Abstract
Chlamydiae are obligate intracellular bacteria that replicate within an inclusion that is trafficked to the peri-Golgi region where it fuses with exocytic vesicles. The host and chlamydial proteins that regulate the trafficking of the inclusion have not been identified. Since Rab GTPases are key regulators of membrane trafficking, we examined the intracellular localization of several green fluorescent protein (GFP)-tagged Rab GTPases in chlamydia-infected HeLa cells. GFP-Rab4 and GFP-Rab11, which function in receptor recycling, and GFP-Rab1, which functions in endoplasmic reticulum (ER)-to-Golgi trafficking, are recruited to Chlamydia trachomatis, Chlamydia muridarum, and Chlamydia pneumoniae inclusions, whereas GFP-Rab5, GFP-Rab7, and GFP-Rab9, markers of early and late endosomes, are not. In contrast, GFP-Rab6, which functions in Golgi-to-ER and endosome-to-Golgi trafficking, is associated with C. trachomatis inclusions but not with C. pneumoniae or C. muridarum inclusions, while the opposite was observed for the Golgi-localized GFP-Rab10. Colocalization studies between transferrin and GFP-Rab11 demonstrate that a portion of GFP-Rab11 that localizes to inclusions does not colocalize with transferrin, which suggests that GFP-Rab11's association with the inclusion is not mediated solely through Rab11's association with transferrin-containing recycling endosomes. Finally, GFP-Rab GTPases remain associated with the inclusion even after disassembly of microtubules, which disperses recycling endosomes and the Golgi apparatus within the cytoplasm, suggesting a specific interaction with the inclusion membrane. Consistent with this, GFP-Rab11 colocalizes with C. trachomatis IncG at the inclusion membrane. Therefore, chlamydiae recruit key regulators of membrane trafficking to the inclusion, which may function to regulate the trafficking or fusogenic properties of the inclusion.
Chlamydiae are major bacterial pathogens of ocular, urogenital, and pulmonary mucosal surfaces (51). Infections caused by Chlamydia trachomatis are the leading cause of bacterially acquired sexually transmitted disease (10), as well as of preventable blindness worldwide (64). In addition, Chlamydia pneumoniae infections are major causes of upper respiratory tract infections and have recently been linked to chronic heart disease (24, 25). Chlamydiae are obligate intracellular bacteria that replicate within a nonacidified vacuole termed an inclusion (26). Within the inclusion, chlamydiae undergo a biphasic developmental cycle that alternates between the infectious metabolically inactive elementary body (EB) and the noninfectious metabolically active reticulate body (40). Although chlamydiae enter nonprofessional phagocytes by multiple mechanisms (reviewed in reference 26), once the chlamydiae are internalized, they actively modify the properties of the nascent vacuole during the first 2 h postinfection, resulting in trafficking of the inclusion to the peri-Golgi region, fusion of the inclusion with a subset of Golgi-derived exocytic vesicles, and avoidance of lysosomal fusion (57). The molecular mechanisms that chlamydiae utilize to control the biogenesis of the vacuole are not known. However, chlamydial gene expression is required (57), suggesting that chlamydial proteins that are secreted into the host cell cytoplasm or incorporated into the inclusion membrane are likely to be important mediators of these properties (49). Although chlamydiae do not appear to traffic through early or late endocytic organelles, transferrin (Tfn)-containing tubular endosomes are intimately associated with both nascent and mature chlamydial inclusions (2, 54, 55, 70).
To facilitate the intracellular trafficking of chlamydiae, chlamydiae are likely to exploit host trafficking pathways via recruitment of components of the host's membrane trafficking machinery to the inclusion membrane. Intracellular membrane trafficking and organelle biogenesis are tightly controlled by a variety of highly conserved membrane and soluble cellular factors, including N-ethylmalemide-sensitive factor (NSF) attachment receptor proteins (v-SNARE and t-SNARE), NSF, soluble NSF attachment proteins, the small GTP binding proteins, ADP-ribosylating factors, and Rab GTPases (reviewed in reference 50). To date, the association of these host factors with the inclusion has not been investigated. However, trafficking of the inclusion is dependent on host microfilaments and microtubules, although differences in dependence on the host cytoskeleton have been observed between different chlamydial species (14, 53). In addition, the intracellular trafficking and the perinuclear localization of the C. trachomatis inclusion are also dependent on Ca2+ (35) and the minus-ended microtubule motor dynein (14). In contrast, the intracellular trafficking of the C. pneumoniae inclusion appears to be dynein independent (14).
Rab GTPases are the largest family of Ras-like small GTPases (45). More than 50 mammalian Rab GTPases have been identified, each localizing to a distinct organelle or organellar domain and function at every known transport step within the cell, including both constitutive and regulated pathways (reviewed in references 16, 46, and 66). Rab GTPases cycle between a cytoplasmic, GDP-bound, inactive state and a membrane-associated, GTP-bound, active state and regulate membrane traffic at multiple steps, including formation of transport vesicles at donor membranes, transport and docking of vesicles, and fusion of vesicles at target membranes. Rab GTPases regulate central roles, such as SNARE recruitment and vesicle tethering, as well as other functions specific to distinct membrane trafficking steps, primarily through specific recruitment of effector molecules (66).
Recently, the selective inclusion or retention of Rab GTPases on vacuolar membranes has been shown to regulate the biogenesis of phagosomes inhabited by several bacteria including Mycobacterium species (71) and Salmonella enterica serovar Typhimurium (39). Because of the importance and specificity imparted by Rab GTPases in membrane trafficking and their role in the maturation of bacterium-containing vacuoles, we sought to determine whether chlamydiae exploit host vesicular trafficking machinery through selective recruitment or exclusion of Rab GTPases to the inclusion membrane. Since inclusions interact with both Tfn-containing recycling endosomes and the Golgi apparatus, we chose to examine the intracellular localization of a collection of Rab GTPases that function in both endocytic and biosynthetic membrane trafficking in chlamydia-infected HeLa cells. In this report, we examined the subcellular localization of Rab GTPases that function in endosomal trafficking (Rab4, Rab5, Rab7, Rab9, and Rab11) (7, 8, 23, 34, 37, 38, 68), Tfn receptor recycling (Rab4 and Rab11) (37, 47, 67), endosome-to-trans-Golgi network (TGN) trafficking (Rab6 and Rab11) (18, 36, 73), endoplasmic reticulum (ER)-to-Golgi and intra-Golgi trafficking (Rab1) (65), and Golgi-to-ER retrograde trafficking (Rab6) (22, 72) and demonstrate that chlamydiae recruit a subset of GFP-Rab GTPases to the inclusion membrane in both a species-dependent and species-independent manner.
MATERIALS AND METHODS
Cell culture and organisms.
Monolayer cultures of HeLa 229 epithelial cells (CCL 1.2; American Type Tissue Culture) were grown in RPMI 1640 (Mediatech, Inc., Herndon, Va.) supplemented with 10% fetal bovine serum (Mediatech) and 10 μg of gentamicin (Invitrogen, Carlsbad, Calif.) per ml at 37°C in an atmosphere of 5% CO2 and 95% humidified air. C. trachomatis LGV 443 (serotype L2), UW-3/CX (serotype D), Chlamydia muridarum (previously referred to as Chlamydia trachomatis mouse pneumonitis biovar), and C. pneumoniae (AR-39) were propagated in HeLa cells and purified by renografin density centrifugation (9). Purified C. pneumoniae was provided by Ted Hackstadt and Kate Wolf (Rocky Mountain Laboratories [RML], National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH], Hamilton, Mont.), while stock cultures of C. trachomatis and C. muridarum were provided by Harlan Caldwell (RML, NIAID, NIH).
Reagents and antibodies.
Mouse antichlamydial lipopolysaccharide (LPS) was generously provided by Harlan Caldwell (RML, NIAID, NIH), and rabbit anti-IncG was prepared as described previously (58). Goat anti-mouse immunoglobulin G (IgG) conjugated to Alexa 568 and Tfn conjugated to Alexa 568 were purchased from Molecular Probes (Eugene, Oreg.), goat anti-mouse IgG conjugated to Cy5 was purchased from Zymed Laboratories, Inc. (South San Francisco, Calif.), and nocodazole was purchased from CalBiochem (San Diego, Calif.).
Plasmid constructions.
