Abstract
Sepsis caused by gram-negative bacteria and that caused by gram-positive bacteria often manifest similar clinical features. We investigated plasma proinflammatory cytokine profiles in patients with sepsis due to gram-positive and gram-negative bacteria and studied the cytokine production and differential gene regulation of leukocytes stimulated ex vivo with Escherichia coli lipopolysaccharide or heat-killed Staphylococcus aureus. Concentrations of tumor necrosis factor alpha, interleukin 1 receptor antagonist (IL-1Ra), IL-8, IL-10, IL-18 binding protein, procalcitonin, and protein C in plasma did not differ between patients with sepsis due to gram-negative and gram-positive bacteria. However, plasma IL-1β, IL-6, and IL-18 concentrations were significantly higher in patients with sepsis due to gram-positive bacteria. Ex vivo stimulation of whole blood with heat-killed S. aureus markedly increased IL-1β and IL-18 levels more than E. coli lipopolysaccharide stimulation. Microarray analysis revealed at least 359 cross-validated probe sets (genes) significant at the P < 0.001 level whose expression discriminated among gram-negative-organism-stimulated, gram-positive-organism-stimulated, and unstimulated whole-blood leukocytes. The host inflammatory responses to gram-negative and gram-positive stimuli share some common response elements but also exhibit distinct patterns of cytokine appearance and leukocyte gene expression.
Despite improvements in hemodynamic monitoring, antibiotics, and other supportive therapies, sepsis remains the most common cause of death within intensive care units (27) and the 13th leading cause of death overall in the United States (10). More than 500,000 people are diagnosed with sepsis each year in the United States, and the observed incidence is still rising (3, 26, 35), with financial costs exceeding $16 billion each year (3).
Although there have been modest successes in treating sepsis in animal models, little advancement has been made in human studies (1, 4, 5, 36). Possible reasons for this failure include heterogeneity of the patient population, improper timing of attempted therapies, and, more recently appreciated, the complex redundancy of molecular pathways of inflammation induced by different microbial pathogens (18, 32). Indeed, there is growing recognition that gram-positive organisms are responsible for an ever-increasing number of septic events (9, 11, 31).
The American College of Physicians and the Society of Critical Care Medicine have adopted consensus definitions of sepsis and the systemic inflammatory response syndrome (SIRS) in an attempt to standardize patient characterization (2). Severe sepsis is a known or suspected microbial infection associated with fever, leukocytosis or leukopenia, hypotension, increased cardiac output, and decreased peripheral vascular resistance (2, 16). Injury severity scoring systems measure clinical parameters to stratify the severity of patient illness, determine level of intervention, and potentially predict patient outcome. However, these scoring systems primarily measure the physiological effect of the infection on the host, not the microbial, biochemical, or genetic mechanism(s) of the organ injury response (23). Similar clinical pictures result from the actions of any number of inflammatory stimuli, both noninfectious and infectious, including different classes of organisms. Specifically, SIRS can be caused by infection with gram-positive or gram-negative bacteria, multiple trauma, ischemia-reperfusion injury, pancreatitis, severe thermal injury, and other noxious stimuli.
The successful use of anticytokine or anti-inflammatory therapies to interrupt the development of the sepsis response requires a more thorough understanding of the initiating mediators involved in its induction. Recent studies on the role of the Toll-like receptor (TLR) family have shown that these cell surface proteins differentially recognize and signal the presence of products of gram-positive and gram-negative bacteria and have highlighted the complexity of the host response to different microbial pathogens (28, 29). Although sepsis and SIRS are mediated in some part by the early production of a variety of proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin 1 (IL-1), IL-12, and IL-18, among others, there is growing evidence to suggest that the patterns of early cytokine and mediator production may be dependent upon the specificity of the microbial pathogens and the host recognition pathways invoked (19, 24, 25).
In the present study, we sought to examine the diversity in host response to gram-negative and gram-positive pathogens by focusing on the differential proinflammatory cytokine responses in the plasma of patients with sepsis due to gram-negative or gram-positive bacteria, as well as assessing ex vivo stimulation of whole blood with gram-negative and gram-positive microbial products. Moreover, an initial genome-wide survey of gene expression patterns was performed on whole blood from three subjects stimulated ex vivo with a representative product from an individual gram-negative or gram-positive organism.
These studies suggest that distinct patterns of cytokine production may be induced by products of different gram-negative and gram-positive microbes. In particular, sepsis due to gram-positive bacteria and exposure of whole blood to heat-killed Staphylococcus aureus appeared to preferentially stimulate the production of members of the IL-1 superfamily, including IL-1β and IL-18. Examination of the early (2-h) genome-wide gene expression patterns of whole-blood leukocytes stimulated with either bacterial lipopolysaccharide (LPS) (Escherichia coli) or heat-killed S. aureus revealed 359 genes whose differential expression discriminated among unstimulated and stimulated whole blood. Several of these differentially expressed genes encoded proinflammatory as well as anti-inflammatory cytokines. Although the physiological responses of the host to sepsis due to gram-negative and gram-positive bacteria may appear similar, these studies suggest that the early inflammatory and immunological response to these pathogens may differ dramatically depending upon the inciting organism.
MATERIALS AND METHODS
Patient enrollment.
Fifty-two patients were enrolled prospectively from 33 different institutions as part of the PAFase (ICOS Inc., Bothell, Wash.) ARDS Prevention Study. The criteria for patient inclusion included the following: age of ≥18 years, clinical evidence of infection within 3 days of enrollment in the study, fulfillment of SIRS criteria, and persistent hypoperfusion attributable to the infection. The SIRS criteria, as established by the consensus statement from the American College of Chest Physicians and the Society of Critical Care Medicine, require that the patient have two of the following: (i) core body temperature of >38°C or <36°C, (ii) tachycardia (>90 beats per min), (iii) tachypnea (>20 breaths per min or an arterial CO2 pressure of <32 mm Hg), (iv) leukocytosis or leukopenia (white blood cell count of >12,000 cells/mm3 or <4,000 cells/mm3) or >10% immature forms (1). Informed consent was provided by the patient or an appropriate surrogate, and the conduct of the study was approved by each individual institutional review board. Permission to receive and analyze blinded samples from these institutions was granted by the Institutional Review Board at the University of Florida. Patients who met any of the following criteria were excluded from the study: (i) radiographic bilateral infiltrates with a ratio of arterial O2 pressure to the fraction of inspired O2 of ≤200 within 72 h of enrollment, (ii) known or suspected infection with human immunodeficiency virus, (iii) Child-Pugh grade C liver dysfunction or known esophageal varices, (iv) Glasgow coma scale of ≤8 arising from a closed head injury, (v) cardiopulmonary arrest within 3 days of enrollment, (vi) administration of pentoxifylline, granulocyte-colony stimulating factor, immunoglobulins, or inhaled nitric oxide within 3 days of enrollment, (vii) administration of corticosteroid doses of ≥1.0 mg of prednisone (or its dose equivalent) per kg of body weight per day or any immunosuppressive therapy within 3 days of enrollment, (viii) participation in a clinical trial within 28 days of enrollment, (ix) positive blood or urine pregnancy test, suspicion of pregnancy, or lactation at the time of enrollment, (x) prior fulfillment of the protocol requirements for inclusion during the present hospitalization, (xi) presence of a “do not resuscitate” order or lack of a commitment of aggressive intensive care, and (xii) prognosis of an underlying disease that limits life expectancy to less than 3 months.
Blood was collected from these patients at the time of enrollment, samples were centrifuged, and the resulting plasma was frozen at −70°C until analysis.
