Abstract
Recent experiments in yeast (Saccharomyces cerevisiae) cells have identified a species-specific region of yeast transcription factor IIB (TFIIB) located at residues 144–157. According to the human TFIIB structure, this region is part of a solvent-exposed helix in the first repeat of the carboxyl-terminal core domain. In this report, we systematically analyze four positions in this region (Lys-147, Cys-149, Lys-151, and Glu-152) that together have been shown previously to be important for yeast TFIIB’s function in vivo. Our experiments suggest that all of these four positions, and in particular positions 151, 149, and 152, are critical for yeast TFIIB’s ability to support cell growth. In addition, we describe an intragenic suppressor screening experiment to identify mutations that reverse, or partially reverse, the temperature-sensitive phenotype of a yeast TFIIB derivative bearing amino acid changes at these four positions to human residues. The suppressor mutations reveal changes at positions 115, 117, and 182 that are located outside the species-specific region of yeast TFIIB, suggesting an extended surface available to interact with other proteins. Finally, we demonstrate that the suppressor mutations restore gene activation in vivo, further supporting the idea that one important function of yeast TFIIB in living cells is to mediate gene activation.
Keywords: Saccharomyces cerevisiae, activators, suppressors
The general transcription factor IIB (TFIIB) is an essential component of the RNA polymerase II transcription machinery in eukaryotes (1). Previous biochemical experiments have demonstrated that it interacts with several other components within the transcription machinery, including RNA polymerase II (2, 3), the TATA box-binding protein (TBP) (4–6), TFIIF (2), and one of the TBP-associated factors (TAF40) (7). It has also been shown to interact with various transcriptional activators (8–13), suggesting that it is involved in the process of transcriptional activation in vitro (1, 14). TFIIB molecules from different eukaryotes share conserved structural motifs and are expected to form similar three-dimensional structures (15, 16). The amino-terminal domain of TFIIB contains a zinc finger motif folded into a zinc ribbon structure (17). The carboxyl-terminal core domain of TFIIB is composed of two imperfect direct repeats that are similarly folded into two subdomains (15, 16). The surfaces of TFIIB suggested to interact with VP16 and TAF40 are located near a cleft formed between these two subdomains (18). One of these surfaces interacts with both VP16 and TAF40 (18), and additionally, with TBP in a DNA–TBP–TFIIB ternary complex structure (16). On the basis of the essential role of TFIIB in transcription initiation and gene regulation, many, if not all, of its solvent-exposed surfaces are likely to interact with other transcription factors—activators, coactivators, or general factors (including RNA polymerase II).
By comparing the yeast Saccharomyces cerevisiae (19) and human (20, 21) forms of TFIIB in yeast cells, we recently identified a species-specific region (residues 144–157) of yeast TFIIB (yTFIIB) (22). This region is part of the solvent-exposed second helix (designated as BH2) of the first direct repeat in the carboxyl-terminal core domain (16). We suggested that four amino acid positions of yTFIIB, Lys-147, Cys-149, Lys-151, and Glu-152, mark the differences between yTFIIB and human TFIIB (hTFIIB) in this region (22). The amino acids in these four positions together have been shown to be important for yTFIIB’s function in both gain-of-function and loss-of-function experiments. An otherwise wild-type yTFIIB (YR1m4) with amino acid changes at these four positions to human residues is functionally impaired and confers a temperature-sensitive phenotype to yeast cells. In addition, yeast cells bearing YR1 m4 but lacking the wild-type yTFIIB differentially affect the expression of genes activated by different activators. These experiments suggest that the species-specific surface of yTFIIB, and in particular, the amino acids at these four positions, may participate in the process of gene activation in vivo (22).
In this paper we further determine the contribution of these four positions to yTFIIB’s function in vivo by systematically analyzing individual and multiple changes in both gain-of-function and loss-of-function experiments. We also identify intragenic suppressors of YR1m4 that reverse, or partially reverse, the temperature-sensitive phenotype. The amino acid positions mutated in these suppressors suggest an extended surface available to interact with other proteins. Finally, we show that three suppressor mutants that were tested have reversed the defect of YR1m4 in gene activation, further supporting the idea that the defect of YR1m4 in fully supporting cell growth is associated with its defect in gene activation. Together, these experiments define an extended functional surface of yTFIIB important for gene activation in living cells.
MATERIALS AND METHODS
Gain-of-Function and Loss-of-Function Experiments.
