global regulator AfsR, which controls the production of certain antibiotics (36). Several distinct serine/threonine protein kinases described in Myxococcus xanthus are involved in fruiting body formation and spore production (43, 62). It is likely that these serine/protein kinases play important roles in signal transduction and adaptive regulation of cellular functions. RsbT and RsbW, a pair of serine/threonine protein kinases, are involved in the transduction of environmental stress signals to the transcription factor σB of Bacillus subtilis (25). IVET analysis of Pseudomonas aeruginosa genes induced during infection identified a novel serine/threonine protein kinase homologous to Pkn1 of M. xanthus (60).
In vivo-expressed genes involved in biosynthesis and metabolism.
Growth in vivo is essential for bacterial pathogenicity. Of numerous genes expressed in vivo by serovar Typhimurium and detected by IVET, the most dominant ones are associated with metabolic functions such as metal acquisition, synthesis of nucleotides and cofactors, membrane modification, and protein targeting (20, 21). Four of twelve genes detected by IVET in an infant mouse model were found to be involved in metabolism and biosynthesis (7). In one STM study of staphylococcal bacteremia of mice, 9 of 24 ive genes were related to biosynthesis and metabolism (37). In the present study, we identified three ive gene candidates assumed to be involved in biosynthesis and metabolism (Table 3).
Among the ive genes identified in the present study is a gene showing ca. 80% identity with the V. cholerae UMP kinase pyrH gene. UMP kinase is an important enzyme for the de novo synthesis of pyrimidine nucleotides. UMP is the precursor for the synthesis of all pyrimidine nucleotides, and UMP kinase (PyrH) catalyzes phosphorylation of UMP to UDP (45). The pyrH has also been demonstrated to be involved in chromosomal partitioning during cell division (63). In addition to its catalytic function in de novo pyrimidine biosynthesis, UMP kinase serves as the sensor of the internal pyrimidine nucleotide pool and regulates the synthase operon carAB of carbamoylphosphate that is required for biosynthesis of arginine and pyrimidines (26).
It is interesting that pyrH is preferentially expressed during the in vivo infection process. In serovar Typhimurium, two subunit carAB genes of carbamoylphosphate synthase, the enzyme catalyzing on earlier step of pyrimidine de novo biosynthesis, were identified by IVET. In Listeria monocytogenes, another de novo pyrimidine biosynthetic gene, pyrE, was expressed preferentially in infected mammalian cells (27). Limited availability of pyrimidines and purines in animal tissues is proposed to be responsible for the in vivo induction of the enzymes required for de novo biosynthesis of them (10, 35). These results suggest that PyrH might serve an attractive new target for antibacterial drugs, since it has no counterpart in eukarya and is essential for in vivo cell growth and division (6).
Surprisingly, the mutation of pyrH, which was identified as an ive gene, resulted in a significant impairment for the growth in HI broth. Previous studies clearly showed that pyrimidine de novo biosynthesis genes are required for in vivo growth of other pathogenic bacteria. In that sense, our experimental result that pyrH was screened as one of the ive genes by IVIAT makes sense. Theoretically, all of the antibodies against in vitro expressed antigens should have been removed by the thorough adsorption procedure. The cells used for the adsorption experiment were cultured at 25°C. It is probable that the pyrH had been expressed at relatively lower level in 2.5% NaCl HI broth at 25°C in comparison with that in the human body milieu. Adsorption with the in vitro-cultured cells and lysates would have left antibodies reactive with PyrH. Another possibility is that the insertional inactivation of chromosomal pyrH might affected the expression of downstream genes by polar effects. The V. vulnificus genome sequence (www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html) showed ribosomal recycling factor, undecaprenyl diphosphate synthase, and CDP-diglyceride synthase genes downstream of pyrH, with the same transcriptional direction. Whether the downstream genes were affected by the insertion of the suicide plasmid and whether the polar effect caused in vitro growth defect require further study.
A putative AICAR transformylase/IMP cyclohydrolase gene (purH) showing ca. 90% identity with that of V. cholerae at the amino acid level was also identified in the present study. PurH is involved in purine nucleotide biosynthesis and catalyzes the final two steps of de novo IMP biosynthesis (64). IMP is the first nucleotide in the de novo purine biosynthetic pathway. STM experiments identified two genes (purD and purL) of serovar Typhimurium (22) and one gene (purL) of S. aureus (37) in the purine de novo biosynthetic pathway to be expressed in vivo. In L. monocytogenes, the expression of purH and purD was increased as much as 100-fold inside mammalian cells compared to in vitro-grown cultures (27). In this regard, de novo purine synthesis in vivo seems to play an important role in the survival and virulence expression of pathogenic bacteria. Salmonella purine auxotrophs showing attenuated in vivo virulence have been used as oral live-vaccine strains (34).
