Abstract
Genome sequencing of C. trachomatis serovar D revealed the presence of three putative open reading frames (ORFs), CT145 (Pkn1), CT673 (Pkn5), and CT301 (PknD), encoding eukaryote-like serine/threonine kinases (Ser/Thr kinases). Two of these putative kinase genes, CT145 and CT301, were PCR amplified from serovar L2, cloned, and sequenced. Predicted translation products of the ORFs showed the presence of conserved kinase motifs at the N terminus of the proteins. CT145 and CT301 (encoding Pkn1 and PknD, respectively) were expressed in Escherichia coli as GST fusion proteins. In vitro kinase assays with Escherichia coli-derived glutathione S-transferase fusion proteins showed autophosphorylation of Pkn1 and PknD, indicating that they are functional kinases. Gene expression analysis of these kinase genes in Chlamydia by reverse transcriptase PCR indicated expression of these kinases at the early mid phase of the developmental cycle. Immunoprecipitated native chlamydial Pkn1 and PknD proteins also showed autophosphorylation in an in vitro kinase assay. Phosphoamino acid analysis by thin-layer chromatography confirmed that Pkn1 and PknD are phosphorylated on both serine and threonine residues. Interaction of Pkn1 and PknD with each other as well as interaction of Pkn1 with inclusion membrane protein G (IncG) was demonstrated by using a bacterial two-hybrid system. These interactions were further suggested by phosphorylation of the proteins in in vitro kinase assays. This report is the first description of the existence of functional Ser/Thr kinases in Chlamydia. The results of these findings should lead to a better understanding of how Chlamydia interact and interfere with host signaling pathways, since kinases represent potential mediators of the intimate host-pathogen interactions that are essential to the intracellular life cycle of Chlamydia.
Protein phosphorylation is one of the principal mechanisms by which extracellular signals are translated into cellular responses. The regulation of eukaryotic signal transduction pathways by phosphorylation and dephosphorylation of serine (Ser), threonine (Thr), and tyrosine (Tyr) residues via protein kinases and protein phosphatases, respectively, has been known for many years and is considered the backbone of regulatory pathways (35). Previously, these kinases and phosphatases were thought to be unique to eukaryotes. In prokaryotes, the phosphorylation system responsible for transduction of the external stimulus to response involves a two-component system consisting of histidine kinase sensors and their response regulators. The histidine kinases were considered to be the predominant class of kinases in bacteria (26). Recent studies and analysis of bacterial genome sequences now available have demonstrated the presence of eukaryote-like Ser/Thr-phosphorylating enzymes in prokaryotes (1, 5, 8, 30, 37). Prokaryotic Ser/Thr protein kinases have been cloned, sequenced, and characterized from Myxococcus (10), Streptomyces (25), Anabaena (36), Yersinia (7), Thermomonospora (15), Mycobacterium (27), and Pseudomonas (22) species during the past several years. Such eukaryote-like Ser/Thr protein kinases have been implicated in three different processes in prokaryotes, namely, regulation of growth and development, stress responses, and pathogenicity. In Streptomyces, Cyanobacterium (Anabaena), and Myxococcus xanthus species, these kinases are involved in the control of stages of development, sporulation, or secondary metabolite production (20, 32, 38, 39, 40). Of the at least four putative Ser/Thr kinases in Pseudomonas aeruginosa, one has been implicated in virulence (24, 33). Note that both a protein-tyrosine phosphatase (YopH) and a Ser/Thr kinase (YpkA) in Yersinia pseudotuberculosis are encoded by the virulence plasmid and have been shown to be injected into the host cell by a type III secretion mechanism (7). A complete-genome analysis of Mycobacterium tuberculosis suggested the presence of 11 eukaryote-like protein kinases. To date, four of the Ser/Thr protein kinases of M. tuberculosis have been cloned and characterized. Although very little is known about their cellular functions, these kinases are proposed to be regulators of metabolic processes, including transcription, cell development, and interaction with host cells (8, 18, 27). Bacteria of the genus Chlamydia have a unique developmental life cycle, during which the bacterium interacts with and exploits host signaling pathways for its own development. The functional significance of Ser/Thr kinases in other bacteria prompted us to investigate these kinases in Chlamydia.