Mammalian expression plasmids containing human green fluorescent protein (GFP)-tagged Rab5 (GFP-Rab5) and GFP-Rab7 were generously provided by Craig Roy (Yale University, New Haven, Conn.) and pEGFP-Rab9 was generously provided by Suzanne Pfeffer (Department of Biochemistry, Stanford University, Stanford, Calif.). pEGFP-Rab11A was constructed by PCR amplification using a 5′ gene-specific primer designed with a 5′ EcoRI site (GAATTCATGGGCACCCGCGACGACGAG) and a 3′ gene-specific primer designed with a 5′ XhoI site (CTCGAGTTAGATGTTCTGACAGCACTGC). The PCR product was digested with EcoRI and XhoI and cloned into the EcoRI and SalI sites of pEGFPC2. pGreenLantern-Rab11A was used as the template and obtained from Craig Roy (Yale University). pEGFP-Rab11B was constructed by PCR amplification using a 5′ gene-specific primer designed with an EcoRI site (GAATTCATGGGGACCCGGGACGACGAG) and a 3′ gene-specific primer designed with a XhoI site (CTCGAGTCACAGGTTCTGGCAGCAGTGC) using a HeLa cDNA library (Clontech, Palo Alto, Calif.) as the template DNA. The PCR product was digested with EcoRI and XhoI and cloned into the EcoRI and SalI sites of pEGFPC2 (Clontech, Palo Alto, Calif.). pEGFP-Rab4A and pEGFP-Rab6A/B were constructed by cloning the coding regions of the respective human gene contained on BamHI/XhoI fragments into the BglII and SalI sites of pEGFPC1 (Clontech) using standard molecular biology techniques. cDNA clones containing each human Rab gene were generously provided by Guthrie cDNA Resource Center, Guthrie Research Institute, Sayre, Pa. (www.cDNA.org). pEGFP-Rab4B was constructed by cloning the coding region of human Rab4B contained within an EcoRI/XhoI fragment (Guthrie cDNA Resource Center) into the EcoRI and SalI sites of pEGFPC2. All PCR amplifications were performed using HiFidelity Platinum Taq polymerase (Invitrogen), and each fusion construct was confirmed by DNA sequencing (BioResource Center, Cornell University, Ithaca, N.Y.).
Eukaryotic transfection.
HeLa 229 cells grown on 12-mm-diameter coverslips (no. 1 thickness) in 24-well plates were washed once in serum-free RPMI 1640 (Mediatech) and transfected with Lipofectamine using a total of 0.4 μg of DNA per well according to the manufacturer's protocol (Invitrogen). For C. trachomatis serovar L2 infections, 24 h posttransfection, the cells were infected at a multiplicity of infection of approximately 1 and incubated for an additional 18 h at 37°C in RPMI 1640 containing 10% fetal bovine serum at 37°C unless otherwise noted (9). For C. trachomatis serovar D, C. muridarum, and C. pneumoniae, the chlamydial inoculum was centrifuged onto transfected HeLa cell monolayers for 1 h at room temperature (RT) at 900 × g, and cycloheximide was added to growth media at a final concentration of 1 μg/ml. Cells were infected for 18 h, except for C. pneumoniae infections, which were allowed to proceed for 44 h.
LSCM.
Cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 60 min at RT. For antibody labeling, fixed cells were permeabilized in PBS containing 0.05% saponin and 0.2% bovine serum albumin for 10 min at RT, and primary and secondary antibodies were incubated in permeabilization buffer sequentially for 60 min each at RT. Coverslips were mounted onto glass slides using Prolong Antifade (Molecular Probes) and viewed by laser-scanning confocal microscopy (LSCM). An Olympus Fluoview 500 confocal laser-scanning imaging system equipped with krypton, argon, and He-Ne lasers on an Olympus IX70 inverted microscope with a PLAPO 60× objective was used (Olympus America, Inc., Melville, N.Y.). Confocal images were processed using Adobe Photoshop 6.0 (Adobe Systems, Inc., Mountain View, Calif.).
Nocodazole treatment of cells.
Cells were transfected and infected as described above. Prior to fixation, cells were treated with 20 μM nocodazole or 0.1% dimethyl sulfoxide for the indicated times at 37°C. After the indicated treatment, cells were fixed in 4% formaldehyde in PBS and labeled with antibodies as described above.
Tfn uptake.
Cells were serum starved for 2 h in RPMI 1640 (Mediatech) at 37°C before the addition of Alexa 568-conjugated Tfn (Molecular Probes) at a final concentration of 10 μg/ml. Cells were incubated with Alexa 568-conjugated Tfn for 60 min, washed once in Hank's balanced salt solution, and fixed for 60 min at RT in PBS containing 4% formaldehyde. Cells were stained with antichlamydial LPS as described above and viewed by LSCM.
RESULTS
GFP-Rab1, GFP-Rab4, GFP-Rab6, and GFP-Rab11 are associated with the C. trachomatis inclusion.
If Rab GTPases function in the maturation, biogenesis, or fusogenicity of the chlamydial inclusion, we reasoned that we would expect to see recruitment of specific Rab proteins to the inclusion membrane in infected cells. Since GFP-tagged Rab fusion proteins expressed in tissue culture cells have been shown to localize to subcellular compartments similar to those of their respective endogenous counterparts (5, 11, 12, 18, 20, 30, 32, 42, 59, 60, 72), we chose to examine the intracellular localization of GFP or enhanced GFP (EGFP) fusion proteins in infected cells. We analyzed the following set of GFP-tagged Rab GTPases: Rab1A, Rab4 (Rab4A and Rab4B), Rab5, Rab6 (Rab6A and Rab6B), Rab7, Rab9, Rab10, and Rab11 (Rab11A and Rab11B) (Table 1). All human Rab genes were fused in frame with EGFP except for Rab5 and Rab7, which were fused in frame with GFP, and all are referred to as GFP-Rab proteins. The collection consisted of Rab proteins that act at distinct steps in the endocytic and exocytic pathways as summarized in the introduction and in Table 1.
TABLE 1.
Rab GTPases examined and sites of action
| Rab GTPase | Site(s) of actiona | Reference(s) |
|---|---|---|
| Rab1 | ER to Golgi, intra-Golgi | 65 |
| Rab4 | EE to PM | 37, 69 |
| Rab5 | PM to EE | 7, 23 |
| Rab6 | Golgi to ER, intra-Golgi, EE to TGN | 22, 36, 72 |
| Rab7 | EE to LE, LE to lysosome | 38 |
| Rab9 | LE to TGN | 34 |
| Rab10 | Golgi-associated | 13 |
| Rab11 | RE to PM, EE to TGN, TGN to PM | 11, 47, 52, 67, 73 |
Abbreviations: EE, early endosome; PM, plasma membrane; LE, late endosome; RE, recycling endosome.
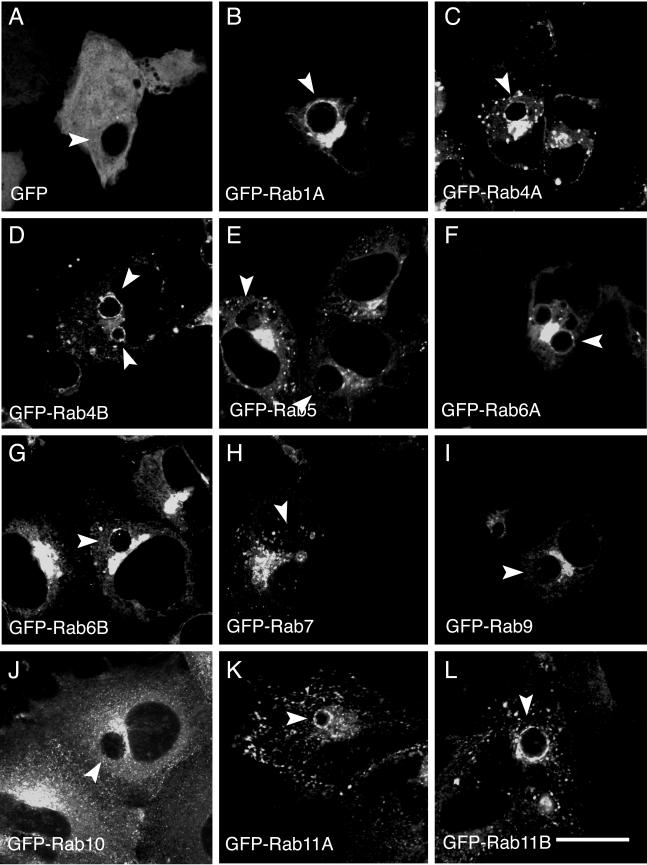
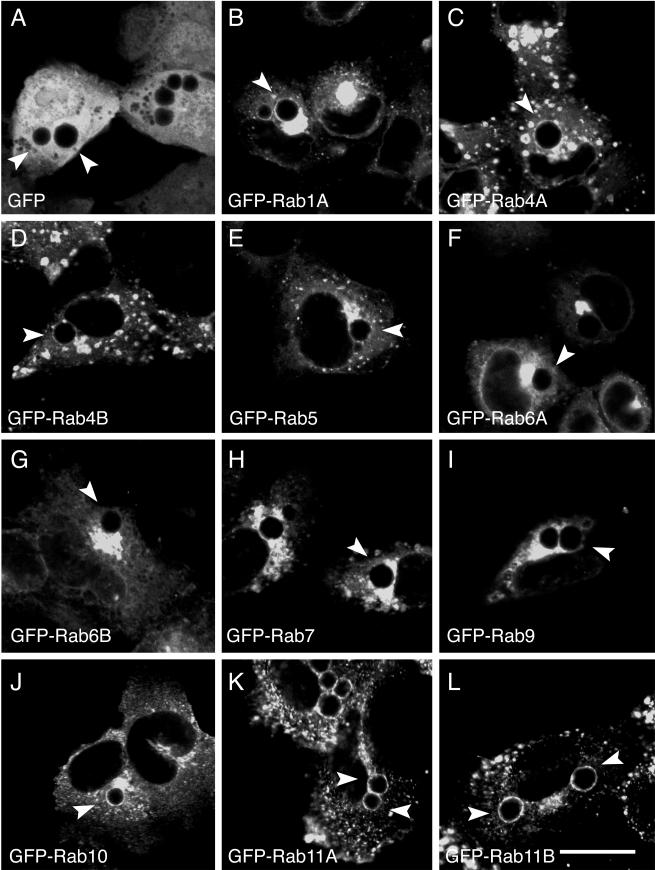