Measurements of inflammatory mediators in plasma.
Levels of TNF-α, IL-1β, IL-1, IL-1 receptor antagonist (IL-1Ra), IL-6, IL-8, IL-10, IL-18, IL-18 binding protein (IL-18BP) (21), procalcitonin, and total protein C in plasma were determined for all patients at baseline (at the time of enrollment prior to drug administration). Workers who performed cytokine measurements were blinded to patient identity and etiology of sepsis. TNF-α, IL-6, IL-8, and IL-10 were measured by enzyme-linked immunosorbent assay (ELISA) using PharmPaks obtained from R&D Systems (Minneapolis, Minn.) according to the manufacturer's instructions. Procalcitonin was determined using a luminescence-based immunoassay from Brahms Diagnostica GmbH (Atlanta, Ga.). The IL-18 ELISA kits were purchased from Medical and Biological Laboratories Co., Ltd. (Watertown, Mass.). IL-18BP levels were determined with a technique developed in the laboratory of Daniela Novick at the Weizmann Institute in Rehovot, Israel, which uses a double-antibody ELISA to calculate free IL-18 concentrations on the basis of the law of mass action (22). This ELISA does not recognize either IL-18 or pro-IL-18, and there is no interference with IL-18BP measurements with increasing concentrations of IL-18 (22). In 34 of the 52 patients where remaining sample was available, total protein C levels were determined by ELISA by using kits provided by Diagnostica Stago, Inc. (Parsippany, N.J.).
Whole blood ex vivo bacterial stimulation.
After informed consent was obtained, venous whole blood was obtained from three healthy young adults by venipuncture into heparinized tubes. For gene expression analyses, venous blood was stimulated with either 1 μg of LPS per ml or 0.1% heat-killed S. aureus Cowan for 2 h at 37°C. Control samples were processed in parallel without stimulation, and each sample was evaluated without pooling of the samples. In one of the three subjects, the blood draw was repeated a total of three times to evaluate the reproducibility of gene expression patterns from the same individual. Because of the volume of blood required, the three separate blood draws were performed 1 week apart. By analyzing replicates from three independent subjects and from a single subject three times, the variation in gene expression due to individual genetic polymorphisms or preexisting conditions (comorbidities) could be distinguished from the variation due to analytical replication. Correlation coefficients for gene expression (for the 12,558 probe sets in the U95Av2 GeneChip) with replicates obtained from the same individual ranged between 0.97 to 0.99 (R2), whereas the same correlation coefficients between different individuals ranged between 0.91 to 0.97.
The blood from each individual collection was divided into three aliquots and either left unstimulated or stimulated for 2 h with LPS or heat-killed S. aureus Cowan. After the 2-h incubation, the whole blood was centrifuged at 400 × g for 10 min at 4°C, and the buffy coat layer was removed. Contaminating erythrocytes were removed by lysis with hypotonic saline and the buffy coat pellet was recovered by centrifugation. Total cellular RNA was isolated from the buffy coat with a commercial kit (RNeasy; Qiagen, Inc.); purity was confirmed by spectrophotometry (A260/A280 ratio), examination of an ethidium bromide stained-RNA agarose gel, and reverse transcription-PCR amplification of a housekeeping gene (Cu/Zn superoxide dismutase) (30).
cRNA synthesis and chip hybridization.
cRNA synthesis was performed by a single investigator starting with 10 μg of total cellular RNA, based on the protocol outlined by Affymetrix, with few modifications. After the mRNA had been selected by hybridization with the primer T7-(dT)24 (Genset Oligos, Genset Corp., La Jolla, Calif.), double-stranded cDNA was synthesized according to a standardized protocol (SuperScript double-stranded cDNA synthesis kit; Invitrogen Corp., Carlsbad, Calif.). cRNA was transcribed in vitro, incorporating biotinylated nucleotides by using a BioArray high-yield RNA transcript labeling kit (T7) (Enzo Life Sciences, Inc., Farmingdale, N.Y.), and hybridized onto Hu95aVer2 oligonucleotide arrays (Affymetrix). Each sample was studied in parallel, and the samples were not pooled. The chips were hybridized for 16 h at 45°C, stained and washed according to the Affymetrix protocol (EukGE-WS2v4) using an Affymetrix fluidics station, and scanned with an Affymetrix scanner.
Microarray data analysis.
Scanned images (.dat files) were analyzed with Affymetrix Microarray Suite version 5. Raw Q scores ranged from 2.2 to 5; average background ranged from 50 to 144. Chip-to-chip normalization was accomplished by using global normalization with an average target gene intensity of 500. Scaling factors ranged from 4 to 18.
Expression filter.
Probe sets that were flagged as absent on all arrays analyzed in this study by the Affymetrix Microarray Suite version 5 software with default settings were removed from the datasets and not included in the analyses reported here. (The signal intensity measurements as detected reflect the degree of hybridization of synthesized cRNA to the probe sets on the microarray chip. These probe sets represent genes or DNA sequences within genes. Some genes are represented by more than one probe set on a given microarray, and hence probe sets are not uniquely correlated to genes. However, for ease of discussion, we use the terms “probe sets” and “genes” interchangeably.)
Variation filter, normalization, and cluster analysis.
The signal intensities of the probe sets remaining after applying the expression filter were ranked according to the coefficient of variation, and the 50% of the data set with the greatest coefficient of variation were then normalized to a mean of 0 and a standard deviation of 1. k-mean determinations and hierarchical cluster analyses were performed with the variance-normalized data set and viewed with the algorithms in the software packages Cluster and TreeView developed by Eisen et al. (14).
Supervised learning, discrimination analysis, and cross validation.
The hybridization signal intensities of the genes passing the initial expression filter were analyzed (in part) with BRB Array Tools 3.01 (developed by Richard Simon and Amy Peng) to identify genes that differentiated among the three treatment classes: unstimulated, stimulated with heat-killed S. aureus Cowan, and stimulated with LPS (P ≤ 0.001). The ability of gene identification to predict treatment class was assessed by “leave-one-out” cross-validation using each of four methods of class prediction: nearest-neighbor prediction, three-nearest-neighbors prediction, linear discriminant analysis, and nearest-centroid analysis.
Blood cytokine analyses from ex vivo stimulation.
Whole blood from three healthy subjects was either unstimulated or stimulated with 1 ng, 1 μg, or 10 μg of E. coli O111:B4 LPS endotoxin (Sigma Fine Chemicals, St. Louis, Mo.) per ml or 0.001, 0.01, or 0.1% heat-killed S. aureus Cowan overnight in a closed system on a wig-wag in a 37°C incubator. At the end of the incubation, the blood was centrifuged at 400 × g for 8 min at 4°C, and the resulting plasma was stored at −70°C for subsequent cytokine analyses.
Statistical analyses.
Microarray data were analyzed as described above. Remaining data were analyzed by either the Mann-Whitney rank sum test, Student's t test, or analysis of variance (ANOVA), as stated in the figure legends and tables. For ANOVA, post hoc multiple-range tests were performed according to the Student-Newman-Keuls method, and significance was designated at the 95% confidence level.
RESULTS
Patient studies.
Of the 52 septic patients studied at baseline, 25 (48.1%) had documented infections with gram-negative organisms and 27 (51.9%) had infections with gram-positive organisms. Table 1 summarizes the microbial source for the two experimental groups. The mean ages were similar, and both groups had a male predominance (Table 2). The mean APACHE II scores and multiple organ dysfunction scores (MODS) were also not different between patients with sepsis due to gram-negative and gram-positive bacteria (Table 2).