The chlamydiae are among the most widespread bacterial pathogens in the world and are responsible for a variety of diseases in different animal species and humans. Chlamydia trachomatis is the leading cause of preventable blindness and is the most common sexually transmitted bacterial species (34). The chlamydiae are obligate intracellular pathogens that undergo a unique biphasic developmental cycle within the host eukaryotic cell. Following internalization, chlamydiae develop and grow within an intracellular vacuole called the inclusion. Within the inclusion, the infectious and metabolically inert extracellular form, the elementary body (EB), differentiates into a noninfectious reticulate body, which is the metabolically active and replicating form of Chlamydia (23). About 20 to 24 h postinfection, the reticulate bodies begin to differentiate into EBs. The developmental cycle is then complete, and infectious EBs are released from the cell after 48 to 72 h. Two major barriers to studying the molecular biology and host-pathogen interactions of Chlamydia are the obligate intracellular nature of bacterial growth and the absence of tools to genetically manipulate Chlamydia. However, recent genomic information and molecular and cellular studies of chlamydial infection have added new insights into the complicated developmental cycle of this bacterium.
Information is rapidly accumulating that shows how Chlamydia, like many other bacteria, exploits and subverts the host signaling and trafficking pathways to support its own developmental cycle. Annotation of the C. trachomatis serovar D genome sequence provided evidence for the presence of three putative open reading frames (ORFs) encoding proteins with significant homology to the eukaryote-like Ser/Thr kinases and one ORF showing homology to the PP-2C-type protein phosphatase (31). These putative ORFs suggest the presence of a functional phosphorylation- and dephosphorylation-based signaling system in Chlamydia. This paper describes the cloning, expression, and molecular characterization of two putative ORFs encoding Ser/Thr kinases from C. trachomatis serovar L2 and shows that the products of these ORFs, Pkn1 and PknD, are functional kinases. Interacting partners of the Pkn1 in Chlamydia were also identified, and interactions were confirmed by using the in vitro kinase assay.
MATERIALS AND METHODS
Materials.
Escherichia coli strain DH5α was used for cloning purposes, and 100 μg of ampicillin per ml was used for the selection of transformants. Trizol reagent (Invitrogen, Carlsbad, Calif.) was used for isolation of total DNA and RNA. Mouse monoclonal antibodies and rabbit polyclonal antibodies directed against phosphoserine (pSer), phosphothreonine (pThr), and phosphotyrosine (pTyr) were obtained from Cell Signaling Technology (Cell Signaling Technology, Beverly, Mass.). Goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase conjugate and goat anti-rabbit IgG-horseradish peroxidase conjugate were obtained from Santa Cruz Biotechnology. An enhanced chemiluminescence detection kit was purchased from Amersham Pharmacia. [γ-32P]ATP used for in vitro kinase assays was purchased from ICN Pharmaceuticals. Rabbit polyclonal antiserum against C. trachomatis IncG was kindly provided by Ted Hackstadt (NIH—Rocky Mountain). The BacterioMatch two-hybrid vector system was purchased from Stratagene.
Infection of L2 cells with C. trachomatis serovar L2.
Monolayer cultures of mouse fibroblast L2 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum at 37°C in an atmosphere of 5% CO2. Mouse fibroblast L2 cell monolayers were infected with C. trachomatis serovar L2 diluted in SPG (250 mM sucrose, 10 mM sodium phosphate, 5 mM l-glutamic acid) at a multiplicity of infection of 1. Infected L2 cells were incubated for 2 to 40 h as indicated in each experiment in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 10 μg of gentamicin per ml, and 10 μg of cycloheximide per ml. EBs were harvested after sonication of the infected cells and purified using discontinuous Renografin gradients as described previously (17).
Cloning and expression of Pkn1 and PknD.
C. trachomatis serovar L2 genomic DNA was prepared using the Qiagen genomic kit (Qiagen). The two ORFs encoding eukaryote-like Ser/Thr protein kinases were amplified from C. trachomatis serovar L2 genomic DNA by using the sequence information available for C. trachomatis serovar D (http://chlamydia-www.berkeley.edu:4231). The primers used for the amplification of ORF145 and ORF301 (encoding Pkn1 and PknD, respectively) from C. trachomatis serovar L2 are listed in Table 1. The amplified products were cloned in pPCR-Script Amp SK(+) (Stratagene) and sequenced. The predicted protein sequence of both putative kinases was analyzed using the multiple alignments for Ser/Thr kinases and catalytic domains.
TABLE 1.