All the mutations were generated by a PCR-mediated site-directed mutagenesis procedure and confirmed by DNA sequencing. The mutant yeast TFIIB genes carried on a TRP1 plasmid were tested in a plasmid shuffle system as described previously (22). Briefly, the TRP1 plasmids were transformed into a yeast strain bearing a copy of the wild-type yeast TFIIB gene carried on a URA3 plasmid but lacking the chromosomal copy. Serially (10-fold) diluted yeast cell cultures were spotted on plates containing 5-fluoroorotic acid (5-FOA) to determine the ability of each yTFIIB derivative to support cell growth. 5-FOA eliminates the wild-type yeast TFIIB gene carried on the URA3 plasmid because it is toxic to cells expressing the URA3 gene. The same serially diluted cultures were spotted on minimal media plates lacking tryptophan and uracil to estimate the total number of cells analyzed.
Immunoblotting Assay.
The inactive derivatives were tagged with the hemagglutinin (HA) epitope and detected by an antibody against HA (HA.11 from Babco, Richmond, CA). The level of actin, detected by a monoclonal antibody (C4) kindly provided by J. Lessard (23), was used as an internal control. See Shaw et al. (22) for further details.
Intragenic Suppressor Screening.
A yeast strain expressing YR1m4 from a TRP1 plasmid but lacking the chromosomal copy of the yTFIIB gene was generated by 5-FOA selection. To completely eliminate the wild-type yeast TFIIB gene carried on the URA3 plasmid, two rounds of 5-FOA selection were performed (this procedure was also used in the retransformation experiments discussed below). A total of 26 independent 10-ml YEPD (yeast extract/peptone/dextrose) liquid cultures (with an estimated total of 3.1 × 109 cells) were grown at 30°C for 1 day, then plated on separate YEPD plates and incubated at 37°C until colonies appeared. We chose to use separate liquid cultures of small volume to minimize the possibility of isolating nonindependent, clonal, suppressor mutants, a problem that appeared in a pilot experiment using a single liquid culture of large volume (data not shown). Most plates had few or no colonies, and a maximum of 3 colonies were picked from each plate. The yeast colonies were then grown in YEPD liquid cultures at 30°C, and plasmid DNA was isolated and amplified in Escherichia coli. These plasmids were retransformed into a fresh plasmid shuffle yeast strain to confirm that the suppression phenotype was linked to the plasmids. For the experiments summarized in Table 1, a total of 38 yeast TFIIB suppressor genes were isolated and sequenced to identify the mutations. In most cases, with the exception of 6 plates, multiple suppressor mutants isolated from the same YEPD plate revealed point mutations leading to different amino acid changes. Not shown in Fig. 4 and Table 1 is a suppressor mutant changing the arginine at position 149 to leucine (R149L), which was isolated only once. Yeast cells bearing this derivative appeared to grow poorly at both 30°C and 37°C but frequently gave rise to fast-growing colonies (data not shown). Also not shown is one candidate gene that did not contain any mutations according to our sequencing data and failed to confer the suppression phenotype after retransformation (data not shown).
Table 1.
Intragenic suppressors of YR1m4
| Mutation | No. of times isolated | Cell growth
|
|
|---|---|---|---|
| At 30°C | At 37°C | ||
| R149C | 13 | ++++ | ++++ |
| R149S | 5 | ++++ | ++++ |
| R149G | 4 | +++ | +++ |
| N152I | 1 | ++++ | +++ |
| R182L | 4 | ++++ | +++ |
| R182S | 3 | ++++ | + |
| R182C | 3 | +++ | + |
| G115R | 1 | +++ | +/− |
| N117Y | 2 | ++++ | +++ |
| YR1m4 | NA | + | − |
| yTFIIB | NA | +++++ | +++++ |
Listed are intragenic suppressor mutants of YR1m4, number of times they were isolated, and their ability to support cell growth at both 30°C and 37°C. The original data on cell growth are shown in Fig. 4. See text for further details. NA, not applicable.