TABLE 1.
Microbial sources in 52 patients with sepsis
| Type and organism | No. of patients with positive blood cultures |
|---|---|
| Gram positive | |
| Staphylococcus aureus | 15 |
| Streptococcus pneumoniae | 3 |
| Streptococcus, β-hemolytic, group A, B, C, F, or G | 3 |
| Bacillus species | 1 |
| Diphtheroids (coryneform bacteria) | 1 |
| Streptococcus viridans group | 1 |
| Streptococcus, group D, unspecified | 1 |
| Other | 2 |
| Gram negative | |
| Klebsiella pneumoniae | 5 |
| Escherichia coli | 4 |
| Bacteroides fragilis | 2 |
| Proteus mirabilis | 2 |
| Acinetobacter baumannii | 1 |
| Acinetobacter calcoaceticus | 1 |
| Enterobacter aerogenes | 1 |
| Haemophilus influenzae | 1 |
| Klebsiella species (other) | 1 |
| Neisseria meningitides | 1 |
| Pseudomonas aeruginosa | 1 |
| Serratia marcescens | 1 |
| Stenotrophomonas | 1 |
| Other | 3 |
TABLE 2.
Demographic data of septic patients
| Parameter | Result for patients with sepsis due to:
|
|
|---|---|---|
| Gram-positive bacteria | Gram-negative bacteria | |
| No. of patients | 27 | 25 |
| Age (yr)a | 54.3 ± 3.4 | 58.1 ± 3.8 |
| No. of male patients (%) | 16 (59.3) | 14 (56.0) |
| APACHE II scorea | 24.9 ± 1.4 | 21.0 ± 1.9 |
| MODS scorea | 7.1 ± 0.6 | 5.6 ± 1.9 |
Means ± standard errors of the means.
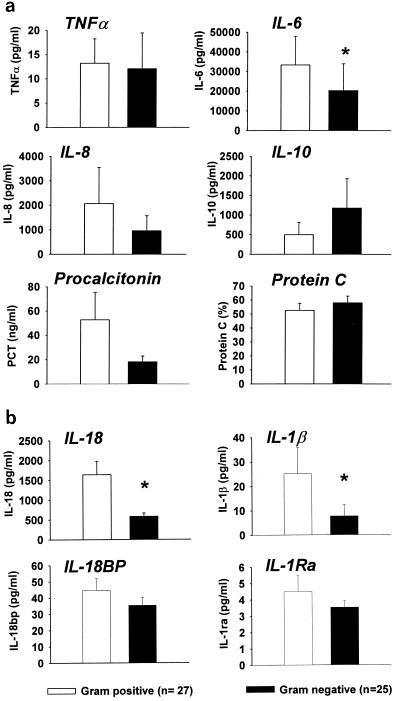
Baseline levels of TNF-α, IL-1Ra, IL-8, IL-10, IL-18BP, procalcitonin, and protein C in plasma were not significantly different between septic patients with gram-positive and gram-negative infections. In contrast, plasma IL-1β, IL-6, and IL-18 concentrations were significantly higher among patients with sepsis due to gram-positive bacteria than patients with sepsis due to gram-negative bacteria (Fig. 1) despite no significant differences in the magnitude of the physiologic response (APACHE II score), the degree of organ injury (MODS score), or other proinflammatory cytokines. Similarly, the concentration of free IL-18 in plasma was significantly higher in patients with sepsis due to gram-positive bacteria than those with sepsis due to gram-negative bacteria (median, 232 pg/ml for gram positive versus 81 pg/ml for gram negative by the Mann-Whitney rank sum test, P < 0.05). These findings, therefore, suggest that the patterns of plasma cytokine appearance may differ between patients with sepsis due to gram-negative and gram-positive bacteria.
FIG. 1.
Plasma cytokine concentrations in patients with sepsis due to gram-negative (n = 25) and gram-positive (n = 27) bacteria. Plasma was obtained at admission to a phase II clinical trial of PAFase prior to administration of test drug or placebo. Measurements of protein C were obtained from 34 of the 52 patients. Values are means ± standard errors of the means. *, P < 0.05 by Student's t test.
Gene expression profiles of leukocytes from whole blood stimulated ex vivo with LPS or S. aureus.
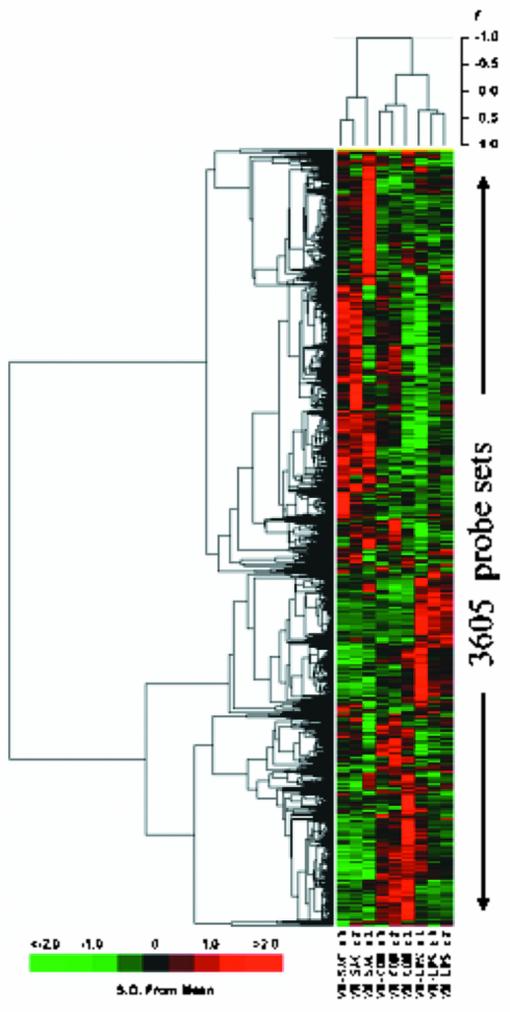
The early global gene expression pattern was evaluated in leukocytes obtained from whole blood stimulated with either LPS or heat-killed S. aureus from three healthy subjects. Probe sets (n = 3,605) whose signal intensities exhibited the greatest variation across the nine arrays (three subjects for each response class: unstimulated, LPS stimulated, and S. aureus stimulated) were hierarchically clustered as shown in Fig. 2. The array dendrogram revealed that each treatment group clustered together, demonstrating excellent reproducibility of the response to the different stimuli. Furthermore, there was greater similarity between expression patterns obtained from the LPS-stimulated and the untreated (control) leukocytes, compared to the pattern observed after S. aureus stimulation.
FIG. 2.
Different patterns of gene expression in human leukocytes after stimulation with LPS and S. aureus. Hierarchical clustering of variance-normalized gene expression data from unstimulated human leukocytes and from leukocytes that had been exposed to either LPS or heat-killed S. aureus for 2 h prior to RNA harvest. Samples were obtained from three subjects. Expression and variation filters were applied to the data set prior to clustering. Probe sets whose hybridization signal intensity was at or below background levels (absent) on all arrays tested were eliminated from further analysis. The resulting data set was culled by ranking on the coefficient of variation and eliminating the bottom half of the data set to remove probe sets whose expression did not vary between the treatment regimens. The gene expression observations were variance normalized to a mean of 0 and a standard deviation of 1, and this normalized data set was subjected to hierarchical cluster analysis with average linkage clustering of the nodes. The variation in gene expression for a given gene is expressed as distance from the mean observation for that gene (S.D., standard deviations). The scale adjacent to the dendrogram is for Pearson's correlationcoefficient. CON, control unstimulated leukocytes; LPS, leukocytes exposed to LPS for 2 h prior to RNA harvest; SAC, leukocytes exposed to S. aureus Cowan for 2 h prior to RNA harvest; r, replicate number.