Primers used to amplify putative kinase ORFs from C. trachomatis serovar L2
| Kinase (ORF) | Primer | Primer sequence |
|---|---|---|
| Pkn1 (145) | Forward | 5′-CGGATCCATGGAAGAGCGAGCCGCAG-3′ |
| Reverse | 5′-GCGGCCGCTTATTTTACATCTTTCGCAC-3′ | |
| PknD (301) | Forward | 5′-CGAATTCTTGCAACGATACGAATTGATTAGGC-3′ |
| Reverse | 5′-GCGGCCGCTTAATCAAAAAAATTGTTCTCC-3′ |
For expression studies, both the kinase ORFs as well as the ORF encoding IncG (CT118) from C. trachomatis serovar L2 were cloned in-frame at the C terminus of the glutathione S-transferase (GST) gene on pGEX-6P-1 (Amersham Pharmacia). The resulting GST fusions of Pkn1, PknD, and IncG were expressed in E. coli strain DH5α. Briefly, bacteria were inoculated into Luria-Bertani medium containing 100 μg of ampicillin per ml and allowed to grow to an optical density at 600 nm of 0.6 as measured at 30°C, and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Bacteria were harvested and resuspended in lysis buffer (30 mM Tris-HCl [pH 7.4], 150 mM NaCl, 25% sucrose, 0.1% Triton X-100, 10 μg each of leupeptin and pepstatin per ml, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and 1 mM phenylmethylsulfonyl fluoride). Bacteria were lysed by sonication, and bacterial debris was pelleted at 20,000 × g for 20 min. The supernatant was then added to washed glutathione Sepharose 4B (Amersham Pharmacia) and mixed gently at 4°C for 1 to 2 h. The beads were centrifuged at 500 × g for 5 min, washed six times with 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl containing 0.1% Triton X-100, and washed three times with kinase buffer without cold ATP (20 mM HEPES [pH 7.6], 20 mM MgCl2, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and 1 mM benzamidine), resuspended in kinase buffer, and stored at 4°C. GST-Pkn1 and GST-PknD bound to agarose beads were directly used for in vitro phosphorylation reactions. For the interaction experiments, the GST fusion proteins were eluted from glutathione Sepharose 4B beads by using reduced glutathione. The eluted proteins were concentrated, and buffer was exchanged for use in the in vitro kinase assays.
Production of antibodies to Pkn1 and PknD.
Peptides specific to Pkn1 (SEQDEHYNELIRLKDSRIC) and PknD (CYEKYRQAYLSMENNFFD) mapping at the C-terminal region of these proteins were synthesized. Antibodies against these peptides were raised in rabbits at BioSource International (Camarillo, Calif.). For both antibody preparations, the fourth bleed was affinity purified. The resulting affinity-purified antibodies were used for Western analysis and immunoprecipitation.
Autophosphorylation of Pkn1 and PknD.
Autophosphorylation of Pkn1 and PknD as well as phosphorylation of substrate myelin basic protein (MBP), a general kinase substrate, was performed with an in vitro kinase assay. GST-Pkn1- and GST-PknD-bound beads were resuspended in 1× kinase buffer containing 5 μM cold ATP and 10 μCi of [γ-32P]ATP. The reactions were incubated at 30°C for 30 min and terminated by the addition of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (250 mM Tris [pH 6.8], 40% glycerol, 20% β-mercaptoethanol, 8% SDS, 0.01% bromophenol blue). Kinase reactions were electrophoresed on an SDS-10% polyacrylamide gel and transferred to nitrocellulose (Bio-Rad) for 2 h and autoradiographed. Kinase assays were performed in the same fashion by using Dynabeads or protein A-agarose- or protein G-agarose bead-bound immunoprecipitates of Pkn1, PknD, and IncG.
Phosphoamino acid analysis by thin-layer chromatography.
To analyze the phosphoamino acids phosphorylated in the in vitro kinase reaction, E. coli-expressed GST-Pkn1 and GST-PknD were autophosphorylated in vitro and subjected to SDS-PAGE and electroblotted onto an Immobilon polyvinylidene difluoride membrane. Autophosphorylated Pkn1 and PknD were subjected to phosphoamino analysis essentially as described previously (4). Briefly, in vitro autophosphorylated Pkn1 and PknD were excised from the polyvinylidene difluoride membrane and subjected to acid hydrolysis at 110°C for 1 h. Hydrolyzed samples were lyophilized and finally dissolved in electrophoresis buffer (10:1:189, glacial acetic acid, pyridine, water [pH 3.5]). An aliquot of hydrolyzed phosphoamino acid along with marker pSer, pThr, and pTyr amino acids was spotted onto a glass-backed cellulose thin-layer chromatography plate. Electrophoresis was carried out for 50 min at 1.3 kV, the plate was dried, and the marker phosphoamino acids were visualized with ninhydrin. The plate was then subjected to autoradiography, and the film was aligned with the ninhydrin staining.