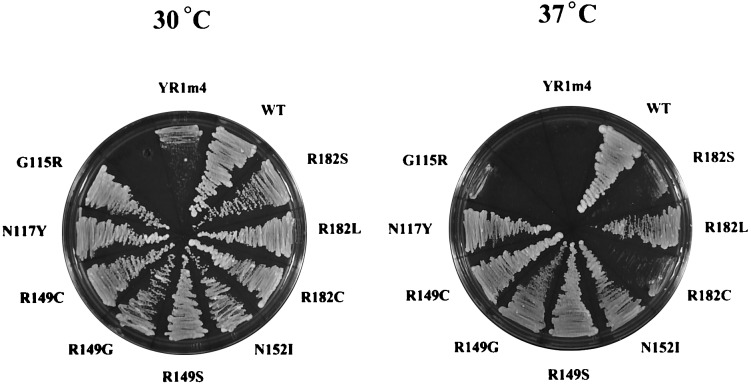
Figure 4.
Yeast cell growth supported by YR1m4 and its intragenic suppressors. Shown are yeast cells grown on YEPD plates at 30°C or 37°C. These cells express the wild-type yTFIIB, YR1m4, or intragenic suppressors isolated in this study. These cell growth results are summarized in Table 1.
β-Galactosidase Assay.
β-Galactosidase assay was performed as described previously (22). Briefly, plasmids carrying the CYC1-lacZ reporter genes under the control of various activators or no activators were used to transform yeast strains. At least three independent yeast transformants were grown in liquid cultures and assayed for β-galactosidase activity.
RESULTS
Gain-of-Function.
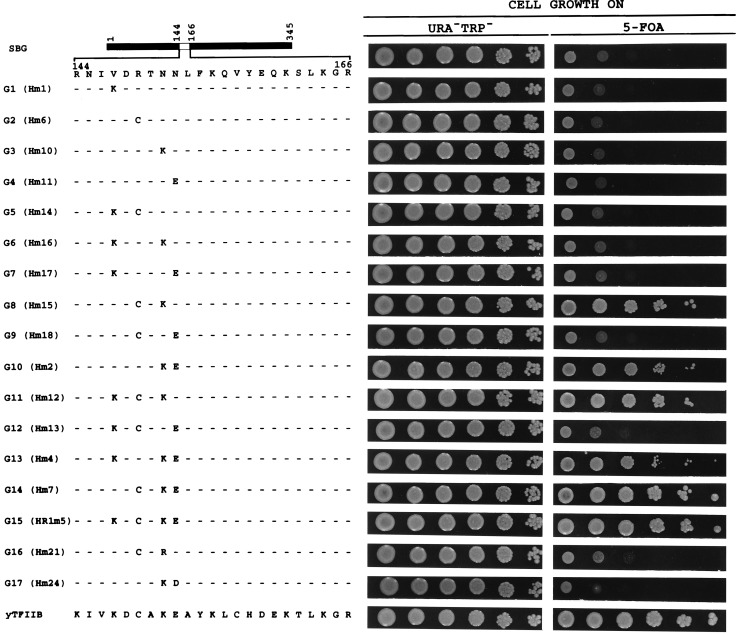
We conducted a systematic gain-of-function study using a completely inactive hybrid yTFIIB protein (SBG) with residues 144–166 replaced by the corresponding hTFIIB sequence (Fig. 1). We analyzed the four amino acid positions (147, 149, 151, and 152) that mark the differences between yTFIIB and hTFIIB in the species-specific region. By site-directed mutagenesis, we generated SBG derivatives bearing individual, double, or triple amino acid changes from human back to yeast residues. For simplicity, each mutant derivative is assigned a number following the letter G (for gain-of-function). The ability of the SBG derivatives to support cell growth was determined in a plasmid shuffle system as described previously (22). Our immunoblotting analysis (Fig. 2) demonstrates that all the inactive yTFIIB derivatives described in this report are stably accumulated in yeast, suggesting that their inability to support cell growth reflects their functional defects in vivo.
Figure 1.
Gain-of-function experiments. Various SBG derivatives were analyzed in a plasmid shuffle system. Serially (10-fold) diluted yeast cultures were spotted on plates containing 5-FOA. The extent of cell growth on 5-FOA plates indicates the ability of the yTFIIB derivative to support cell growth (see ref. 22 and text for further details). The same serially diluted cultures were also spotted on plates lacking tryptophan and uracil to estimate the total number of cells analyzed. The actual names of the plasmids expressing the derivatives are indicated in the parentheses.
Figure 2.