A putative acetohydroxyacid isomeroreductase gene (ilvC) showing ca. 50% identity with a V. cholerae homologue at amino acid level was also identified. IlvC catalyzes the conversion of acetolactate and acetohydroxybutyrate to the dihydroxy acids that are subsequently used for the synthesis of the branched-chain amino acids such as leucine, valine, and isoleucine (57). The same enzyme also catalyzes the reduction of α-ketopantoate to pantoate, an intermediate in the biosynthesis of coenzyme A that serves as the predominant acyl group carrier in cells (24). Branched-chain amino acids account for 20% of the total amino acids in Escherichia coli (44). The biosynthetic pathway leading to the branched-chain amino acids is of interest for the development of novel antimicrobial agents. Inhibition of the IlvC resulted in some antimicrobial effects against Mycobacterium tuberculosis (14). Identification of IlvC as an ive antigen raises the probability of the branched-chain amino acid biosynthetic pathway as a new therapeutic target for bacterial infections. Of note was that transcriptional activator of ilvC was also identified as an ive gene in the present study.
In vivo-induced genes involved in secretion.
Nearly all bacterial virulence factors are located on the bacterial surface or are secreted (11). To this end, bacteria have evolved a wide spectrum of mechanisms for the secretion of virulence factors. The present study identified two novel ive genes that are predicted to be involved in the secretion mechanisms (Table 3).
The first is a putative Sec-independent protein translocase gene (tatB), which has 75% identity with a V. cholerae homologue at the amino acid level. The Tat (twin-arginine translocation) system is a bacterial protein export pathway with the remarkable ability to transport prefolded proteins across the cytoplasmic membrane (51). TatB is a cytoplasmic membrane-embedded protein that interacts with TatA and forms a large membrane-bound complex, together with TatC (52). Proteins exported by the Tat apparatus function predominantly in respiratory and photosynthetic electron transport chains and are vital for many types of bacterial energy metabolism (4). The V. vulnificus metabolic requirements in vivo are likely significantly different from the requirements for in vitro culture. Thus, the Tat system might play an important role in translocating novel enzymes required for energy metabolism when the organism is replicating in human hosts.
Another novel putative protein secretion factor, PAS, was also identified. The PAS factor was originally identified in Vibrio alginolyticus and potentiates the secretion of periplasmic β-lactamase and alkaline phosphatase into media when expressed in E. coli (56). PAS factor is a small protein of 76 amino acids that facilitates the excretion of periplasmic proteins. The mechanism of how the PAS factor induces excretion of periplasmic proteins is poorly studied to date. The PAS factor might play significant roles in facilitating excretion of unidentified factors required for in vivo survival and pathogenesis of V. vulnificus.
In vivo-expressed transcriptional regulator genes.
IVIAT identified two putative transcriptional regulator genes, the ilvY and hlyU homologues, of V. vulnificus (Table 3). IlvY is a LysR-type transcriptional regulator. The ilvY gene forms an operon with its regulatory target gene ilvC. IlvY positively regulates ilvC transcription in an inducer-dependent manner while it negatively regulates its own transcription, irrespective of inducers. Substrates of IlvC, α-acetolactate, and α-acetohydroxybutyrate serve as inducers of the ilvYC operon (49). It is interesting that both genes of the ilvYC operon were identified by IVIAT through the present study. V. vulnificus should experience a strong induction condition for the operon while infecting humans.
HlyU upregulates expression of the hemolysin, HlyA, in V. cholerae. The transcriptional activator HlyU belongs to a small regulatory protein family, including NolR of Rhizobium meliloti, SmtB of Synechococcus sp. strain PCC 7942, and ArsR of Staphylococcus aureus, that contain a helix-turn-helix motif (61). V. vulnificus produces a potent hemolysin/cytolysin (VvhA) sharing a homologous domain with HlyA of V. cholerae (63). VvhA shows strong cytotoxicity and induces guanylate cyclase to cause vasodilation through a novel mechanism (15, 28). VvhA was not produced in the hlyU mutant CMM1710. Using a chromosomal vvhA::lacZ reporter, we could not detect any transcriptional activity of vvhA in CMM1710 (data not shown). Interestingly, the production of the elastolytic protease, another exotoxin produced by V. vulnificus, was also significantly decreased in the mutant (29). HlyU thus appears to be one of the master regulators of in vivo virulence expression in V. vulnificus. The HlyU protein itself and the genes under its control would serve as important targets in developing a new-paradigm therapy against V. vulnificus.
Other in vivo-expressed genes with no known function.
Of the ive genes identified by IVIAT, two matched hypothetical ORFs of V. cholerae. One showed 74% identity with the V. cholerae putative lipoprotein gene, yaeC, whose function has yet not been determined (Table 3). The other hypothetical ORF showed 56% identity with the VC1606 hypothetical protein of V. cholerae. Depending on the methods used, 25 to 50% of the screened ive genes showed no match to any previously characterized genes in GenBank (20, 22, 37).