To identify the subcellular localization of each GFP-Rab GTPase, HeLa cells were transiently transfected with DNA containing each respective GFP-Rab protein. Cells were also transfected with a plasmid expressing only GFP (Fig. 1A). Twenty-four hours posttransfection, HeLa cells were infected with C. trachomatis serovar L2 at a multiplicity of infection of approximately 1, and at 18 h postinfection, the intracellular localization of each GFP-Rab fusion protein was analyzed by LSCM. Chlamydial inclusions were identified by labeling with antichlamydial LPS (data not shown). Colocalization experiments with well-characterized cellular markers of endocytic organelles and the Golgi apparatus demonstrated that each GFP-Rab protein localized to its appropriate subcellular localization (data not shown). No differences in localization patterns were observed between isoforms of the same Rab protein. As shown in Fig. 1, in C. trachomatis serovar L2-infected cells, a subset of GFP-Rab proteins localize specifically to the chlamydial inclusion. Although adjacent to the inclusion, GFP-Rab5 (Fig. 1E), GFP-Rab7 (Fig. 1H), GFP-Rab9 (Fig. 1I), and GFP-Rab10 (13) (Fig. 1J) do not associate specifically with the inclusion. In contrast, GFP-Rab1A (Fig. 1B), GFP-Rab4A and GFP-Rab4B (Fig. 1C and D), GFP-Rab6A and GFP-Rab6B (Fig. 1F and G), and GFP-Rab11A and GFP-Rab11B (Fig. 1K and L) are enriched at the periphery of the inclusion in a discrete rim-like staining pattern indistinguishable from the intracellular staining patterns of chlamydial inclusion membrane proteins (49) and Fig. 8. Because not all GFP-Rab proteins analyzed localize circumferentially to the inclusion, the inclusion association that we detect between GFP-Rab1, GFP-Rab4, GFP-Rab6, and GFP-Rab11 is unlikely to be the result of an interaction with GFP. The failure to detect localization of GFP-Rab5, GFP-Rab7, and GFP-Rab9 is consistent with the absence of classical endocytic markers within the inclusion (29, 55, 70). Of the GFP-Rab proteins that are associated with the inclusion, GFP-Rab4 and GFP-Rab11 localize primarily to pericentriolar recycling endosomes (52, 60, 67, 69), and GFP-Rab1 and GFP-Rab6 localize to early Golgi compartments (18, 44, 65).
FIG.1.
Localization of GFP-Rab GTPases in C. trachomatis serovar L2-infected HeLa cells. HeLa cells were transiently transfected with DNA containing the indicated GFP-tagged Rab GTPases (B to L) or with pEGFPC1 (A). At 24 h posttransfection, the cells were infected with C. trachomatis serovar L2, and at 18 h postinfection, the cells were fixed and viewed by LSCM. Arrowheads indicate chlamydial inclusions, as determined by indirect immunofluorescence staining with antichlamydial LPS (data not shown). Specific associations between GFP-Rab1A, GFP-Rab4 (GFP-Rab4A and GFP-Rab4B), GFP-Rab6 (GFP-Rab6A and GFP-Rab6B), and GFP-Rab11 (GFP-Rab11A and GFP-Rab11B) and the C. trachomatis serovar L2 inclusion were detected, as shown by rim-like fluorescence staining around the entire periphery of the inclusion. Bar, 10 μm.
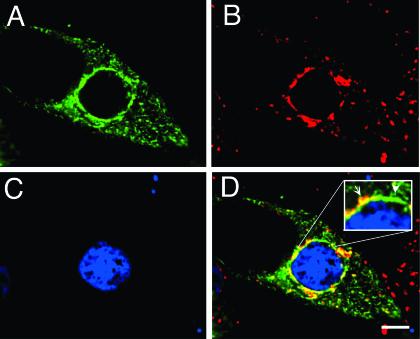
FIG. 8.
GFP-Rab11A partially colocalizes with IncG at the inclusion membrane. Transiently transfected HeLa cells expressing GFP-Rab11A were infected with C. trachomatis serovar L2 for 18 h. Cells were fixed, permeabilized, stained with polyclonal anti-C. trachomatis IncG, and incubated with goat anti-rabbit IgG conjugated to Alexa 594. GFP-Rab11A colocalizes with IncG at many regions along the inclusion membrane, as indicated by the yellow fluorescence, but there are regions that are IncG positive but Rab11A negative. In addition, Rab11A partially colocalizes to the IncG-laden fibers that stain and that extend from the inclusion membrane. The arrowheads indicate representative areas where GFP-Rab11A and IncG colocalize along the fibers. Two inclusions, at different focal planes, are visible within the cell, and each is indicated with an asterisk. (A) GFP-Rab11A (green), (B) IncG (red), and (C) panels A and B merged. Bar, 5 μm.
GFP-Rab proteins are associated with chlamydial inclusions in both a species-independent and species-dependent fashion.
To date, only one host protein, 14-3-3β, a phosphoserine binding protein that regulates cell cycle control, mitogenic signal transduction, and apoptotic cell death, has been shown to interact with the chlamydial inclusion membrane (21, 56). 14-3-3β directly interacts with C. trachomatis IncG, and its association with the chlamydial inclusion is species specific, since it is associated only with C. trachomatis and C. muridarum (mouse pneumonitis biovar) inclusions and not with Chlamydia psittaci or C. pneumoniae inclusions (56).
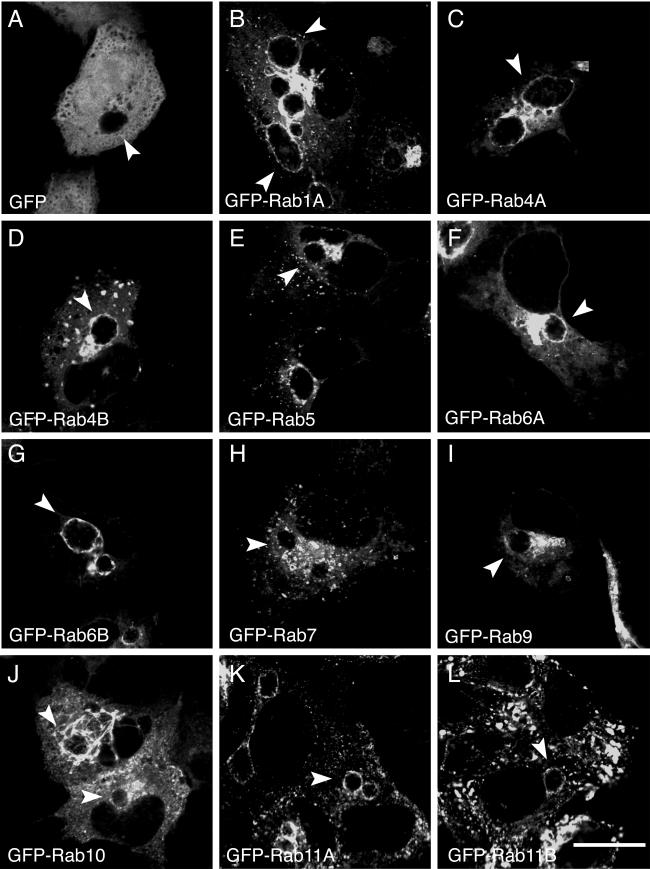
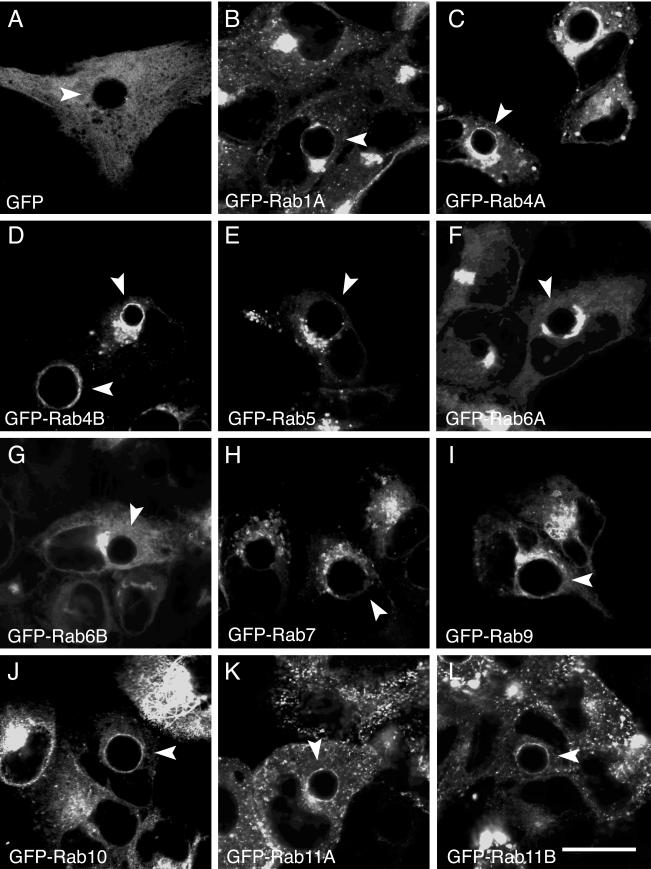
To determine the specificity of interaction for each GFP-Rab fusion protein, we examined their intracellular localization in HeLa cells infected with the following representative chlamydial strains: C. trachomatis serovar D (Fig. 2), C. muridarum (Fig. 3), and C. pneumoniae AR39 (Fig. 4). As in the previous experiment, HeLa cells transiently expressing each GFP-Rab protein were infected with the indicated chlamydial serovars and grown in the presence of cycloheximide to permit growth of C. trachomatis serovar D and C. pneumoniae. The presence of cycloheximide in the growth media did not alter the localization of any GFP-Rab protein in C. trachomatis serovar L2-infected HeLa cells (data not shown). Due to the slower growth of C. pneumoniae, C. pneumoniae infections were allowed to proceed for 44 h. Similar to the results obtained with serovar L2, no colocalization was observed between GFP-Rab5 (Fig. 2E, 3E, and 4E), GFP-Rab7 (Fig. 2H, 3H, and 4H), or GFP-Rab9 (Fig. 2I, 3I, and 4I) and any chlamydial inclusion, while GFP-Rab1A (Fig. 2B, 3B, and 4B), GFP-Rab4A and GFP-Rab4B (Fig. 2C and D, 3C and D, and 4C and D) and GFP-Rab11A and GFP-Rab11B (Fig. 2K and L, 3K and L, and 4K and L) localized specifically to all chlamydial inclusions. In contrast, GFP-Rab6A and GFP-Rab6B localized only to C. trachomatis serovar L2 (Fig. 1F and G) and serovar D inclusions (Fig. 2F and G) but not to C. muridarum (Fig. 3F and G) or C. pneumoniae (Fig. 4F and G) inclusions. The opposite was observed for GPF-Rab10 (Fig. 1J, 2J, 3J, and 4J). GFP-Rab recruitment to inclusions may be temporally regulated with respect to the chlamydial developmental cycle. Therefore, because the C. pneumoniae developmental cycle differs dramatically in duration from that of C. trachomatis species, we examined the intracellular localization of GFP-Rab6 in C. pneumoniae-infected HeLa cells at different times postinfection (18 to 96 h postinfection). Consistent with what was observed at 44 h postinfection, GFP-Rab6 was not recruited to the inclusion at any time point examined (data not shown). Therefore, a subset of GFP-Rab GTPases is recruited to chlamydial inclusions in both a species-dependent and species-independent fashion (summarized in Table 2).