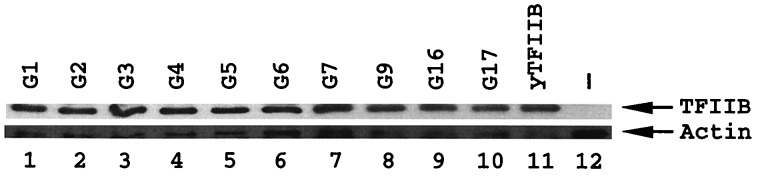
Immunoblotting assay. Inactive yTFIIB hybrids shown in Fig. 1 were detected in an immunoblotting assay as described previously (22). Lane 12 represents a negative control with no tagged yTFIIB protein expressed in yeast cells. The results shown here clearly demonstrate that all these inactive derivatives were stably accumulated in yeast cells, suggesting that their inability to support cell growth reflects their functional defects rather than their intracellular protein levels.
Fig. 1 summarizes our gain-of-function experiments. None of the SBG derivatives with individual amino acid changes from human back to yeast residues (G1–G4) was able to support cell growth. Among all the derivatives with double changes (G5–G10), only G8 and G10 supported cell growth significantly. Both of these derivatives include a change at position 151 from asparagine to the native yeast residue lysine, indicating the importance of this position in yTFIIB function. All the derivatives bearing triple changes (G11–G14), except G12, which has a human residue at position 151, supported cell growth efficiently. Derivative G14, similar to the derivative bearing all four amino acid changes [G15; previously called HR1m5 (22)], supported cell growth as efficiently as the wild-type yTFIIB. Taken together, these results are consistent with the idea that, among the four positions systematically tested in this report, position 151 is most critical for yTFIIB’s function in vivo, followed by positions 149 and 152, and finally position 147 (also see Fig. 3).
Figure 3.
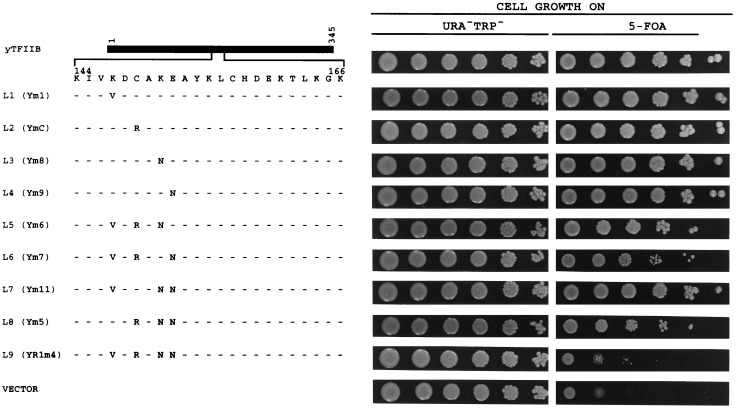
Loss-of-function experiments. These loss-of-function mutants were assayed in a manner similar to that for Fig. 1. See legend to Fig. 1 for further details.
Two amino acids at the three most critical positions in the species-specific region of yTFIIB, Lys-151 and Glu-152, are charged. To determine whether their respective charges or amino acid side chains are important for yTFIIB’s function in vivo, we generated two additional derivatives, G16 and G17. In these two double-mutation derivatives, amino acids at positions 151 and 152 are changed to arginine and aspartic acid, respectively, rather than to the native yeast residues lysine and glutamic acid (G8 and G10), thus introducing identical charges but different amino acid side chains at these positions. The results obtained from G16 and G17, in comparison with those from G8 and G10, respectively, strongly suggest that the amino acid side chains at positions 151 and 152, rather than their charges alone, are important for yTFIIB’s function in vivo. These results are consistent with the idea that the species-specific region of yTFIIB is involved in protein–protein interactions.
Loss-of-Function.
In a loss-of-function experiment (Fig. 3), we changed, either individually or in triplets, the amino acids at the four positions of yTFIIB to human residues. Again, for simplicity, each mutant derivative is assigned a number following the letter L (for loss-of-function). The ability of these derivatives to support cell growth was determined in a similar plasmid shuffle system, and the results are summarized in Fig. 3. yTFIIB derivatives bearing individual amino acid changes (L1–L4) do not show any detectable decrease in the protein’s ability to support cell growth. In addition, none of the derivatives bearing triple changes (L5–L8) showed any dramatic decrease in the protein’s ability to support cell growth. The results obtained with the triple mutation derivatives of yTFIIB suggested to us that double-mutation derivatives would have little effect on the protein’s function in vivo and, therefore, we did not generate and test all the possible double-mutation derivatives. Consistent with our previous results (22), the derivative with all four amino acids changed (L9; previously called YR1m4) is functionally impaired. Together, these results suggest that changes in all four of the positions of yTFIIB are required to generate a severe loss-of-function phenotype. It should be noted that the gain-of-function and loss-of-function experiments were conducted in two different contexts and their results were not expected to be simple mirror images of one another. Our experiments clearly demonstrate that more amino acid changes are required to inactivate the wild-type yTFIIB than to regain the function of the inactive hybrid yTFIIB derivative SBG (Figs. 1 and 3).