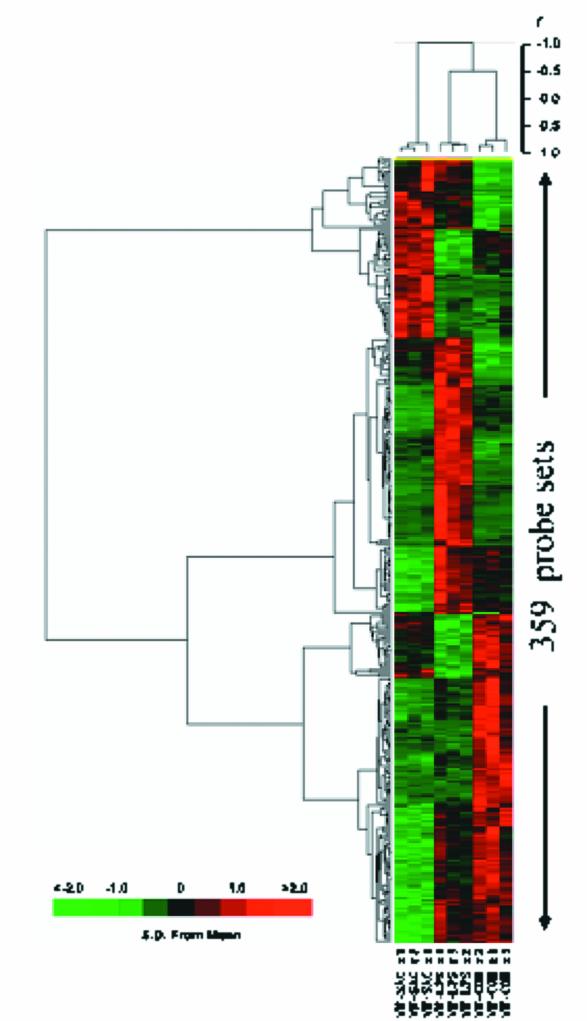
BRB Array Tools was used to detect genes that displayed differential hybridization signal intensities among the three treatment classes (unstimulated, LPS-stimulated, or S. aureus-stimulated leukocytes). There were 359 probe sets that differentiated among the treatment classes (P ≤ 0.001) (Fig. 3). Leave-one-out cross validation was used to establish the ability of probe sets significant at the P < 0.001 level to function as classifiers of treatment response (Table 3). In every case, the class identity of the array left out of the training set was correctly predicted by each of the four prediction models: nearest-neighbor prediction, three-nearest-neighbors prediction, linear discriminant analysis, and nearest-centroid analysis.
FIG. 3.
Hierarchical clustering of hybridization signal intensity (variance normalized gene expression) of probe sets significant at the P ≤ 0.001 level in unstimulated leukocytes and leukocytes that had been exposed to either LPS or heat-killed S. aureus for 2 h prior to RNA harvest. Samples were obtained from three subjects. A total of 359 genes were identified by using BRB Array Tools at the P < 0.001 level. Hierarchical clustering reveals that the LPS-stimulated and unstimulated samples were more similar than the S. aureus-stimulated samples. Also see Table 3.
TABLE 3.
Leave-one-out cross validationa
| Array | Array ID | Class labelb | No. of genes in classifier | Validation by:
|
|||
|---|---|---|---|---|---|---|---|
| Linear discriminant analysis | Nearest-neighbor prediction | 3-Nearest-neighbors prediction | Nearest-centroid Analysis | ||||
| 1 | CON r1 | CON | 185 | Yes | Yes | Yes | Yes |
| 2 | CON r2 | CON | 152 | Yes | Yes | Yes | Yes |
| 3 | CON r3 | CON | 185 | Yes | Yes | Yes | Yes |
| 4 | LPS r1 | LPS | 209 | Yes | Yes | Yes | Yes |
| 5 | LPS r2 | LPS | 194 | Yes | Yes | Yes | Yes |
| 6 | LPS r3 | LPS | 154 | Yes | Yes | Yes | Yes |
| 7 | SAC r1 | SAC | 295 | Yes | Yes | Yes | Yes |
| 8 | SAC r2 | SAC | 170 | Yes | Yes | Yes | Yes |
| 9 | SAC r3 | SAC | 182 | Yes | Yes | Yes | Yes |
Leave-one-out cross validation demonstrated the ability of probe sets significant at the P < 0.001 level to function as classifiers of treatment response. In every case, the class identity of the array left out of the training set was correctly predicted by each of the four prediction models. See also Fig. 3.
CON, control unstimulated leukocytes; SAC, leukocytes exposed to S. aureus Cowan for 2 h prior to RNA harvest.
To examine more closely the response to microbial stimulation, and to eliminate differences in gene expression due to subject-to-subject variability, gene expression profiles were also examined from three independent replicates of the experiment with blood obtained from the same donor. The data were processed and subjected to similar methods of unsupervised and supervised analyses, as described above.
When gene expression patterns were investigated from leukocytes obtained from a single individual, the gene expression pattern in response to the LPS and S. aureus stimuli were similar to the patterns seen from the three independent subjects. As expected, more probe sets were found to be significantly different among the treatment groups when obtained from replicates of the same individual (n = 758) than were seen with three independent subjects (n = 359), of which 169 probe sets were concordant. Once again, the gene expression pattern in response to LPS was more similar to the pattern seen in controls than to S. aureus stimulation. Leave-one-out cross validation established the ability of the significant probe sets to act as classifiers of treatment response.
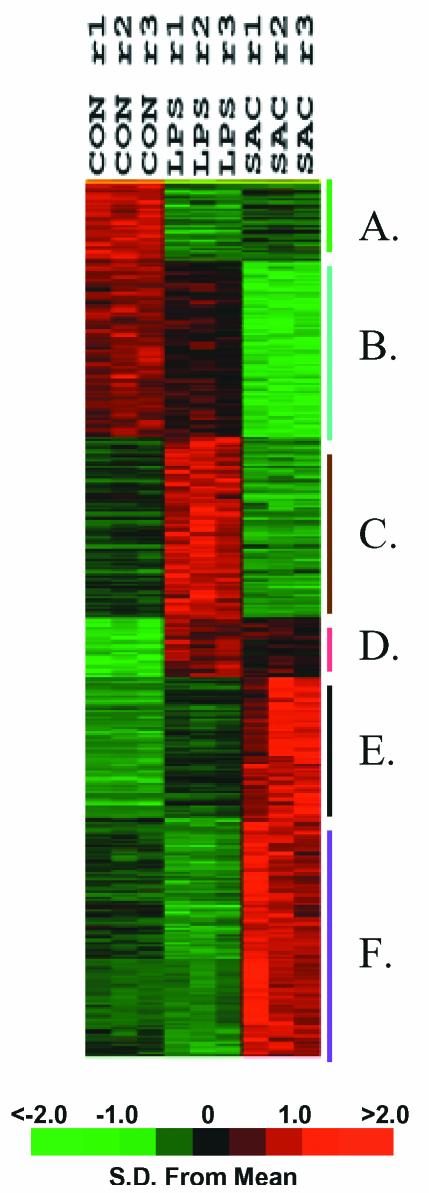
Figure 4 shows the k-means clustering pattern of these 758 probe sets. Among these, 223 probe sets (29.4%) whose signals decreased after stimulation with either LPS or S. aureus (bins A and B), with the probe sets in bin B having less expression attenuation with LPS stimulation than those represented in bin A. Bin C harbors 155 genes (20.4%) whose signal intensity was increased after LPS stimulation and downregulated with S. aureus stimulation (see Table 3 in the supplementary data at http://www.surgery.ufl.edu/Research/Active_Research_Projects/LPS-SAC_research_project.htm). Bins D and E hold 172 genes (22.7%) that exhibit increased expression from stimulation with either LPS or S. aureus. Bin F contains 208 genes (27.4%) that increased in signal intensity after S. aureus stimulation (see Table 4 in the supplementary data).