FIG.2.
Localization of GFP-Rab GTPases in C. trachomatis serovar D-infected HeLa cells. HeLa cells were transiently transfected with DNA containing the indicated GFP-tagged Rab GTPases (B to L) or pEGFPC1 (A). At 24 h posttransfection, the cells were infected with C. trachomatis serovar D, and at 18 h postinfection, the cells were fixed and viewed by LSCM. Arrowheads indicate chlamydial inclusions, as determined by indirect immunofluorescence staining with antichlamydial LPS (data not shown). Specific associations between GFP-Rab1A, GFP-Rab4 (GFP-Rab4A and GFP-Rab4B), GFP-Rab6 (GFP-Rab6A and GFP-Rab6B), and GFP-Rab11 (GFP-Rab11A and GFP-Rab11B) and C. trachomatis serovar D inclusion were detected, as shown by rim-like fluorescence staining around the entire periphery of the inclusion. Bar, 10 μm.
FIG.3.
Localization of GFP-Rab GTPases in C. muridarum-infected HeLa cells. HeLa cells were transiently transfected with DNA containing the indicated GFP-tagged Rab GTPases (B to L) or pEGFPC1 (A). At 24 h posttransfection, the cells were infected with C. muridarum, and at 18 h postinfection, the cells were fixed and viewed by LSCM. Arrowheads indicate chlamydial inclusions, as determined by indirect immunofluorescence staining with antichlamydial LPS (data not shown). Specific associations between GFP-Rab1A, GFP-Rab4 (GFP-Rab4A and GFP-Rab4B), GFP-Rab10, and GFP-Rab11 (GFP-Rab11A and GFP-Rab11B) and the C. muridarum inclusion were detected, as shown by rim-like fluorescence staining around the entire periphery of the inclusion. Bar, 10 μm.
FIG.4.
Localization of GFP-Rab GTPases in C. pneumoniae AR39-infected HeLa cells. HeLa cells were transiently transfected with DNA containing the indicated GFP-tagged Rab GTPases (B to L) or pEGFPC1 (A). At 24 h posttransfection, the cells were infected with C. pneumoniae strain AR39, and at 44 h postinfection, the cells were fixed and viewed by LSCM. Arrowheads indicate chlamydial inclusions, as determined by indirect immunofluorescence staining with antichlamydial LPS (data not shown). Specific associations between GFP-Rab1A, GFP-Rab4 (GFP-Rab4A and GFP-Rab4B), GFP-Rab10, and GFP-Rab11 (GFP-Rab11A and GFP-Rab11B) and the C. pneumoniae inclusion were detected, as shown by rim-like fluorescence staining around the entire periphery of the inclusion. Bar, 10 μm.
TABLE 2.
Summary of GFP-Rab interactions with chlamydial inclusions
| GFP-Rab | GFP-Rab interaction with chlamydial inclusiona
|
|||
|---|---|---|---|---|
| CT L2 | CT D | MoPn | Cpn | |
| GFP-Rab5 | − | − | − | − |
| GFP-Rab7 | − | − | − | − |
| GFP-Rab9 | − | − | − | − |
| GFP-Rab1 | + | + | + | + |
| GFP-Rab4 | + | + | + | + |
| GFP-Rab11 | + | + | + | + |
| GFP-Rab6 | + | + | − | − |
| GFP-Rab10 | − | − | + | + |
Interaction with inclusions (+) or no association with chlamydial inclusions (−) of C. trachomatis serovar L2 (CT L2), C. trachomatis serovar D (CT D), C. muridarum (formerly C. trachomatis mouse pneumonitis biovar) (MoPn), and C. pneumoniae (Cpn). Interactions were determined by LSCM.
The association of GFP-Rab GTPases with chlamydial inclusions is microtubule independent.
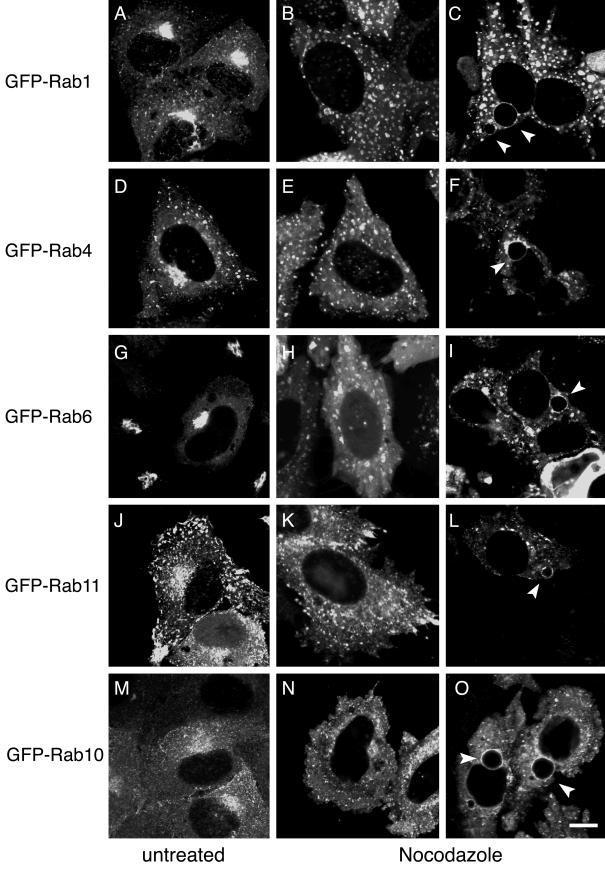
The pericentriolar localization and morphology of recycling endosomes and the Golgi apparatus are dependent on an intact microtubule network (15, 63). Since chlamydiae also traffic to and replicate in the pericentriolar region of the host cell, the association of pericentriolar Rab proteins (Rab4 and Rab11) and Golgi-localized Rab proteins (Rab1, Rab6, and Rab10) with inclusions may simply reflect their presence in the same region of the cell. Therefore, if GFP-Rab GTPases are associated with inclusions due to the physical proximity of each Rab protein to the inclusion, then dispersal of pericentriolar recycling endosomes and the Golgi apparatus should redistribute the inclusion-associated GFP-Rabs to the cytoplasm. On the other hand, if there is a specific interaction between any of the GFP-Rab proteins and the inclusion, then those GFP-Rabs should remain associated with the inclusion even in the absence of an intact microtubule network. To test this, we analyzed the intracellular localization of each GFP-Rab protein after treatment with the microtubule-destabilizing drug nocodazole in HeLa cells infected with C. trachomatis serovar L2 (GFP-Rab1, GFP-Rab4, GFP-Rab6, and GFP-Rab11) or C. pneumoniae (GFP-Rab10) (Fig. 5). HeLa cells were transiently transfected with DNA containing each fusion protein, and 24 h posttransfection, cells were mock infected or infected with the indicated chlamydial strain. After confirming by fluorescence microscopy that each GFP-Rab protein was specifically localized to the inclusion, cells were treated for 3 h with nocodazole. Microtubules were completely dissembled during this incubation period as determined by indirect immunofluorescence microscopy using a monoclonal antitubulin antibody (data not shown). In uninfected nocodazole-treated cells, the intracellular localization of each of the GFP-Rab proteins was dramatically altered in comparison to untreated cells (Fig. 5). Each of the endosome-localized GFP-Rab proteins, GFP-Rab4 (Fig. 5D and E) and GFP-Rab11 (Fig. 5J and K), and the Golgi-localized Rabs, GFP-Rab1 (Fig. 5A and B), GFP-Rab6 (Fig. 5G and H), and GFP-Rab10 (Fig. 5M and N) were dispersed throughout the cytoplasm. In infected cells, the majority of each GFP-Rab protein remained associated with the inclusion despite the absence of intact microtubules as indicated by the distinct rim-like staining pattern surrounding the chlamydial inclusion (Fig. 5C, F, I, L, and O). Identical results were obtained in C. muridarum-infected cells expressing GFP-Rab10 (data not shown). Therefore, intact microtubules are not required to maintain the association of GFP-Rab1, GFP-Rab4, GFP-Rab6, or GFP-Rab11 with C. trachomatis serovar L2 inclusions or of GFP-Rab10 with C. pneumoniae or C. muridarum inclusions. These data suggest that a stable interaction exists between each GFP-Rab fusion protein and the inclusion membrane and that maintenance of each interaction is not dependent on the pericentriolar localization of either recycling endosomes or the Golgi apparatus.