Analysis of gene expression by RT-PCR.
Gene transcription of both kinase ORFs at different phases of the C. trachomatis developmental cycle was analyzed by reverse transcriptase PCR (RT-PCR). Genome equivalents of C. trachomatis in each sample were quantitated at different time points postinfection (2, 6, 12, 20, and 36 h) along with uninfected controls by quantitative competitive PCR essentially performed as described previously (6). Briefly, L2 monolayers were infected with a multiplicity of infection of 1, and whole-culture RNA and DNA were isolated sequentially using the Trizol reagent (Invitrogen). Quantitative competitive PCR was performed by amplifying the Hsp60 gene (groEL) from total genomic DNA isolated from infected cultures with known amounts of exogenously added competitor plasmid DNA. pAV4, a plasmid containing a truncated Hsp60 gene fragment of C. trachomatis, which will amplify a small-size fragment compared to the fragment amplified from the genomic copy with the same primer pair, was used for quantitation purposes. Genomes were quantified for each sample by comparing integrated density values for agarose gel-resolved PCR products. Gene transcription of Pkn1 and PknD was assayed by RT-PCR by using gene-specific primers using the RNA corresponding to an equal number of genome equivalents in each sample at the different time periods of infection.
Preparation of Chlamydia lysates, immunoprecipitation, and immunoblotting.
C. trachomatis serovar L2-infected host cells were harvested 40 h postinfection, and EBs were purified through a 30% Renografin gradient. EB lysates were prepared by resuspending the purified EBs in lysis buffer containing 10 mM Tris-HCl (pH 7.5), 10% glycerol, 1% Triton X-100, 10 μg each of leupeptin and pepstatin per ml, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and 1 mM phenylmethylsulfonyl fluoride, and the lysates were incubated on ice for 1 h. Lysates were briefly sonicated and centrifuged at 12,000 × g for 30 min. Cleared supernatant was collected and used for immunoprecipitation. Pkn1 and PknD were immunoprecipitated from Chlamydia lysates by using affinity-purified antipeptide antibodies against Pkn1 and PknD. Briefly, the chlamydial lysate was precleared by using normal rabbit IgG, normal mouse IgG, and protein A-agarose beads. Affinity-purified anti-Pkn1, anti-PknD, or anti-pSer, anit-pThr, and anti-pTyr antibodies were added to the precleared lysates and incubated overnight at 4°C. Immunocomplexes were precipitated by adding protein A-agarose or protein G-agarose and rocking for 2 to 4 h or overnight at 4°C and collected by centrifugation. Immunoprecipitated pellets were washed several times with 1% Triton X-100 in 10 mM Tris buffer (pH 7.5). In addition, immunoprecipitations were also carried out using protein A conjugated to magnetic beads (Dynabeads) essentially as recommended by the manufacturer (Dynal). Finally, immunocomplexes were resuspended in 1× SDS-PAGE buffer, boiled for 5 min, and proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were blocked in Tris-buffered saline (20 mM Tris-HCl [pH 7.6], 140 mM NaCl) containing 0.1% (vol/vol) Tween 20 with 5% bovine serum albumin or skim-milk powder. Anti-Pkn1 antibody was used at a dilution of 1:2,000, whereas anti-PknD antibody was used at 1:1,000 dilution. Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology) was used at a dilution of 1:2,000. The enhanced chemiluminescence detection system (Amersham Pharmacia) was used according to the manufacturer's directions. Immunoprecipitates of Pkn1 and PknD were also used in the kinase assay. For immunoprecipitation of IncG, total lysates of Chlamydia-infected L2 cells were prepared. IncG was immunoprecipitated from these lysates by using anti-IncG rabbit polyclonal antisera in the same manner as mentioned above. Immunoprecipitated material was directly used for in vitro kinase assay.
Interactions in bacterial two-hybrid system.