FIG. 4.
k-means clustering of hybridization signal intensity (gene expression) differences of 758 probe sets significant at the P ≤ 0.001 level in unstimulated leukocytes and leukocytes that had been exposed to either LPS or heat-killed S. aureus for 2 h prior to RNA harvest. Samples were obtained from a single subject with three replicates. The gene expression observations were variance normalized and subjected to k-means clustering to six bins, labeled A through F. The variation in gene expression for a given gene is expressed as distance from the mean observation for that gene (S.D., standard deviations). CON,control unstimulated leukocytes; LPS, leukocytes exposed to LPS for 2 h prior to RNA harvest; SAC, leukocytes exposed to S. aureus Cowan for 2 h prior to RNA harvest; r, replicate number.
Among the 758 predictor genes, 378 genes were upregulated with LPS stimulation. Of these, 180 displayed a greater-than-twofold increase with LPS stimulation over their respective unstimulated expression values. Hence, 198 probe sets were significantly upregulated with LPS stimulation, but the magnitude of the change was less than the traditional twofold cutoff. Similarly, there were 418 genes upregulated with S. aureus stimulation, and of these, 289 were increased more than twofold. Only 87 genes were commonly upregulated twofold and statistically significant (P ≤ 0.001) with both gram-positive and gram-negative stimulation (see Table 5 in the supplementary data). Not unexpectedly, many genes whose expression was commonly increased by the two inflammatory stimuli included those encoding TNF-α, IL-1β, IL-6, IL-1Ra, CD69 (an early T-cell antigen), and several members of the chemokine family, including Groβ, Groγ, and several small inducible cytokines. In addition, several genes involved in NF-κB activation were upregulated uniformly.
Likewise, of these 758 predictor genes, 88 were reduced more than twofold following LPS stimulation while S. aureus stimulation reduced the hybridization signal intensity of 247 genes more than twofold. Forty-three genes displayed more than a twofold reduction in hybridization signal intensities after either LPS or S. aureus stimulation. Commonly down-regulated genes included those encoding caspase 8, IL-8 receptor β, and a transforming growth factor receptor.
Ex vivo whole-blood stimulation.
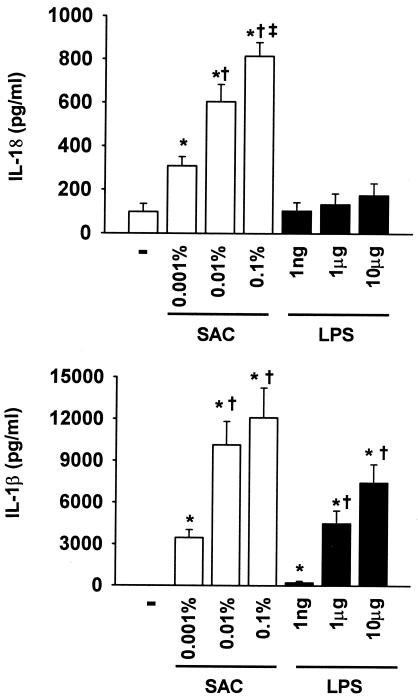
To determine more directly whether products of gram-negative and gram-positive microbes induce a varied proinflammatory cytokine response, heparinized whole blood was stimulated ex vivo overnight with increasing concentrations of either E. coli LPS or heat-killed S. aureus Cowan, and plasma cytokine release into the plasma was determined. As shown in Fig. 5, both LPS and heat-killed S. aureus produced dose-dependent increases in the production of proinflammatory cytokines. However, the magnitude of the increase in IL-1β and IL-18 release was dramatically lower in whole blood stimulated with LPS than in whole blood stimulated with gram-positive S. aureus. In contrast, the concentrations of IL-18BP produced in response to ex vivo stimulation with LPS and S. aureus were comparable (data not shown). These findings are consistent with the baseline plasma measurements from septic patients, suggesting that the patterns of blood cytokine production differ with the inducing organism and that a gram-positive organism, such as S. aureus, induces IL-1β and IL-18 production to a greater extent than bacterial LPS.
FIG. 5.
IL-1β and IL-18 production from whole blood stimulated overnight with increasing concentrations of LPS or heat-killed S. aureus. Heparin-anticoagulated whole blood was obtained from three healthy volunteers and incubated with 100 ng, 1 μg, or 10 μg of E. coli LPS/ml or 0.01, 0.1, or 1% (wt/vol) heat-killed S. aureus Cowan (SAC) overnight at 37°C. The following morning, plasma was separated from the cells, and plasma cytokine concentrations were determined. The concentrations of IL-1β and IL-18 were markedly higher with S. aureus stimulation than with LPS (P < 0.05 by ANOVA and the Student-Newman-Keuls multiple-range test). In contrast, TNF-α, IL-6, and IL-18BP production was unaffected (data not shown). *, P < 0.05 versus control (unstimulated); ‡, P < 0.05 versus low-dose SAC or LPS (0.001% and 1 ng, respectively); †, P < 0.05 versus 0.01% SAC.
DISCUSSION
Sepsis is defined as a systemic, physiological response to serious infection. The current scoring systems for assessing the magnitude of the physiological insult and the degree of organ injury examine the host response at a single point in time without reference to the underlying immunological or inflammatory response or to the inciting noxious stimulus, whether microbial or resulting from another type of tissue injury, e.g., multiple trauma, thermal injury, or pancreatitis. In contrast, recent efforts have focused on the immunological and genetic forces that underlie the organ injury when sepsis develops.
Although the conventional wisdom of septic shock argues that the products of the microbial infection initiate an exaggerated or uncontrolled release of proinflammatory mediators that lead to cardiovascular failure, inadequate tissue perfusion, and irreversible organ injury, treatment of septic patients with anticytokine or anti-inflammatory agents has produced only modest benefits (1, 4, 5, 36). The basic premise for the widespread testing of these agents was based on a large body of preclinical studies, as well as on a frequent finding of elevated concentration of cytokines in the blood of these patients.
The specific role of gram-positive pathogens in the pathogenesis of septic shock has received less research emphasis in large part because of earlier conceptions that gram-positive microbes were less frequently involved in the pathogenesis of human sepsis syndromes and that host responses to gram-positive pathogens and their underlying pathological mechanisms of action were similar to responses to endotoxin or to gram-negative pathogens. The present study, however, and the recent data of others challenge this dogma. First, gram-positive pathogens have become progressively more frequent causes of sepsis, and the prevalence of sepsis due to gram-positive bacteria has either matched or exceeded that of sepsis due to gram-negative bacteria in a number of recent reports (25). More importantly, there is growing appreciation that the host recognition pathways for gram-negative and gram-positive microbial products are markedly different and therefore likely to induce a variant host response. Unlike gram-negative bacteria, which are recognized primarily by their shed or membrane-associated endotoxin or LPS, the gram-positive microbes are recognized by the host through contact with membrane peptidoglycans, lipoteichoic acid, or soluble extracellular toxins (12).