Intragenic Suppressors.
Our previous experiments have demonstrated that yeast cells bearing YR1m4 (named L9 in Fig. 3) but lacking the wild-type yTFIIB completely fail to grow at 37°C (Fig. 4). We were interested in determining whether intragenic suppressor mutations could reverse, or partially reverse, this temperature-sensitive phenotype. Spontaneous mutations of this mutant yeast TFIIB gene were isolated that enabled yeast cells to grow at 37°C to various extents. Plasmids carrying the mutant yeast TFIIB genes were purified from these yeast cells and then transformed into a fresh yeast strain to confirm that the suppression phenotype was linked to the plasmids (see Materials and Methods for further details). The mutant yeast TFIIB genes were then sequenced to identify the suppressor mutations. Each suppressor gene was found to contain a single base pair change. Table 1 summarizes the results, illustrating the amino acid changes and number of times each change was identified. The ability of each intragenic suppressor to support cell growth at both 30°C and 37°C is shown in Fig. 4 and summarized in Table 1. The suppressor mutants are named according to the amino acid changes for simplicity, but it should be noted that the starting protein, YR1m4 itself, is a mutant bearing four changes at positions 147, 149, 151, and 152 from yeast to human amino acids. It should be emphasized that, as demonstrated previously (22), YR1m4 is a stable protein in yeast cells and that the suppressor mutants are not likely to simply change the stability of the protein. We note that many mutations were isolated independently more than once, suggesting that our genetic screening recovered most of the possible suppressor mutations.
The intragenic suppressor experiment revealed amino acid changes at five positions, two of which had been mutated originally in YR1m4, as expected. First, Arg-149 was changed to, with a decreasing ability to support cell growth, cysteine, serine, and glycine. Second, Asn-152 was found to be changed to isoleucine. It should be noted that a reversion of Asn-152 to the native yeast residue, glutamic acid, requires more than a single base pair change and, therefore, was not isolated in our screening. Mutants changing either Asn-151 or Val-147 back to the native yeast residue, lysine, were not isolated in our screening because such mutants generated by site-directed mutagenesis (L6 and L8, respectively, in Fig. 3) remained temperature sensitive (not shown).
The remaining three positions of amino acid changes revealed by the intragenic suppressor experiment are located outside the four residues originally mutated in YR1m4. Arg-182 was changed to, with a decreasing ability to support cell growth, leucine, serine, and cysteine. In addition, Gly-115 and Glu-117 were found to be changed to arginine and tyrosine, respectively. (The hTFIIB residues corresponding to Gly-115, Glu-117, and Arg-182 of yTFIIB are asparagine, arginine, and glutamine, respectively.) The implication of these intragenic suppressor results is further addressed in Discussion. A careful analysis of the results shown in Fig. 4 and Table 1 indicates that, interestingly, several suppressor mutations (e.g., G115R, R182S, and R182C) resulted in a much more improved cell growth phenotype at 30°C than at 37°C. It is possible that these mutant proteins’ abilities to support cell growth at 30°C and 37°C may reflect slightly different aspects of their functional properties in vivo.
Restoring Gene Activation Function.