Interactions in the BacterioMatch two-hybrid vector system (Stratagene) were analyzed by cloning chlamydial kinase genes for use as bait in pBT. A C. trachomatis serovar L2 library was constructed in pTRG vector. The library was screened using Pkn1 and PknD as bait separately per the manufacturer's instructions. Interactions were established using a screening based on resistance to various concentrations of carbenicillin by activation of an ampicillin resistance gene. A second reporter gene, β-galactosidase, expressed from the same activatable promoter was also utilized to validate the bait and target interaction.
Nucleotide sequence accession numbers.
The nucleotide sequence data for Pkn1 and PknD from C. trachomatis serovar L2 were deposited in GenBank and assigned accession numbers AY148435 and AY148436, respectively.
RESULTS
DNA sequence analysis of putative kinase ORFs CT451 and CT301.
Sequence analysis of the C. trachomatis serovar D genome (26) revealed the presence of three putative ORFs, CT145 (Pkn1), CT673 (Pkn5), and CT301 (PknD), that show homology to eukaryote-like Ser/Thr kinases. However, a closer analysis of the amino acid sequence encoded by CT673 indicates that the original annotation was probably incorrect and that Pkn5 is not a kinase (see Discussion). Therefore, in this study two ORFs, CT145 and CT301 encoding for Pkn1 and PknD, respectively, were characterized. The corresponding homologs of these kinases are also present in C. pneumoniae strain CWL029 and in other species of Chlamydia. Comparison of the C. trachomatis serovar D and C. pneumoniae CWL029 putative kinase ORFs showed a 58.17% identity between CT145 (Pkn1) and CPn0148 and a 48.29% identity between CT301 (PknD) and CPn0095 at the amino acid level. Homology of the putative chlamydial kinases with the kinase domain of eukaryote and prokaryote Ser/Thr kinases was confined to the N-terminal region of the ORFs. The C-terminal regions of these putative Chlamydia Ser/Thr kinase ORFs showed no significant homology to any protein in the database.
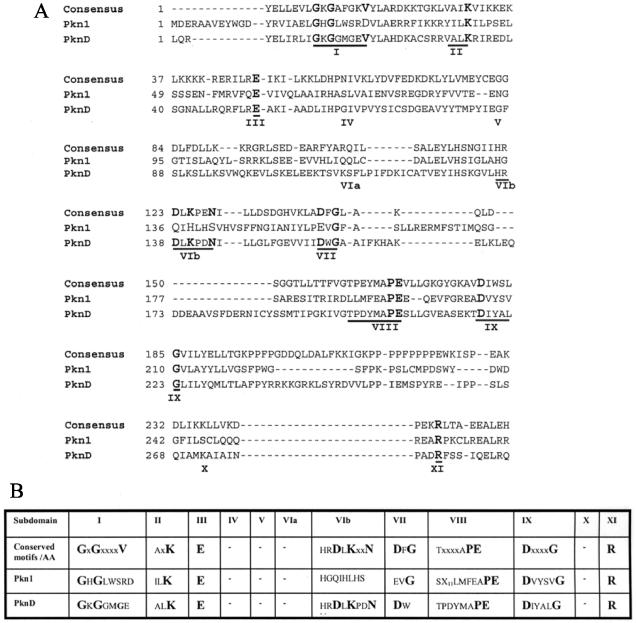
ORFs encoding putative kinases Pkn1 and PknD were amplified from C. trachomatis serovar L2 using primers designed from the published C. trachomatis D sequence (Table 1). The amplified products were cloned into pPCR-Script Amp SK(+) and sequenced. The predicted coding sequences of the C. trachomatis serovar L2 pkn1 (encoding a protein of 615 amino acids) and pknD (936 amino acids) genes showed 99% homology at the amino acid level with their respective homologs from C. trachomatis serovar D. The degree of absolute conservation is relatively small among eukaryotic protein kinases, while prokaryotic candidate kinases demonstrate small deviations from the eukaryotic prototype. The eukaryotic and prokaryotic kinase domains contain 12 conserved subdomains numbered I to V, VIa, VIb, and VII to XI, ranging in size from a single amino acid to stretches of 10 or more residues (9). An amino acid alignment of Pkn1 and PknD with the consensus N terminus of Ser/Thr kinases of eukaryotes is shown in Fig. 1A. To facilitate analysis of kinase domains in chlamydial kinases, a comparison of the conserved subdomains and amino acids present in the consensus kinase domain is also presented in Fig. 1B. The subdomain I to IV constitutes the nucleotide-binding domain. The key feature of subdomain I is the presence of conserved middle Gly residues in the motif G-x-G-x-x-G-x-V. Pkn1 and PknD have conserved Gly in subdomain I. The conserved Lys residue of subdomain II (which interacts with α and β phosphoryl groups of ATP and helps to anchor and orient ATP for catalysis) and the invariant Glu residue of subdomain III are present in both chlamydial ORFs. Subdomain VIb is the catalytic loop and contains a highly conserved motif, H-R-D-L-K-x-x-N, which is absolutely conserved in PknD but not in Pkn1. PknD possesses a conserved subdomain VII (D-x-G), whereas Pkn1 has a Glu instead of an Asp residue. The A-P-E motif in subdomain VIII is called the P + 1 loop because it is the docking site for the P + 1 residue of the substrate. It is very well conserved among the kinases and was also present in both of the putative chlamydial kinases. The Asp residue in subdomain IX and the Arg residue in subdomain XI are conserved in all the kinases in prokaryotes. Subdomain XI stabilizes the protein substrate binding during phosphorylation. The chlamydial proteins contain both the conserved Asp residue in subdomain IX and the conserved Arg in subdomain XI.