In the present study, we chose to compare whole-blood responses to a gram-negative bacterial LPS and heat-killed S. aureus, rather than comparing individual elements of the gram-positive microbe. This decision was based on the observation that bacterial LPS frequently recapitulates the shock-producing properties of gram-negative bacteria, whereas administration of gram-positive peptidoglycans and lipoteichoic acid in themselves do not produce shock and organ injury (12). Furthermore, the responses to these purified gram-positive products are not necessarily comparable and often do not mimic the response to the whole pathogen. For example, Wang et al. demonstrated using a much more limited microarray that blood monocyte gene expression responses to S. aureus and purified peptidoglycans differed (34). Similarly, the whole-blood cytokine responses to peptidoglycans and lipoteichoic acid are quantitatively and temporally different (33). Hence, we chose to compare the stimulatory effects of LPS and heat-killed S. aureus. We recognize, however, that the findings reported here cannot necessarily be extrapolated to the response to different gram-negative or gram-positive pathogens or their constituents. Clearly, the response to microbial pathogens varies depending upon the specific pathogen or microbial product, its concentration, and the duration of the exposure. The findings here, however, illustrate the potential differences in the expression patterns of whole-blood leukocytes in response to two disparate, but common, stimulants.
The discovery of the TLR family has provided unequivocal evidence that the recognition pathways for gram-negative and gram-positive pathogens are different. Although not fully resolved, there is accumulating consensus that endotoxin of gram-negative bacteria is primarily recognized through the CD14-TLR4 cell surface complex, whereas peptidoglycans and lipoteichoic acid are recognized by TLR2 and, to a more limited extent, TLR4. The intracellular signaling pathways for TLR4 and TLR2 are generally similar, as both receptors are members of the IL-1 receptor superfamily and employ IL-1 receptor-associated kinases. However, TLR4 is apparently unique in that intracellular signaling can occur through both MyD88 (a TLR-associated adapter protein)-dependent and -independent pathways (17).
There has been some recognition that the host responses to gram-negative and gram-positive pathogens can vary, although the majority of the studies were performed on isolated cell populations with purified products from gram-positive and gram-negative pathogens. Almost 10 years ago, Bjork and colleagues examined cytokine production in individual peripheral blood mononuclear cells stimulated with endotoxin or Staphylococcus enterotoxin A and showed divergent cytokine responses (6). Although endotoxin produced a potent early proinflammatory cytokine response characterized by production of TNF-α, IL-1α, IL-1β, and IL-8, T-cell cytokines such as TNFβ, IL-2, IL-4, and gamma interferon were not detected. In contrast, it was shown that enterotoxin A induced a different cytokine pattern, with TNF-α, IL-1β, TNFβ, IL-4, and gamma interferon production predominating (6). The production of IL-8, IL-6, and TNF did not discriminate between infections with gram-positive and gram-negative bacteria (6).
In the present study, using ex vivo whole blood stimulation, we reached comparable conclusions with the observation that increased IL-1β and IL-18 production, not TNF-α, IL-8, or IL-10, discriminated between gram-positive (heat-killed S. aureus) and gram-negative (E. coli LPS) stimulants. The ability of heat-killed S. aureus to induce IL-18 production is consistent with a recent study by Bocker et al. (7). The present study suggests that concentrations of these two cytokines (as well as IL-6, to a lesser extent) in plasma may discriminate among patients presenting with sepsis due to gram-negative and gram-positive bacteria. Although the total number of patients studied was relatively small (n = 52), the concentrations of IL-1β and IL-18 in plasma were nearly two- to threefold higher in patients with sepsis due to gram-positive bacteria than in those with sepsis due to gram-negative bacteria. In this regard, the study extends the earlier work of Oberholzer et al. (24) demonstrating significantly elevated plasma IL-18 concentrations in patients with sepsis compared to severely injured trauma patients and healthy humans. Furthermore, those authors found that septic patients who died and patients with septic shock exhibited higher levels of IL-18 than survivors and septic patients without shock. In addition, septic patients with gram-positive organisms had significantly higher plasma IL-18 levels than patients with gram-negative organisms (24). In unpublished findings, Lungstras-Butler et al. observed in patients with necrotizing fasciitis of predominantly gram-positive bacterial origin that circulating levels of IL-18 and IL-1β were markedly higher in patients who died than in those who survived (C. A. Dinarello, personal communication).
We can extend these findings to a larger, independent patient population and confirm that despite these different IL-18 and IL-1β responses, the plasma IL-18BP concentrations were not different in patients with sepsis due to gram-positive and gram-negative bacteria. IL-18BP is a constitutively expressed and secreted protein that binds specifically to mature IL-18 and neutralizes its activity. We observed no difference in IL-18BP levels in the plasma of patients with sepsis due to gram-negative and gram-positive bacteria, nor did either LPS or heat killed S. aureus induce an ex vivo IL-18BP release from whole blood (data not shown). Furthermore, free IL-18 was higher among patients with gram-positive sepsis than those with gram-negative sepsis. Taken together, these findings suggest that the host response to a gram-positive pathogen like S. aureus is a preferential induction of IL-18 in the absence of any increased production of one of its natural inhibitors, such as IL-18BP.
To further elucidate the differential host response to gram-negative and gram-positive stimulation, we employed Affymetrix microarrays to globally explore gene expression patterns in whole blood in response to ex vivo LPS and S. aureus stimulation. Samples were obtained from three healthy subjects, and for one of the subjects, the analyses were repeated three times. In this manner, the variation in gene expression between and within healthy donors could be evaluated. Although microarray technology is relatively new, it has been used to compare responses by inflammatory cells to various stimuli. Nau et al. evaluated gene expression responses to macrophages cultured with gram-positive, gram-negative, and mycobacterial stimuli (20). Of the genes they studied, 977 exhibited altered expression due to the action of at least one of these stimulants but only 132 were commonly upregulated among the eight stimuli tested (20). Similarly, Boldrick et al. stimulated blood mononuclear cells with LPS and diverse heat-killed bacteria and found over 200 genes to be induced by each bacterial challenge (8).
We sought to evaluate the effects of bacterial stimulation on human whole blood, not isolated cells, and found a similar broad genomic response. The responses to LPS and S. aureus stimulation in samples from three separate subjects and from the same subject three times were comparable. When the changes in expression in a single subject analyzed three times were examined, there were 642 genes twofold upregulated or downregulated by stimulation with gram-positive or gram-negative organisms. However, the expression of 758 genes distinguished the three treatment groups with a high degree of statistical significance (P ≤ 0.001), and of these, only 172 were upregulated by exposure to both gram-positive and gram-negative organisms (Fig. 4, bins D and E). The induction of these genes is consistent with what Nau and colleagues have defined as a “shared activation program” primarily responsible for transforming the leukocyte into “a cell primed to interact with its environment and mount an immune response” (20). Our data perhaps showed fewer common response elements in part due to the method of statistical analysis chosen, which identified genes that were capable of discrimination among the three experimental treatment classes.