FIG.5.
Association of GFP-Rab proteins with chlamydial inclusions is maintained in the absence of microtubules. Transiently transfected HeLa cells were mock infected (A, B, D, E, G, H, J, K, M, and N) or infected with C. trachomatis serovar L2 for 18 h (C, F, I, and L) or with C. pneumoniae for 44 h (O). Cells were treated with 20 μM nocodazole for 3 h to disassemble microtubules, fixed, permeabilized, and stained with antichlamydial LPS (data not shown). Although treatment with nocodazole disrupts the microtubule network and causes dispersal of recycling endosomes and the Golgi apparatus, GFP-Rab1, GFP-Rab4, GFP-Rab6, GFP-Rab11, and GFP-Rab10 remain associated with the inclusion. Arrowheads indicate inclusions. Bar, 5 μm.
Next, we examined whether microtubules were required for the initial association with the chlamydial inclusion. For this experiment, we analyzed only GFP-Rab11A. To determine whether GFP-Rab11A's initial association with the inclusion was dependent on microtubules, microtubules were depolymerized prior to chlamydial infection. Transfected cells were treated with nocodazole for 3 h prior to chlamydial infection and during the subsequent 8-h infection period. Cells were subsequently fixed and labeled with antichlamydial LPS. As previously reported, in the absence of microtubules, chlamydiae remain dispersed throughout the cytoplasm in nonfused vacuoles (53) (Fig. 6). However, even under these conditions, GFP-Rab11A is still associated with individual EB-containing vacuoles, demonstrating that GFP-Rab11A's initial association with the inclusion is also microtubule independent and that localization of the chlamydial vacuole at the pericentriolar region of the host cell is not required.
FIG. 6.
Association of GFP-Rab11A with the chlamydial inclusion is independent of microtubules. Transiently transfected HeLa cells expressing GFP-Rab11A were treated with nocodazole (D to F) or dimethyl sulfoxide (A to C) for 3 h prior to chlamydial infection to depolymerize microtubules. Cells were then infected with C. trachomatis serovar L2 at a multiplicity of infection of approximately 20. The cells were infected for 8 h in the presence of nocodazole, fixed in formaldehyde, permeabilized, labeled with antichlamydial LPS, and incubated with goat anti-mouse IgG conjugated to Alexa 594 (red). (A and D) GFP-Rab11A (green), (B and E) antichlamydial LPS (red), (C) panels A and B merged, (F) panels D and E merged. In the complete absence of microtubules, GFP-Rab11A is still recruited to individual chlamydia-containing vacuoles (D to F). The arrowhead indicates Golgi-localized chlamydial vacuoles, and arrows indicate examples of dispersed chlamydial vacuoles. Bar, 5 μm.
The association of GFP-Rab11A with the inclusion is independent of the Tfn-containing endosome association with the inclusion.
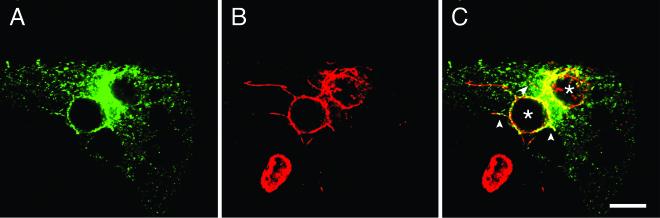
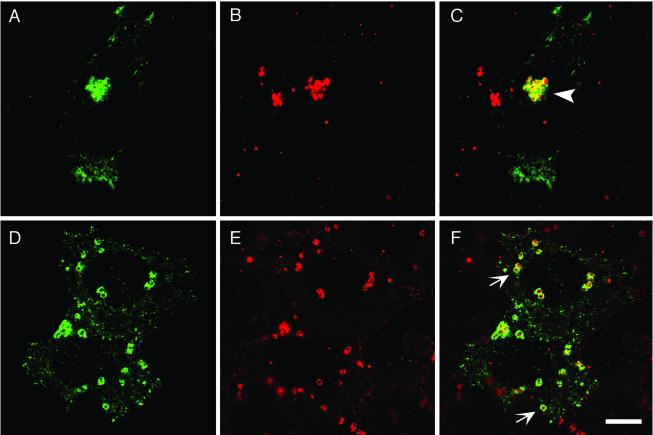
Although Rab11 preferentially localizes to Tfn-containing pericentriolar recycling endosomes in nonpolarized tissue culture cells, Rab11-positive Tfn-negative endosomes have been reported (67). Because Tfn-containing tubular endosomes are closely associated but do not fuse with either early or late chlamydial inclusions (2, 54, 55, 70), we wanted to determine whether GFP-Rab11A is associated with the chlamydial inclusion via its association with Tfn-containing recycling endosomes. To address this question, we compared the intracellular localization of fluorescently labeled Tfn (Alexa 568-labeled Tfn) with that of GFP-Rab11A in C. trachomatis serovar L2-infected HeLa cells. Transiently transfected infected cells were serum starved for 2 h, incubated for 60 min at 37°C in the presence of Tfn, fixed, and labeled with antichlamydial LPS. Labeled cells were then viewed by LSCM (Fig. 7). A steady-state distribution of endosomes containing fluorescent Tfn is obtained under these labeling conditions. In infected cells, in addition to the Tfn localized in the pericentriolar region, Tfn also localized to discrete sites adjacent to the inclusion membrane (55) (Fig. 7). Although Tfn and EGFP-Rab11A partially colocalize at the inclusion membrane, a portion of the inclusion-associated GFP-Rab11A does not colocalize with Tfn (Fig. 7). These observations demonstrate that not all inclusion-associated GFP-Rab11A is associated with Tfn-containing endosomes.
FIG. 7.
GFP-Rab11A partially colocalizes with Tfn-containing endosomes at the periphery of the chlamydial inclusion. Transiently transfected HeLa cells expressing GFP-Rab11A were infected with C. trachomatis serovar L2 for 18 h. Recycling endosomes were labeled with Alexa 568-labeled Tfn by serum starving cells for 2 h and incubated with Alexa 568-labeled Tfn for 60 min at 37°C. Cells were fixed, permeabilized, stained with antichlamydial LPS, and incubated with goat anti-mouse IgG conjugated to Cy5 (blue). In the inset in panel D, the arrowhead indicates Rab11A-positive Tfn-negative regions around the inclusion and the arrow indicates Rab11A-positive Tfn-positive regions. (A) GFP-Rab11A (green), (B) Alexa 568-labeled Tfn (red), (C) antichlamydial LPS (blue), (D) panels A, B, and C merged. Bar, 5 μm.
GFP-Rab11A partially colocalizes with C. trachomatis IncG at the inclusion membrane.