In conclusion, we identified 12 ive genes by using IVIAT. Among them, mutants of pyrH, purH, and hlyU homologues showed significantly decreased virulence in an animal model. These ive genes have not yet been implicated in the virulence of V. vulnificus. Such a finding would provide a more holistic understanding of the pathogenesis of V. vulnificus infection. These results also clearly demonstrate the suitability of IVIAT for identifying genes associated with V. vulnificus virulence in vivo. The products of the three ive genes might serve new as specific targets for antimicrobial therapy and vaccine development for V. vulnificus infections.
Of note was that 11 ive genes identified in the present study were supposed to encode intracellular proteins. Only the Vv-ive-11 gene was predicted to encode a putative lipoprotein (Table 3). Except for hlyU (Vv-ive-10), the other 10 ive genes did not appear to encode authentic virulence factors. Bacterial virulence factors are generally secreted or exposed on surface and hence are more easily recognized by the immune system (11, 12). In this regard, we expected IVIAT would identify those authentic virulence factors preferentially in vivo. Against our expectation, most of the ive antigens seemed to be associated with survival and growth in vivo: i.e., chemotaxis and signaling (MCP, GGDEF protein, and serine/threonine protease), biosynthesis and metabolism of nucleic acids and amino acids (PyrH, PurH, IlvC, and IlvY), and secretion of proteins (TatB and PAS factor). These antigens should have been exposed to the immune system after the lysis of V. vulnificus cells during infection.
The estimated genome size of V. vulnificus is ca. 5.1 Mb. For the present study, we screened only ∼60,000 clones or ca. 45% of a pET30abc expression library at a 99% confidence level, with a 1-kb mean insert size, three reading frames, and two directions for every expressed protein. More extensive IVIAT screening with bigger expression libraries would guarantee the discovery of many more ive genes involved in in vivo virulence expression. Our success with V. vulnificus encourages us to investigate the application of IVIAT to other septicemia-causing pathogens for screening critical in vivo-expressed virulence genes. The in vivo roles of other ive genes identified in the present study should be analyzed by measuring their expression levels in appropriate experimental models that reproduce the conditions of actual human infection.
Acknowledgments
This study was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health and Welfare (HMP-00-B-20200-0014) and by the 2001 National Research Laboratory grant from the Ministry of Science and Technology of the Republic of Korea.
Editor: V. J. DiRita
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. Sonnhammer. 1999. Pfam 3.1: 1,313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 6.Bucurenci, N., L. Serina, C. Zaharia, S. Landais, A. Danchin, and O. Barzu. 1998. Mutational analysis of UMP kinase from Escherichia coli. J. Bacteriol. 180:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 9.Ditta, G., D. C. Stanfield, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, B. B., and S. Falkow. 1989. Common themes in microbial pathogenicity. Microbiol. Rev. 53:210-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 14.Grandoni, J. A., P. T. Marta, and J. V. Schloss. 1998. Inhibitors of branched-chain amino acid biosynthesis as potential antituberculosis agents. J. Antimicrob. Chemother. 42:475-482. [DOI] [PubMed] [Google Scholar]
- 15.Gray, L. D., and A. S. Kreger. 1985. Purification and characterization of extracellular cytolysin produced by Vibrio vulnificus. Infect. Immun. 48:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handfield, M., and R. C. Levesque. 1999. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol. Rev. 23:69-91. [DOI] [PubMed] [Google Scholar]
- 17.Handfield, M., L. J. Brady, A. Progulske-Fox, and J. D. Hillman. 2000. IVIAT: a novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 8:336-339. [DOI] [PubMed] [Google Scholar]
- 18.Hautefort, I., and J. C. D. Hinton. 2000. Measurement of bacterial expression in vivo. Phil. Trans. R. Soc. London B. Biol. Sci. 255:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Slazberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heithoff, D. M., C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel, and M. J. Mahan. 1997. Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. USA 94:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heithoff, D. M., C. P. Conner, and M. J. Mahan. 1997. Dissecting the biology of a pathogen during infection. Trends Microbiol. 5:509-513. [DOI] [PubMed] [Google Scholar]
- 22.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 23.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackowski, S. 1996. Cell cycle regulation of membrane phospholipid metabolism. J. Biol. Chem. 271:20219-20222. [DOI] [PubMed] [Google Scholar]
- 25.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma (b) in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kholti, A., D. Charlier, D. Gigot, N. Huysveld, M. Roovers, and N. Glansdorff. 1998. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J. Mol. Biol. 280:571-582. [DOI] [PubMed] [Google Scholar]
- 27.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE, and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585-597. [DOI] [PubMed] [Google Scholar]