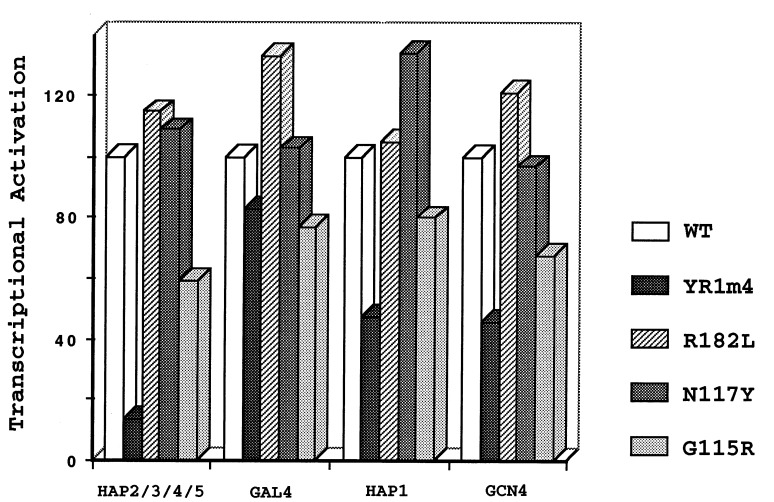
Our previous experiments suggested that the defect of YR1m4 in fully supporting cell growth is due to the protein’s defect in mediating gene activation (22). First, YR1m4, a protein stably accumulated in yeast cells, did not grossly affect transcription start sites. In addition, the expression of genes activated by different activators was differentially affected in cells bearing YR1m4. Specifically, while transcription activated by GAL4, as well as nonactivated transcription, remained similar in yeast cells bearing either the wild-type or the mutant yTFIIB, the expression of a reporter gene activated by HAP2/3/4/5 was dramatically decreased in cells bearing this mutant yTFIIB protein. We analyzed three intragenic suppressor mutants, R182L, N117Y, and G115R, to determine whether the defect in mediating gene activation was reversed. Fig. 5 shows that both R182L and N117Y completely restored the expression of the target gene activated by HAP2/3/4/5. The activation by other activators, HAP1 and GCN4, was also increased to wild-type levels. Gene activation was partially restored in yeast cells bearing G115R (Fig. 5), an intragenic suppressor mutant that only partially reversed the temperature-sensitive phenotype (Fig. 4). These results strongly suggest that the defects of YR1m4 in mediating gene activation and in fully supporting cell growth are associated with each other.
Figure 5.
Restoring the gene activation function. CYC1-lacZ reporter genes under the control of various activators or no activators were assayed in yeast cells bearing the wild-type yTFIIB, YR1m4, or the intragenic suppressor mutants R182L, N117Y, and G115R. Transcriptional activation by each activator, calculated by dividing the activated levels of expression by the nonactivated levels of transcription, is shown as percentage of activation obtained in the wild-type yTFIIB strain. The results for YR1m4 are according to Shaw et al. (22).
DISCUSSION
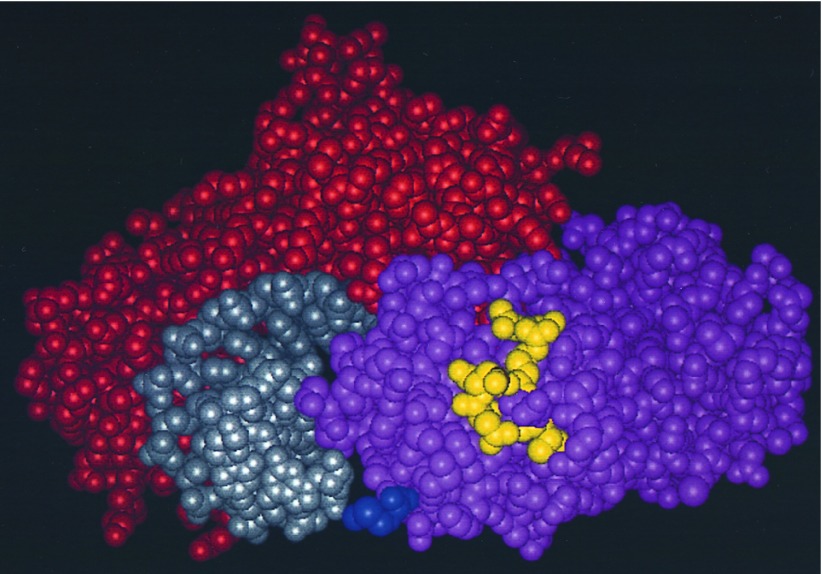
The experiments described in this report suggest an extended surface of yTFIIB important for its function in vivo. Both gain-of-function and loss-of-function experiments suggest that four amino acid positions in the species-specific region of yTFIIB (Lys-147, Cys-149, Lys-151, and Glu-152), and in particular positions 151, 149, and 152, are important for the protein’s ability to support cell growth. According to the hTFIIB structure (16), these amino acid positions are located on the solvent-exposed side of the second helix (BH2) in the first repeat of the carboxyl-terminal core domain (Fig. 6). One of the amino acid positions revealed in our intragenic suppressor analysis, Lys-182, is located at the end of the third helix (BH3). The hTFIIB structure information suggests that this solvent-exposed residue of yTFIIB and the four amino acids in the species-specific region form an extended surface (Fig. 6).