The chlamydial inclusion membrane comprises a large family of highly variable chlamydial inclusion membrane proteins, termed Inc proteins (3, 49). The carboxy-terminal domains of many Inc proteins are exposed to the host cell cytoplasm, which may permit interaction with host cytoplasmic proteins (49, 56). Therefore, Rab GTPases may be recruited to the inclusion through direct interactions with chlamydial inclusion membrane proteins. To investigate this, we compared the intracellular localization of GFP-Rab11A to the localization of a well-characterized C. trachomatis inclusion membrane protein, IncG (58), by LSCM. As shown in Fig. 8, some colocalization between the inclusion-associated GFP-Rab11A and IncG was observed, as indicated by the yellow immunofluorescence staining suggesting a close association of GFP-Rab11A with the inclusion membrane. However, GFP-Rab11A and IncG did not colocalize completely. In addition, in many infected cells, chlamydial Inc-laden fibers extend from the inclusion membrane (6). As shown in Fig. 8, GPF-Rab11A colocalizes to some of these IncG-laden fibers. A greater degree of colocalization with these fibers is seen in cells expressing GFP-Rab11AQ70L, a constitutively active GTPase-deficient Rab11 allele (data not shown). The function of these Inc-laden fibers is not known, but it has been suggested that chlamydial antigens may be trafficked to the plasma membrane along these fibers, since several intracellular chlamydial antigens have been shown to colocalize with these fibers under certain growth conditions (6, 76). This is the first report of a host protein that localizes to Inc-laden fibers.
GFP-Rab11A is associated with C. trachomatis inclusions early during the chlamydial developmental cycle.
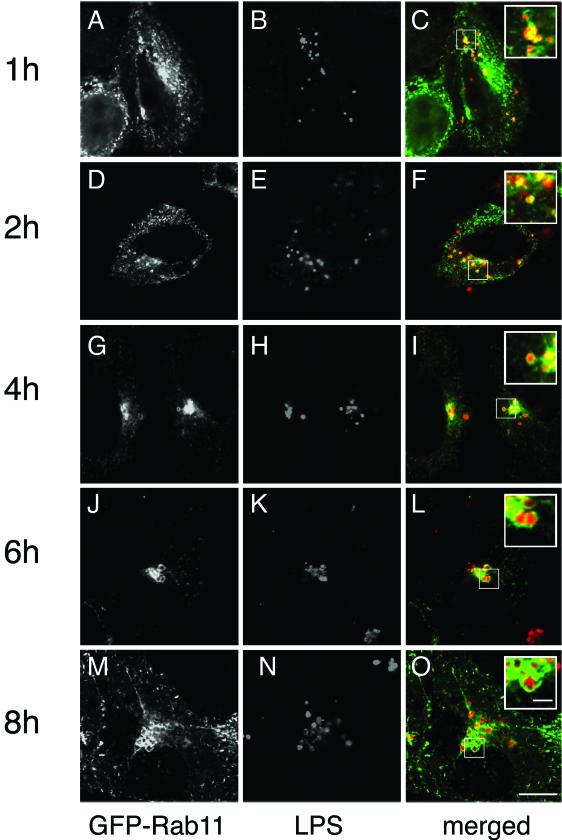
As mentioned above, the recruitment of Rab proteins to chlamydial inclusions may be regulated temporally with respect to the chlamydial developmental cycle, and this in turn may influence the role that each Rab protein may play during chlamydial infection. As the first step in determining the stage during the chlamydial developmental cycle that GPF-Rab11A is recruited to the chlamydial inclusions, we examined the intracellular localization of GFP-Rab11A in C. trachomatis-infected HeLa cells during the initial 8 h postinfection. HeLa cells expressing GFP-Rab11A were infected at a multiplicity of infection of approximately 25, and at different times postinfection, the intracellular localization of GFP-Rab11A was determined by LSCM. Beginning as early as 1 h postinfection, GFP-Rab11A colocalizes with individual chlamydial vacuoles (Fig. 9A, B, and C), and by 4 h postinfection, a distinct rim of GFP-Rab11A is observed surrounding each chlamydial vacuole (Fig. 9G, H, and I). GFP-Rab11A remains associated with the chlamydial vacuoles throughout the time course of the experiment and can be seen associated with chlamydial vacuoles that have not yet been trafficked to the pericentriolar region of the cell.
FIG.9.
Temporal analysis of GFP-Rab11A recruitment to C. trachomatis inclusions. HeLa cells transiently expressing GFP-Rab11A were infected at a multiplicity of infection of approximately 20. At different times postinfection (shown to the left of the figure), the cells were fixed and stained with antichlamydial LPS. The cells were then incubated with goat anti-mouse IgG conjugated to Alexa 594 (red) and viewed by LSCM. Beginning as early as 1 h postinfection, colocalization of EBs and GFP-Rab11A is observed. At each time point, insets show representative vacuoles colocalizing with GFP-Rab11A. (A, D, G, J, and M) GFP-Rab11A, (B, E, H, K, and N) antichlamydial LPS, (C, F, I, L, and O) GFP-Rab11A (green) and antichlamydial LPS (red) merged. Bars, 10 μm (O) and 2 μm (inset in panel O).
DISCUSSION
The specific host and chlamydial factors that mediate the biogenesis or intracellular trafficking of the chlamydial inclusion remain largely undefined. Here, we report a specific and intimate association of a subset of eukaryotic GFP-Rab GTPases with mature chlamydial inclusions. Specific associations between GFP-Rab4 and GFP-Rab11 isoforms (which function sequentially in Tfn receptor recycling) and GFP-Rab1 (which functions in ER-to-Golgi and intra-Golgi trafficking) were detected with all chlamydial inclusions tested. In contrast, association of the Golgi-localized Rab GFP-Rab6 was observed only with C. trachomatis inclusions, while association of another Golgi-localized Rab, GFP-Rab10, was observed only with C. pneumoniae and C. muridarum inclusions. Since not all GFP-Rab proteins tested associated with the inclusion, the interactions that we observed are specific and not likely due to GFP itself. These data suggest that chlamydiae selectively recruit critical components of the host Rab membrane trafficking machinery to the inclusion in both a species-dependent and species-independent fashion.
We examined the intracellular distribution of Rab GTPases in infected HeLa cells that overexpressed amino-terminal GFP-tagged Rab GTPases. In this transient-transfection system, no toxic effects were observed, and each GFP-Rab localized to subcellular compartments similar to those of their endogenous counterparts (5, 11, 12, 18, 20, 30, 32, 42, 59, 60, 72) (data not shown). This approach will aid in future functional studies by allowing the use of activated, GTP-restricted, and dominant interfering inactive mutants and will also facilitate the examination of Rab-inclusion interactions in viable cells through the use of time-lapse video microscopy. These observations demonstrate that GFP-Rab GTPases, when overexpressed, are able to interact with the chlamydial inclusion, but they do not demonstrate that endogenous Rab GTPases behave in a similar fashion. Attempts to localize endogenous Rab GTPases in infected HeLa cells by indirect immunofluorescence microscopy using antibodies specific to Rab4, Rab6, and Rab11 were unsuccessful. The inability to detect endogenous Rab proteins in infected cells may be due to the fact that the levels of endogenous Rab proteins recruited to the inclusion are too low to detect by indirect immunofluorescence microscopy using the available antibodies. Therefore, although overexpressed GFP-tagged Rab proteins localize to subcellular compartments similar to those of their endogenous counterparts and are thought to function similarly, the role of endogenous Rab GTPases in infected cells should be examined further in the future. However, consistent with a possible role for Rab6A during chlamydial infection, Rab6A is transcriptionally induced in C. trachomatis-infected HeLa cells (77). In addition, Rab4, Rab6, and Rab11 isoforms are ubiquitously expressed, which suggests that they are expressed in chlamydia-infected cells in vivo.
It has been shown previously that the chlamydial inclusion does not interact with the host's degradative endosomal or lysosomal pathway as demonstrated by the absence of classical endocytic markers, such as Tfn, Tfn receptor, early endosomal antigen 1 (EEA1), cation-independent M6PR, and lysosomal glycoprotein Lamp1 (29, 54, 55, 61, 70). The failure to detect specific associations between the endosomal Rabs, GFP-Rab5, GFP-Rab7, and GFP-Rab9, and the inclusion is consistent with these previous data. However, since we examined only cells infected for 18 h, we cannot discount transient interactions with the inclusion. This is unlikely, given that even as early as 5 min postinfection, EEA1 does not associate with the inclusion membrane (54). Thus, chlamydiae appear to bypass the early endocytic pathway completely.
Even though no fusion between endosomes and the inclusion has been detected, an intimate association of Tfn-containing tubular endosomes with inclusions has been described (2, 54, 55, 70). Tfn endosomal association with the inclusion is dependent on early chlamydial gene expression and occurs by 2 h postinfection. Both GFP-Rab4 and GFP-Rab11, which are recruited to all inclusions, are primarily associated with recycling endosomes in nonpolarized epithelial cells (60, 67). However, not all inclusion-associated GFP-Rab11A colocalizes with Tfn or its receptor. Because recycling endosomes are heterogeneous in nature (62), these data suggest that inclusion-associated GFP-Rab11 or GFP-Rab4 may be associated with different populations of recycling endosomes, including those that are Tfn positive and those that are Tfn negative (67). Alternatively, GFP-Rab4 or GFP-Rab11, as well as the other inclusion-associated GFP-Rab proteins, may be anchored within the inclusion membrane. We cannot distinguish between these two possibilities by LSCM. Therefore, immunoelectron microscopy should be done to clarify whether each GFP-Rab protein is associated with the inclusion via an intimate association of nonfused closely juxtaposed vesicles, as was demonstrated for Tfn-containing vesicles (51), or anchored within the inclusion membrane. Rab5, Rab4, and Rab11 regulate sequential steps along the recycling pathway and are localized to distinct domains on early and recycling endosomes (60). Therefore, the absence of GFP-Rab5 and the association of GFP-Rab4 and GFP-Rab11 demonstrate that inclusions are associated with markers or endosomal domains that are characteristic of late steps in the recycling pathway.