More surprising, however, was the divergence of the response to bacterial product stimulation. k-means clustering of the same data set revealed 155 probe sets to be upregulated with LPS stimulation and downregulated with S. aureus Cowan stimulation (Fig. 4, bin C; see also Table 3 in the supplementary data) and 208 that were upregulated with Staphylococcus stimulation and downregulated with LPS (Fig. 4, bin F; see also Table 4 in the supplementary data). As expected, the genes upregulated by LPS included genes involved in proinflammatory signaling, such as those encoding interferon-regulatory factors, interferon-stimulated proteins, mitogen-activated protein kinase-activating proteins, platelet-activating factor receptor, protein C kinase, and signal transduction genes such as NF-κB and I-κB. This pattern of gene expression may have reflected the blood monocyte being the primary target of LPS in whole blood rather than the T lymphocyte. In contradistinction to plasma cytokine measurements, the expression of IL-18 was upregulated with stimulation by gram-negative organisms.
In contrast, there were several T-cell genes specifically upregulated to a greater extent with S. aureus than with LPS, including those for granzyme A, an activated T-cell nuclear factor, and a T-cell receptor. These responses are not generally unexpected, given that T-cell activation occurs rapidly in response to S. aureus. However, there were marked increases in the expression of several of the heat-shock proteins and 52 different ribosomal proteins, which were not seen with LPS stimulation.
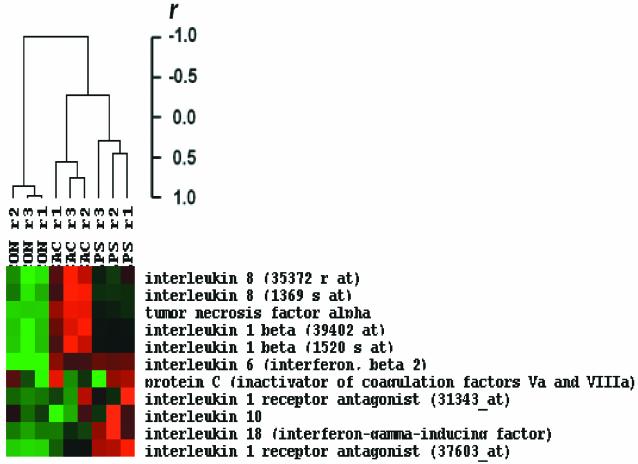
When the expression data from whole-blood leukocytes were queried for the genes of proteins measured within the plasma of septic patients or following ex vivo stimulation, there were some good correlations for specific cytokines, and other correlations were notably lacking. As shown in Fig. 6, the expression of the genes encoding the pro- and anti-inflammatory cytokines that were measured in the plasma were increased by both gram-negative and gram-positive stimuli compared to unstimulated controls. However, in many cases, the magnitude of the increase did not necessarily correlate with the increase in protein concentration seen in the plasma or in ex vivo-stimulated whole blood. For example, IL-8, IL-1β, and TNF-α gene expression was increased further by S. aureus Cowan than by E. coli LPS, but only IL-1β levels were increased with the gram-positive organism. Similarly, LPS appeared to induce whole-blood leukocyte expression of IL-18 to a greater extent than S. aureus, whereas the protein responses were markedly greater with the gram-positive organism.
FIG. 6.
Hierarchical clustering of hybridization signal intensity (gene expression) differences for pro- and anti-inflammatory cytokine probe sets. The gene expression observations from the single subject with three replicates were variance normalized and subjected to hierarchical clustering. The variation in gene expression for a given gene is expressed as the distance from the mean observation for that gene (S.D., standard deviations). Several genes were represented by more than one probe set on the Affymetrix U95aVer2 chip, and the different probe sets are identified by their Affymetrix identification numbers. CON, control unstimulated leukocytes; LPS, leukocytes exposed to LPS for 2 h prior to RNA harvest; SAC, leukocytes exposed to S. aureus Cowan for 2 h prior to RNA harvest; r, replicate number.
These disparate results between gene expression and protein production reflect a general limitation of experimental approaches based on mRNA expression, since they do not consider posttranscriptional regulation. It is possible that this finding of diminished IL-18 gene expression upon stimulation with gram-positive bacteria relative to that with gram-negative bacteria reflects the importance of posttranscriptional modification of IL-18 protein. IL-18 and IL-1β, for that matter, require processing from a higher-molecular-weight intracellular species to the biologically active secreted protein (13). Regulation at this level of IL-18 processing may be rate limiting for its secretion and plasma appearance.
In addition, an overall discrepancy between leukocyte gene expression and plasma cytokine concentrations may occur because whole-blood leukocytes are not necessarily the primary source for many of the cytokines in systemic circulation. Although TNF-α expression in blood leukocytes was higher following S. aureus stimulation than LPS stimulation, TNF-α levels in the blood of patients with sepsis due to gram-positive bacteria were not elevated compared to levels in patients with sepsis due to gram-negative bacteria. It may well be that blood leukocytes are not the primary source of TNF-α in the circulation of septic patients. We showed, for example, that more than 60% of the TNF-α in the peripheral blood of patients following an in vivo endotoxin challenge originated from the visceral organs and the hepatic venous circulation, rather than the systemic circulation (15).
What these studies do underscore, however, is the incompleteness of our fundamental understanding of the host response to gram-negative and gram-positive pathogens. Although the results are limited by the study of a single gram-positive and gram-negative stimulant, they do reveal that there are common elements in the responses to these pathogens, including the release of several proinflammatory cytokines, including TNF-α, IL-6, and some chemokines in the blood of septic patients. In response to both in vivo and ex vivo stimulation, however, the host responses to the two different classes of pathogens are probably as disparate as they are similar. We may have been inappropriately reassured that the responses were similar based on the comparable clinical presentations, as well as a limited number of cytokines in the blood, including IL-6 and TNF-α. Although IL-1β and IL-18 are two clear examples of where the host responses to LPS and heat killed S. aureus differ significantly, the gene expression data suggest that this may be only the beginning. Examining the global gene expression pattern suggests that the responses are considerably more diverse than originally predicted and that the genes which make up the shared activation program are relatively limited in number and are characterized by many of the previously known mediators, TNF-α, IL-6, chemokines, etc. If anything, these studies emphasize that future research should focus on furthering our understanding of the regulation of pathogen-specific genes and how their products contribute to the host response. These complexities in the host response to different pathogens will make it more difficult to formulate biological response modifiers for patients with sepsis with broad action against various classes of pathogens and the unique host responses they elicit.
Acknowledgments
This study was supported in part by grants GM-40586 (L.L.M.), AI-15614 (C.A.D.), and HL-68743 (C.A.D.) awarded by the National Institutes of Health (UPSHS). A.O. was supported in part by a grant from the Swiss National Science Foundation. R.J.F. was supported in part by a training grant (T32 GM-80721) from the National Institute of General Medical Sciences in burn and trauma research.