FIG. 1.
Sequence alignment of the kinase domains of Pkn1 and PknD. (A) Amino acid sequence alignment between the consensus motifs of prototype eukaryotic Ser/Thr kinases and the N-terminal domains of the ORFs encoding putative protein Ser/Thr kinases Pkn1 and PknD of C. trachomatis serovar L2. The regions corresponding to subdomains I to XI proposed for the Ser/Thr kinases are underlined. The conserved amino acids within each different motif are in boldface type. The residues are marked on the left. (B) Comparison of conserved motifs and amino acids in the kinase domain of Pkn1 and PknD. Conserved amino acids in each motif are in boldface type.
Expression of chlamydial ORFs in E. coli.
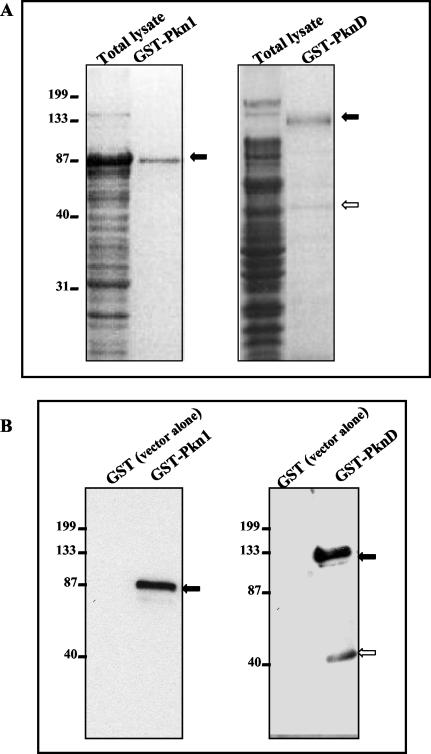
The presence of conserved kinase motifs in the putative chlamydial kinases prompted us to analyze these proteins for functional activity. C. trachomatis serovar L2 ORFs encoding Pkn1 and PknD were cloned as fusions with GST, expressed, and analyzed by SDS-PAGE. As shown in Fig. 2A, Coomassie blue staining revealed proteins of 90 to 95 kDa for GST-Pkn1 (predicted molecular mass is 70 kDa plus 27 kDa for GST) and 130 to 135 kDa and a faint band at 45 to 50 kDa for GST-PknD (predicted molecular mass is 107 kDa plus 27 kDa for GST). The sizes of the products obtained as GST fusion proteins were in good agreement with the expected molecular masses. GST purified from bacterial lysates of E. coli containing the vector plasmid without insert revealed only a 27-kDa protein (data not shown). Western blot analysis using mouse monoclonal anti-GST antibodies revealed that the purified proteins were GST fusion proteins (data not shown). The lower-molecular-mass (45 to 50 kDa) protein band present along with the full-length GST-PknD on the Coomassie blue-stained gel was not recognized by anti-GST antibodies. These results led us to believe that it is either a contaminant protein copurifying with full-length GST-PknD or a processed or cleaved product of the GST-PknD fusion protein devoid of the GST tag.
FIG. 2.