Recruitment of Rab proteins to the inclusion may regulate the transport of the nascent inclusion to the peri-Golgi region, since isoforms of both Rab6 (36) and Rab11 (73) regulate transport from early endosomes to the TGN. Consistent with this, Rab proteins interact with several different molecular motors, such as myosin Vb (32), cytoplasmic dynein light chain (5), and rabkinesin 6 (17). However, to function in this manner, Rab proteins would have to be recruited to the inclusion early during development. At least for GFP-Rab11A, we detect association with chlamydia-containing vacuoles as early as 1 h postinfection, a point during chlamydial development when chlamydia-containing vacuoles are in the process of being trafficked to the peri-Golgi region (57).
One of the unique characteristics of all chlamydial inclusions is their ability to fuse with Golgi-derived vesicles containing NBD-sphingomyelin, endogenously synthesized from 6{N-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)amino]caproylsphingosine}(C6-NBD-ceramide) (27, 48, 74) in a process that is dependent on early chlamydial gene expression (57). The recruitment of Golgi-localized Rab GTPases to the inclusion may provide a mechanism for delivery and fusion of exocytic vesicles with the inclusion. Since fusion with exocytic vesicles is a property shared by all chlamydial inclusions, Rab1, which is associated with all inclusions, may regulate this trafficking step. Alternatively, chlamydiae may exploit functionally redundant pathways regulated by either Rab6 or Rab10. On the other hand, the roles of Rab1 and Rab6 during chlamydial infection might not be related to their Golgi trafficking function but might instead be related to Rab6's role in endosome-to-TGN trafficking or Rab1's proposed role in transcytosis in polarized epithelial cells (31). Chlamydiae also obtain other molecules from the host, including eukaryotic glycerophospholipids and cholesterol (75). Therefore, Rab GTPases may also be involved in regulating acquisition of these compounds. Consistent with this, Rab11 has recently been shown to be involved in regulating cholesterol trafficking and homeostasis (30). Finally, because of Rab11's association with the Inc-laden fibers, Rab11 may play a role in trafficking chlamydial antigens along these fibers, thus providing a mechanism for the appearance of chlamydial antigens at the plasma membrane (6, 76).
The distinct rim-like intracellular localization patterns of the GFP-Rab GTPases, the colocalization of Rab11A with IncG, and the fact that each GFP-Rab protein remains associated with the inclusion even after microtubules are disassembled and both pericentriolar recycling endosomes and the Golgi apparatus are redistributed strongly support a specific interaction with the inclusion membrane. These interactions may be mediated directly through an interaction between each Rab protein and an inclusion membrane protein, as was recently demonstrated between 14-3-3 and IncG (56). Alternatively, Rab recruitment to the inclusion may be mediated through Rab effector interactions, since Rab1 (1, 41), Rab4 (5, 33, 43), Rab6 (17), and Rab11 (28, 78) interact with a variety of effector molecules.
The differential association of GFP-Rab GTPases to inclusions containing different chlamydial species highlights the fact that individual inclusions interact differently with their hosts. On the basis of comparative genomic analysis, the protein compositions of chlamydial inclusion membranes are thought to differ dramatically for different chlamydial species (3). The exposure of a different complement of inclusion membrane proteins to the host cytosol creates the potential for extreme diversity in host-pathogen interactions. For instance, GFP-Rab6 may associate only with C. trachomatis inclusions, because it interacts with an inclusion membrane protein that is present and exposed only in C. trachomatis inclusions. Although recent data have suggested that differences in tissue tropism and disease expression for different chlamydial species may be due in part to genetic differences found within a specific region of the chlamydial genome termed the plasticity zone (4, 19), the differential association of Rab GTPases and 14-3-3 species-specific interactions with chlamydial inclusions suggest that different host interactions with the inclusion may also play significant roles.
Although we have not yet determined the biological consequences of Rab recruitment to the inclusion, the association of GFP-tagged Rab GTPases involved in both recycling and biosynthetic pathways to the inclusion demonstrates for the first time that chlamydiae recruit key regulators of membrane trafficking to the inclusion in both a species-specific and species-independent fashion. These data lend support to the idea that chlamydiae, by modulating the activity of host factors that function in vesicle trafficking, control the biogenesis of the inclusion. Further characterization through the expression of dominant interfering and constitutively active Rab mutants should help to elucidate the roles that Rab GTPases may play during chlamydial infection and determine whether recruitment of Rab GTPases is essential for the intracellular survival of chlamydia.
Acknowledgments
We thank Craig Roy for providing GFP-Rab5, GFP-Rab7, and GFP-Rab11 constructs, Suzanne Pfeffer for providing the GFP-Rab9 construct, Ted Hackstadt and Kate Wolf for providing purified C. pneumoniae, and Harlan Caldwell for providing C. trachomatis and C. muridarum stocks and antichlamydial LPS antiserum. We also thank Barbara Butcher, Hélène Marquis, Ruth Collins, and David Russell for critical reading of the manuscript.
Editor: B. B. Finlay
REFERENCES
- 1.Allan, B. B., B. D. Moyer, and W. E. Balch. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science 289:444-448. [DOI] [PubMed] [Google Scholar]
- 2.Al-Younes, H. M., T. Rudel, and T. F. Meyer. 1999. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., R. S. Griffiths, V. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structural motif predictor of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35-48. [DOI] [PubMed] [Google Scholar]
- 4.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielli, A., P. O. Thornquist, A. G. Hendrick, R. Finn, K. Fitzgerald, and M. W. McCaffrey. 2001. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem. Biophys. Res. Commun. 281:1141-1153. [DOI] [PubMed] [Google Scholar]
- 6.Brown, W. J., Y. A. Skeiky, P. Probst, and D. D. Rockey. 2002. Chlamydial antigens colocalize within IncA-laden fibers extending from the inclusion membrane into the host cytosol. Infect. Immun. 70:5860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucci, C., R. G. Parton, I. H. Mather, H. Stunnenber, K. Simons, B. Hoflack, and M. Zerial. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70:715-728. [DOI] [PubMed] [Google Scholar]
- 8.Bucci, C., P. Thomsen, P. Nicoziani, J. McCarthy, and B. van Deurs. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11:467-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1997. Chlamydia trachomatis genital tract infections-United States, 1995. Morb. Mortal. Wkly. Rep. 46:193-198. [PubMed] [Google Scholar]
- 11.Chen, W., Y. Feng, D. Chen, and A. Wandinger-Ness. 1998. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9:3241-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, W., and A. Wandinger-Ness. 2001. Expression and functional analyses of Rab8 and Rab11a in exocytic transport from trans-Golgi network. Methods Enzymol. 329:165-175. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Y. T., C. Holcomb, and H. P. Moore. 1993. Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc. Natl. Acad. Sci. USA 90:6508-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clausen, J. D., G. Christiansen, H. U. Holst, and S. Birklund. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 25:441-449. [DOI] [PubMed] [Google Scholar]
- 15.Cole, N. B., and J. Lippincott-Schwartz. 1995. Organization of organelles and membrane traffic by microtubules. Curr. Opin. Cell Biol. 7:55-64. [DOI] [PubMed] [Google Scholar]
- 16.Collins, R. N., and P. Brennwald. 2000. Rab proteins. GTPases. Front. Mol. Biol. 24:137-175. [Google Scholar]
- 17.Echard, A., F. Jollivet, O. Martinez, J. J. Lacapere, A. Rousselet, I. Janoueix-Lerosey, and B. Goud. 1998. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 279:580-585. [DOI] [PubMed] [Google Scholar]
- 18.Echard, A., F. J. M. Opdam, H. J. P. C. de Leeuw, F. Jollivet, P. Savelkoul, W. Hendriks, J. Voorberg, B. Goud, and J. A. M. Fransen. 2000. Alternative splicing of the human Rab6A gene generates two close but functionally different isoforms. Mol. Biol. Cell 11:3819-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Y., B. Press, W. Chen, J. Zimmerman, and A. Wandinger-Ness. 2001. Expression and properties of Rab7 in endosome function. Methods Enzymol. 329:175-187. [DOI] [PubMed] [Google Scholar]
- 21.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 22.Girod, A., B. Storrie, J. C. Simpson, L. Johannes, B. Goud, L. M. Roberts, J. M. Lord, T. Nilsson, and R. Pepperkok. 1999. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat. Cell Biol. 1:423-430. [DOI] [PubMed] [Google Scholar]