- 28.Kook, H., S. E. Lee, Y. H. Baik, S. S. Chung, and J. H. Rhee. 1996. Vibrio vulnificus hemolysin dilates rat thoracic aorta by activating guanylate cyclase. Life Sci. 59:41-47. [DOI] [PubMed] [Google Scholar]
- 29.Kothary, M. H., and A. S. Kreger. 1987. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J. Gen. Microbiol. 133:1783-1791. [DOI] [PubMed] [Google Scholar]
- 30.Kreger, A., L. DeChatelet, and P. Shirley. 1981. Interaction of Vibrio vulnificus with human polymorphonuclear leukocytes: association of virulence with resistance to phagocytosis. J. Infect. Dis. 144:244-248. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. E., S. Y. Kim, S. J. Shin, H. S. Kim, J. H. Shin, S. H. Choi, S. S. Chung, and J. H. Rhee. 1998. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J. Clin. Microbiol. 36:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, S. E., S. H. Shin, S. Y. Kim, Y. R. Kim, D. H. Shin, S. S. Chung, Z. H. Lee, J. Y. Lee, K. C. Jeong, S. H. Choi, and J. H. Rhee. 2000. Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J. Bacteriol. 182:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 34.Levine, M. M., D. Herrington, J. R. Murphy, J. G. Morris, G. Losonsky, B. Tall, A. A. Lindberg, S. Svenson, S. Baqar, M. F. Edwards, and B. A. D. Stocker. 1987. Safety, infectivity, immunogenicity, and in vivo stability of two attenuated auxotrophic mutant strains of Salmonella typhi, 541Ty and 543Ty, as live oral vaccines in humans. J. Clin. Investig. 79:888-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto, A., S. K. Hong, H. Ishizuka, S. Horinouchi, and T. Beppu. 1994. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene 146:47-56. [DOI] [PubMed] [Google Scholar]
- 37.Mei, J., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteremia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 38.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motley, S. T., and S. Lory. 1999. Functional characterization of a serine/threonine protein kinase of Pseudomonas aeruginosa. Infect. Immun. 67:5386-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mougel, C., and I. B. Zhulin. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26:582-584. [DOI] [PubMed] [Google Scholar]
- 43.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1991. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67:995-1006. [DOI] [PubMed] [Google Scholar]
- 44.Neidhart, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-28. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 45.Neuhard, J., and R. A. Kelln. 1996. Biosynthesis and conversion of pyrimidines, p. 580-599. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 46.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 47.Reddy, G. P., U. Hayat, C. Abeygunawardana, C. Fox, A. C. Wright, D. R. Maneval, Jr., C. A. Bush, and J. G. Morris, Jr. 1992. Purification and determination of the structure of capsular polysaccharide of Vibrio vulnificus MO6-24. J. Bacteriol. 174:2620-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed, L. J., and H. Muench. 1938. A simple method of estimating the fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 49.Rhee, K. Y., D. F. Senear, and G. W. Hatfield. 1998. Activation of gene expression by a ligand-induced conformational change of a protein-DNA complex. J. Biol. Chem. 273:11257-11266. [DOI] [PubMed] [Google Scholar]
- 50.Ross, R., R. Mayer, and R. Benziman. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:33-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sargent, F., N. R. Stanley, B. C. Berks, and T. Palmer. 1999. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073-36082. [DOI] [PubMed] [Google Scholar]
- 53.Simpson, L. M., and J. D. Oliver. 1983. Siderophore production by Vibrio vulnificus. Infect. Immun. 41:644-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glyxinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Testa, J., L. W. Daniel, and A. S. Kreger. 1984. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect. Immun. 45:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokugawa, K., M. Kakitani, T. Ishii, K. Nakamura, H. Masaki, and T. Uozumi. 1994. A novel protein secretion factor from a Vibrio species which operates in Escherichia coli. J. Biotechnol. 35:69-76. [DOI] [PubMed] [Google Scholar]
- 57.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 58.Urabe, H., and H. Ogawara. 1995. Cloning, sequencing, and expression of serine/threonine kinase-encoding genes from Streptomyces coelicolor A3(2). Gene 153:99-104. [DOI] [PubMed] [Google Scholar]
- 59.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 60.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, S. G., S. R. Attridge, and P. A. Manning. 1993. The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol. Microbiol. 9:751-760. [DOI] [PubMed] [Google Scholar]
- 61a.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769-1773. [DOI] [PMC free article] [PubMed]
- 62.Wu, J., N. Ohta, J. L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto, K., A. C. Wright, J. B. Kaper, and J. G. Morris, Jr. 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to V. cholerae El Tor hemolysin 62. gene. Infect. Immun. 58:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zalkin, H., and P. Nysgaard. 1996. Biosynthesis of purine nucleotides, p. 561-579. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.