Figure 6.
An extended functional surface of yTFIIB. Shown is a space-filling model of the DNA–TBP–hTFIIB ternary complex structure (16), viewed from downstream, with DNA, TBP, and hTFIIB colored in gray, red, and pink, respectively. Highlighted (in yellow) are the amino acid residues of hTFIIB (Val-135, Arg-137, Asn-139, Asn-140, and Gln-170) corresponding to Lys-147, Cys-149, Lys-151, Glu-152, and Arg-182 of yTFIIB. These residues form an extended surface available to interact with other proteins. Along the path of the highlighted amino acids, the residue corresponding to Arg-182 of yTFIIB is located at the top, followed by those corresponding to Cys-149, Glu-152, Lys-151, and Lys-147 of yTFIIB, respectively. Also highlighted (in blue at the bottom) is the artificially engineered methionine residue attached to the amino terminus (residue 113) of the carboxyl-terminal core domain of hTFIIB. This methionine corresponds to Asn-124 of yTFIIB, which is 8 and 6 residues away from Gly-115 and Asn-117 of yTFIIB, respectively, and is in proximity to the highlighted patch of residues (in yellow). We favor the idea that Gly-115 and Asn-117 also participate in the formation of a further extended functional surface important for gene activation in vivo (see text for further discussions).
No structural information is currently available for the region surrounding Gly-115 and Gln-117 of yTFIIB. The hTFIIB amino acid positions corresponding to Gly-115 and Gln-117 of yTFIIB are only 8 and 6 residues, respectively, away from the amino terminus of the hTFIIB core domain whose structure has been solved (16) (Fig. 6). Previous studies suggest that this region is a flexible and extended linker between the amino-terminal zinc finger domain and the carboxyl-terminal core domain (24). On the basis of the anticipated proximity to each other, positions 115 and 117, together with BH2 and position 182 at the end of BH3, are likely to form a further extended surface (Fig. 6).
We currently favor the idea that all the amino acid positions discussed in this report form an extended functional surface. These residues (with the possible exception of positions 115 and 117) are clearly located on the surface (Fig. 6) according to the hTFIIB structure (16). Mutations at these positions, unlike those involving residues located in the interior of the protein, are less likely to induce a gross conformational alteration. In addition, none of these amino acids of yTFIIB is located near the corresponding hTFIIB sequences involved in an intramolecular interaction and a conformational change induced by VP16 (25). It should be noted that, although we favor the extended surface model, our current experiments cannot rule out other possibilities. For example, on the basis of the proximity between BH2 and BH3, a change at position 182 could potentially shift or reposition BH2 slightly, thus compensating for the defects in BH2. In the crystal structure (16), Arg-169 of hTFIIB, a conserved amino acid corresponding to Arg-181 of yTFIIB, makes contacts with TBP. It is therefore also formally possible that mutations at Arg-182 of yTFIIB may influence the protein’s ability to interact with TBP. Furthermore, our current results cannot rule out the possibility that positions 115 and 117 in the linker region may directly interact with BH2 in vivo.
Our previous experiments suggest that the defect of YR1m4 in fully supporting cell growth may be caused by its defect in mediating gene activation (22). Consistent with this idea, we demonstrate in this report that the intragenic suppressors of YR1m4, R182L and N117Y, completely restore the ability of yTFIIB to mediate gene activation in vivo. In addition, G115R, a suppressor mutant that partially reverses the temperature-sensitive phenotype, partially restores gene activation. These experiments suggest that the extended surface of yTFIIB identified in this report is important for gene activation in vivo.
We suggest that the extended functional surface of yTFIIB participates in the process of gene activation by interacting with other proteins. As discussed in the Introduction, most surfaces of yTFIIB are likely to participate in intricate protein–protein interactions. The experiments shown in Fig. 1 (G16 and G17) demonstrate that amino acid side chains at positions 151 and 152, rather than their charges alone, are important for yTFIIB’s function in vivo, supporting the idea that the surface revealed by our current study is involved in protein–protein interaction. We note that some of our intragenic suppressor mutants have changed positions 149 and 152 to residues that are not native to yTFIIB (see Table 1), suggesting that these positions can tolerate a subset of, but not any, amino acid residues. We also note that the amino acid positions revealed in this study may not represent the entire functional surface of yTFIIB because other solvent-exposed residues important for protein–protein interactions may be conserved and/or invariable.