- 23.Gorvel, J. P., P. Chavrier, M. Zerial, and J. Gruenberg. 1991. rab5 controls early endosome fusion in vitro. Cell 64:915-925. [DOI] [PubMed] [Google Scholar]
- 24.Grayston, J. T., M. B. Aldous, A. Easton, S. P. Wang, C. C. Kuo, L. A. Campbell, and J. Altman. 1993. Evidence that Chlamydia pneumoniae causes pneumonia and bronchitis. J. Infect. Dis. 168:1231-1235. [DOI] [PubMed] [Google Scholar]
- 25.Grayston, J. T., and L. A. Campbell. 1999. The role of Chlamydia pneumoniae in atherosclerosis. Clin. Infect. Dis. 28:993-994. [DOI] [PubMed] [Google Scholar]
- 26.Hackstadt, T. 1999. Cell biology, p. 101-138. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 27.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 92:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hales, C. M., R. Griner, K. C. Hobdy-Henderson, M. C. Dorn, D. Hardy, R. Kumar, J. Navarre, E. K. L. Chan, L. A. Lapierre, and J. R. Goldenring. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067-39075. [DOI] [PubMed] [Google Scholar]
- 29.Heinzen, R. A., M. A. Scidmore, D. D. Rockey, and T. Hackstadt. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holtta-Vuori, M., K. Tanhuanpaa, W. Mobius, P. Somerharju, and E. Ikonen. 2002. Modulation of cellular cholesterol transport and homeostasis by Rab11. Mol. Biol. Cell 13:3107-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin, M., L. Saucan, M. G. Farquhar, and G. E. Palade. 1996. Rab1a and multiple other Rab proteins are associated with the transcytotic pathway in rat liver. J. Biol. Chem. 271:30105-30113. [DOI] [PubMed] [Google Scholar]
- 32.Lapierre, L. A., R. Kumar, C. M. Hales, J. Navarre, S. G. Bhartur, J. O. Burnette, J. D. W. Provance, J. A. Mercer, M. Bahler, and J. R. Goldenring. 2001. Myosin Vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12:1843-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsay, A. J., A. G. Hendrick, G. Cantalupo, F. Senic-Matuglia, B. Goud, C. Bucci, and M. W. McCaffrey. 2002. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 277:12190-12199. [DOI] [PubMed] [Google Scholar]
- 34.Lombardi, D., T. Soldati, M. A. Riederer, Y. Goda, M. Zerial, and S. R. Pfeffer. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12:677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majeed, M., M. Gustafsson, E. Kihlstrom, and O. Stendalh. 1993. Roles of Ca2+ and F-actin in intracellular aggregation of Chlamydia trachomatis in eukaryotic cells. Infect. Immun. 61:1406-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallard, F., B. L. Tang, T. Galli, D. Tenza, A. Saint-Pol, X. Yue, C. Antony, W. Hong, B. Goud, and L. Johannes. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCaffrey, M. W., A. Bielli, G. Cantalupo, S. Mora, V. Roberti, M. Santillo, F. Drummond, and C. Bucci. 2001. Rab4 affects both recycling and degradative endosomal trafficking. FEBS Lett. 495:21-30. [DOI] [PubMed] [Google Scholar]
- 38.Meresse, S., J. P. Gorvel, and P. Chavrier. 1995. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 108:3349-3358. [DOI] [PubMed] [Google Scholar]
- 39.Meresse, S., O. Steele-Mortimer, B. B. Finlay, and J. P. Gorvel. 1999. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 18:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulder, J. W. 1985. Comparative biology of intracellular parasitism. Microbiol. Rev. 49:298-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyer, B. D., B. B. Allan, and W. E. Balch. 2001. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2:268-276. [DOI] [PubMed] [Google Scholar]
- 42.Moyer, B. D., J. Matteson, and W. E. Balch. 2001. Expression of wild-type and mutant green fluorescent protein-Rab1 for fluorescence microscopy analysis. Methods Enzymol. 329:6-14. [DOI] [PubMed] [Google Scholar]
- 43.Nagelkerken, B., E. Van Anken, M. Van Raak, L. Gerez, K. Mohrmann, N. Van Uden, J. Holthuizen, L. Pelkmans, and P. Van Der Sluijs. 2000. Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem. J. 346:593-601. [PMC free article] [PubMed] [Google Scholar]
- 44.Opdam, F. J., A. Echard, H. J. Croes, J. A. van den Hurk, R. A. van de Vorstenbosch, L. A. Ginsel, B. Goud, and J. A. Fransen. 2000. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 113:2725-2735. [DOI] [PubMed] [Google Scholar]
- 45.Pereira-Leal, J. B., and M. C. Seabra. 2000. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301:1077-1087. [DOI] [PubMed] [Google Scholar]
- 46.Pfeffer, S. R. 2001. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11:487-491. [DOI] [PubMed] [Google Scholar]
- 47.Ren, M., G. Xu, J. Zeng, C. De Lemos-Charandini, M. Adesnik, and D. Sabatini. 1998. Hydrolysis of GTP on Rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rockey, D. D., E. R. Fischer, and T. Hackstadt. 1996. Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect. Immun. 64:4269-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 50.Rothman, J. E., and F. T. Wieland. 1996. Protein sorting by transport vesicles. Science 272:227-234. [DOI] [PubMed] [Google Scholar]
- 51.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 52.Schlierf, B., G. H. Fey, J. Hauber, G. M. Hocke, and O. Rosorius. 2000. Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res. 259:257-265. [DOI] [PubMed] [Google Scholar]
- 53.Schramm, N., and P. B. Wyrick. 1995. Cytoskeletal requirements in Chlamydia trachomatis infection in host cells. Infect. Immun. 63:324-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 57.Scidmore, M. A., D. D. Rockey, E. R. Fischer, R. A. Heinzen, and T. Hackstadt. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753-765. [DOI] [PubMed] [Google Scholar]
- 59.Short, B., C. Preisinger, J. Schaletzky, R. Kopajtich, and F. A. Barr. 2002. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr. Biol. 12:1792-1795. [DOI] [PubMed] [Google Scholar]
- 60.Sonnichsen, B., S. De Renzis, E. Nielsen, J. Rietdorf, and M. Zerial. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149:901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taraska, T., D. M. Ward, R. S. Ajioka, P. B. Wyrick, S. R. Davis-Kaplan, C. H. Davis, and J. Kaplan. 1996. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect. Immun. 64:3713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teter, K., G. Chandy, B. Quinones, K. Pereyra, T. Machen, and H.-P. H. Moore. 1998. Cellubrevin-targeted fluorescence uncovers heterogeneity in the recycling endosomes. J. Biol. Chem. 273:19625-19633. [DOI] [PubMed] [Google Scholar]
- 63.Thyberg, J., and S. Moskalewski. 1999. Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 246:263-279. [DOI] [PubMed] [Google Scholar]
- 64.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W. H. O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 65.Tisdale, E. J., J. R. Bourne, R. Khosravi-Far, C. J. Der, and W. E. Balch. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuvim, M. J., R. Adachi, S. Hoffenberg, and B. F. Dickey. 2001. Traffic control: Rab GTPases and the regulation of interorganellar transport. News Physiol. Sci. 16:56-61. [DOI] [PubMed] [Google Scholar]
- 67.Ullrich, O., S. Reinsch, S. Urbe, M. Zerial, and R. G. Parton. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Sluijs, P., M. Hull, P. Webster, P. Male, B. Goud, and I. Mellman. 1992. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70:729-740. [DOI] [PubMed] [Google Scholar]
- 69.van der Sluijs, P., M. Hull, A. Zahraoui, A. Tavitian, B. Goud, and I. Mellman. 1991. The small GTP-binding protein rab4 is associated with early endosomes. Proc. Natl. Acad. Sci. USA 88:6313-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Ooij, C., G. Apodaca, and J. Engel. 1997. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect. Immun. 65:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Via, L. E., D. Deretic, R. J. Ulmer, N. S. Hibler, L. A. Huber, and V. Deretic. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272:13326-13331. [DOI] [PubMed] [Google Scholar]
- 72.White, J., L. Johannes, F. Mallard, A. Girod, S. Grill, S. Reinsch, P. Keller, B. Tzschaschel, A. Echard, B. Goud, and E. H. Stelzer. 1999. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 147:743-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilcke, M., L. Johannes, T. Galli, V. Mayau, B. Goud, and J. Salamero. 2000. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151:1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolf, K., and T. Hackstadt. 2001. Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell. Microbiol. 3:145-152. [DOI] [PubMed] [Google Scholar]
- 75.Wylie, J. L., G. M. Hatch, and G. McClarty. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179:7233-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyrick, P. B., J. Choon, S. T. Knight, D. Goyeau, E. S. Stuart, and A. B. MacDonald. 1994. Chlamydia trachomatis antigens on the surface of infected human endometrial epithelial cells. Immunol. Infect. Dis. 4:131-141. [Google Scholar]
- 77.Xia, M., R. E. Bumgarner, M. F. Lampe, and W. E. Stamm. 2003. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J. Infect. Dis. 187:424-434. [DOI] [PubMed] [Google Scholar]
- 78.Zeng, J., M. Ren, D. Gravotta, C. De Lemos-Charandini, M. Lui, H. Erdjument-Bromage, P. Tempst, G. Xu, T. H. Shen, T. Morimoto, M. Adesnik, and D. Sabatini. 1999. Identification of a putative effector protein for Rab11 that participates in transferrin recycling. Proc. Natl. Acad. Sci. USA 96:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]