Editor: F. C. Fang
REFERENCES
- 1.Abraham, E. 1999. Why immunomodulatory therapies have not worked in sepsis. Intensive Care Med. 25:556-566. [DOI] [PubMed] [Google Scholar]
- 2.American College of Chest Physicians/Society of Critical Care Medicine. 1992. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit. Care Med. 20:864-874. [PubMed] [Google Scholar]
- 3.Angus, D. C., W. T. Linde-Zwirble, J. Lidicker, G. Clermont, J. Carcillo, and M. R. Pinsky. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29:1303-1310. [DOI] [PubMed] [Google Scholar]
- 4.Baue, A. E. 1997. Multiple organ failure, multiple organ dysfunction syndrome, and systemic inflammatory response syndrome. Why no magic bullets? Arch. Surg. 132:703-707. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, G. R., J. L. Vincent, P. F. Laterre, S. P. LaRosa, J. F. Dhainaut, A. Lopez-Rodriguez, J. S. Steingrub, G. E. Garber, J. D. Helterbrand, E. W. Ely, and C. J. Fisher, Jr. 2001. Efficacy and safety of recombinant human activated protein C for severe sepsis. N. Engl. J. Med. 344:699-709. [DOI] [PubMed] [Google Scholar]
- 6.Bjork, L., J. Andersson, M. Ceska, and U. Andersson. 1992. Endotoxin and Staphylococcus aureus enterotoxin A induce different patterns of cytokines. Cytokine 4:513-519. [DOI] [PubMed] [Google Scholar]
- 7.Bocker, U., T. Manigold, J. M. Watson, M. V. Singer, and S. Rossol. 2001. Regulation of Staphylococcus aureus-mediated activation of interleukin-18 in peripheral blood mononuclear cells. Eur. Cytokine Netw. 12:631-638. [PubMed] [Google Scholar]
- 8.Boldrick, J. C., A. A. Alizadeh, M. Diehn, S. Dudoit, C. L. Liu, C. E. Belcher, D. Botstein, L. M. Staudt, P. O. Brown, and D. A. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bone, R. C. 1994. Gram-positive organisms and sepsis. Arch. Intern. Med. 154:26-34. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1990. Increase in National Hospital Discharge Survey rates for septicemia—United States. 1979-1987. Morb. Mortal. Wkly. Rep. 39:31-34. [PubMed] [Google Scholar]
- 11.Cockerill, F. R., III, J. G. Hughes, E. A. Vetter, R. A. Mueller, A. L. Weaver, D. M. Ilstrup, J. E. Rosenblatt, and W. R. Wilson. 1997. Analysis of 281,797 consecutive blood cultures performed over an eight-year period: trends in microorganisms isolated and the value of anaerobic culture of blood. Clin. Infect. Dis. 24:403-418. [DOI] [PubMed] [Google Scholar]
- 12.De Kimpe, S. J., M. Kengatharan, C. Thiemermann, and J. R. Vane. 1995. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proc. Natl. Acad. Sci. USA 92:10359-10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinarello, C. A. 2001. Novel targets for interleukin 18 binding protein. Ann. Rheum. Dis. 60(Suppl. 3):iii18-iii24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong, Y. M., M. A. Marano, L. L. Moldawer, H. Wei, S. E. Calvano, J. S. Kenney, A. C. Allison, A. Cerami, G. T. Shires, and S. F. Lowry. 1990. The acute splanchnic and peripheral tissue metabolic response to endotoxin in humans. J. Clin. Invest. 85:1896-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry, D. E. 2000. Sepsis syndrome. Am. Surg. 66:126-132. [PubMed] [Google Scholar]
- 17.Kaisho, T., K. Hoshino, T. Iwabe, O. Takeuchi, T. Yasui, and S. Akira. 2002. Endotoxin can induce MyD88-deficient dendritic cells to support T(h)2 cell differentiation. Int. Immunol. 14:695-700. [DOI] [PubMed] [Google Scholar]
- 18.Krishnagopalan, S., and R. P. Dellinger. 2001. Innovative therapies for sepsis. BioDrugs 15:645-654. [DOI] [PubMed] [Google Scholar]
- 19.Muller-Alouf, H., J. E. Alouf, D. Gerlach, J. H. Ozegowski, C. Fitting, and J. M. Cavaillon. 1994. Comparative study of cytokine release by human peripheral blood mononuclear cells stimulated with Streptococcus pyogenes superantigenic erythrogenic toxins, heat-killed streptococci, and lipopolysaccharide. Infect. Immun. 62:4915-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nau, G. J., J. F. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novick, D., S. H. Kim, G. Fantuzzi, L. L. Reznikov, C. A. Dinarello, and M. Rubinstein. 1999. Interleukin-18 binding protein: a novel modulator of the Th1 cytokine response. Immunity 10:127-136. [DOI] [PubMed] [Google Scholar]
- 22.Novick, D., B. Schwartsburd, R. Pinkus, D. Suissa, I. Belzer, Z. Sthoeger, W. F. Keane, Y. Chvatchko, S. H. Kim, G. Fantuzzi, C. A. Dinarello, and M. Rubinstein. 2001. A novel IL-18BP ELISA shows elevated serum IL-18BP in sepsis and extensive decrease of free IL-18. Cytokine 14:334-342. [DOI] [PubMed] [Google Scholar]
- 23.Oberholzer, A., C. Oberholzer, R. M. Minter, and L. L. Moldawer. 2001. Considering immunomodulatory therapies in the septic patient: should apoptosis be a potential therapeutic target? Immunol. Lett. 75:221-224. [DOI] [PubMed] [Google Scholar]
- 24.Oberholzer, A., U. Steckholzer, M. Kurimoto, O. Trentz, and W. Ertel. 2001. Interleukin-18 plasma levels are increased in patients with sepsis compared to severely injured patients. Shock 16:411-414. [DOI] [PubMed] [Google Scholar]
- 25.Opal, S. M., and J. Cohen. 1999. Clinical gram-positive sepsis: does it fundamentally differ from gram-negative bacterial sepsis? Crit. Care Med. 27:1608-1616. [DOI] [PubMed] [Google Scholar]
- 26.Parrillo, J. E. 1993. Pathogenetic mechanisms of septic shock. N. Engl. J. Med. 328:1471-1477. [DOI] [PubMed] [Google Scholar]
- 27.Parrillo, J. E., M. M. Parker, C. Natanson, A. F. Suffredini, R. L. Danner, R. E. Cunnion, and F. P. Ognibene. 1990. Septic shock in humans. Advances in the understanding of pathogenesis, cardiovascular dysfunction, and therapy. Ann. Intern. Med. 113:227-242. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi, O., and S. Akira. 2001. Toll-like receptors; their physiological role and signal transduction system. Int. Immunopharmacol. 1:625-635. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 30.Tannahill, C. L., K. Fukuzuka, T. Marum, Z. Abouhamze, S. L. MacKay, E. M. Copeland III, and L. L. Moldawer. 1999. Discordant tumor necrosis factor-alpha superfamily gene expression in bacterial peritonitis and endotoxemic shock. Surgery 126:349-357. [PubMed] [Google Scholar]
- 31.Valles, J., C. Leon, F. Alvarez-Lerma, et al. 1997. Nosocomial bacteremia in critically ill patients: a multicenter study evaluating epidemiology and prognosis. Clin. Infect. Dis. 24:387-395. [DOI] [PubMed] [Google Scholar]
- 32.Vincent, J. L. 1998. Search for effective immunomodulating strategies against sepsis. Lancet 351:922-923. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J. E., P. F. Jorgensen, M. Almlof, C. Thiemermann, S. J. Foster, A. O. Aasen, and R. Solberg. 2000. Peptidoglycan and lipoteichoic acid from Staphylococcus aureus induce tumor necrosis factor alpha, interleukin 6 (IL-6), and IL-10 production in both T cells and monocytes in a human whole blood model. Infect. Immun. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Z. M., C. Liu, and R. Dziarski. 2000. Chemokines are the main proinflammatory mediators in human monocytes activated by Staphylococcus aureus, peptidoglycan, and endotoxin. J. Biol. Chem. 275:20260-20267. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler, A. P., and G. R. Bernard. 1999. Treating patients with severe sepsis. N. Engl. J. Med. 340:207-214. [DOI] [PubMed] [Google Scholar]
- 36.Zeni, F., B. Freeman, and C. Natanson. 1997. Anti-inflammatory therapies to treat sepsis and septic shock: a reassessment. Crit. Care Med. 25:1095-1100. [DOI] [PubMed] [Google Scholar]