It remains to be determined what protein(s) interacts with the extended functional surface of yTFIIB. One model (22) proposes that some transcriptional activators can interact (either directly or through some intermediaries) with this surface of yTFIIB, thus recruiting the protein, either by itself (26) or as part of the holoenzyme (27), to the transcription machinery. Based on the observation that this surface of yTFIIB faces downstream, rather than upstream of the TATA box (16), a second model (22) proposes that it could interact with other general transcription factors, in particular TFIIF and RNA polymerase II. These two possibilities are not mutually exclusive. Similar to the surface of hTFIIB that interacts with TBP, TAF40, and VP16 (18), the extended surface of yTFIIB discussed in this report could in principle interact with multiple proteins, either simultaneously or consecutively. It will be interesting to determine whether and how the interaction events of yTFIIB with different proteins may influence each other during the process of gene activation.
Acknowledgments
We thank S. Burley, R. Ebright, J. Zhang, M. Barton, D. Wiginton, R. Ganschow, and members of this laboratory for discussions and/or comments on the manuscript; D. Nikolov and S. Burley for providing the DNA–TBP–hTFIIB ternary complex structure coordinates; J. Lessard for providing the antibody against actin; and B. Aronow, J. Howard, P. Rosevear, and M. Sussman for assistance in creating Fig. 6. We also thank anonymous reviewers for helpful suggestions. J.M. is a recipient of an American Cancer Society Junior Faculty Research Award. D.J.C is supported in part by National Institutes of Health Teratology Training Grant ES07051. M.J.D. was a recipient of a National Institutes of Health Postdoctoral Fellowship. This work was supported in part by a grant from the National Institutes of Health to J.M. and a Ruth Lyons Cancer Research Challenge Grant from the University of Cincinnati to J.M. and M.J.D.
ABBREVIATIONS
- TFIIB
transcription factor IIB
- yTFIIB
yeast TFIIB
- hTFIIB
human TFIIB
- TBP
TATA box-binding protein
- TAF
TBP-associated factor
- 5-FOA
5-fluoroorotic acid
References
- 1.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 2.Ha I, Roberts S, Maldonado E, Sun X, Kim L-U, Green M, Reinberg D. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 3.Tschochner H, Sayre M H, Flanagan P M, Feaver W J, Kornberg R D. Proc Natl Acad Sci USA. 1992;89:11292–11296. doi: 10.1073/pnas.89.23.11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S, Hahn S, Guarente L, Sharp P A. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita S, Hisatake K, Kokubo T, Doi K, Roeder R G, Horikoshi M, Nakatani Y. Science. 1993;261:463–466. doi: 10.1126/science.8332911. [DOI] [PubMed] [Google Scholar]
- 6.Hisatake K, Roeder R, Horikoshi M. Nature (London) 1993;363:744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- 7.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 8.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colgan J, Wampler S, Manley J L. Nature (London) 1993;362:549–553. doi: 10.1038/362549a0. [DOI] [PubMed] [Google Scholar]
- 10.Kim T K, Roeder R G. Proc Natl Acad Sci USA. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Nature (London) 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y-S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 13.Roberts S R E, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 14.Sharp P A. Nature (London) 1991;351:16–18. doi: 10.1038/351016d0. [DOI] [PubMed] [Google Scholar]
- 15.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 16.Nikolov D B, Chen H, Halay E D, Usheva A A, Kisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhu W, Zeng Q, Colangelo C M, Lewis L M, Summers M, Scott R A. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 18.Hori R, Pyo S, Carey M. Proc Natl Acad Sci USA. 1995;92:6047–6051. doi: 10.1073/pnas.92.13.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto I, Ware D E, Hampsey M. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 20.Ha I, Lane W S, Reinberg D. Nature (London) 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 21.Malik S, Hisatake K, Suminoto H, Horikoshi M, Roeder R. Proc Natl Acad Sci USA. 1991;88:9553–9557. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw S P, Wingfield J, Dorsey M J, Ma J. Mol Cell Biol. 1996;16:3651–3657. doi: 10.1128/mcb.16.7.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lessard J L. Cell Motil Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- 24.Barberis A, Muller C, Harrison S, Ptashne M. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts S G E, Green M R. Nature (London) 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y-S, Green M R. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 27.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]