Expression and purification of Pkn1 and PknD in E. coli. E. coli DH5α cells harboring pGEX-6P-1/Pkn1 or PknD were grown in Luria-Bertani medium and induced with 1 mM IPTG. Total lysates of E. coli expressing fusion proteins were purified to homogeneity using GST beads. GST-bound proteins were mixed with SDS-PAGE buffer and subjected to electrophoresis. (A) Total lysates and GST-purified preparations of Pkn1 and PknD after Coomassie blue staining. (B) Immunoblot analysis of GST fusion proteins with anti-Pkn1 and anti-PknD peptide antibodies. Both gels include E. coli expressing GST alone as a negative control. Solid arrows in each panel indicate the expected size of the GST fusion proteins. The open arrow in both panels represents the cleaved protein fragment of full-length PknD detected in Coomassie blue-stained protein gels and by immunoblot analysis. Positions of molecular mass markers (in kilodaltons) are indicated on the left.
As described in Materials and Methods, antibodies were generated against peptides mapping within the C terminus of Pkn1 and PknD. Affinity-purified antipeptide antibodies against Pkn1 and PknD were tested in immunoblots with GST-Pkn1 and GST-PknD and were found to be specific and did not react with fractions prepared from E. coli containing the expression vectors without inserts (Fig. 2B). The specificity of the antipeptide antibodies was further confirmed with peptide inhibition assays (data not shown). It is important that immunoblots of E. coli-derived GST-PknD with anti-PknD peptide antibody recognized a lower-molecular-mass (45 to 50 kDa) protein band (Fig. 2B). These results lead us to believe that the lower-molecular-mass protein band in E. coli-derived GST-PknD preparations is in fact a cleaved and processed C-terminal fragment of PknD rather then a contaminant protein.
Autophosphorylation of Pkn1 and PknD.
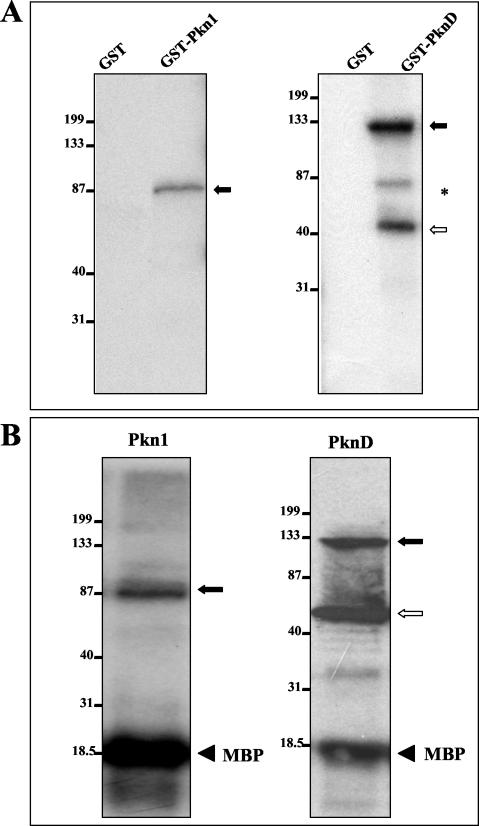
Autophosphorylation activity of Pkn1 and PknD was examined by an in vitro kinase assay using GST fusion proteins expressed in E. coli and [γ-32P]ATP as phosphoryl donor. As shown in Fig. 3A, kinase assays revealed that Pkn1 and PknD were capable of autophosphorylation. For GST-Pkn1, only one phosphorylation product of ∼90 to 95 kDa (the size of the GST-Pkn1 fusion protein) was observed. Proteins of 130 to 135 and 45 to 50 kDa were phosphorylated in the in vitro kinase assay with GST-PknD. GST-Pkn1 and GST-PknD were also able to phosphorylate MBP, a conventional in vitro substrate for Ser/Thr kinases (Fig. 3B). Taken together, the above observations clearly indicate that the kinase domains of Pkn1 and PknD have enzymatic activity in E. coli and that the proteins are autophosphorylated.
FIG. 3.
Autophosphorylation of E. coli-expressed GST-Pkn1 and GST-PknD. Purified GST-Pkn1 and GST-PknD fusion protein beads were directly used in the in vitro kinase assays. Kinase assays were performed as described in Materials and Methods, and proteins were separated by SDS-PAGE. After electrophoresis, the gel was transferred onto a nitrocellulose membrane and autoradiographed. (A) Solid arrows represent the expected phosphorylated products. The open arrow represents phosphorylation of the PknD protein fragment detected in the Coomassie blue-stained protein gel shown in Fig. 2A. *, a nonspecific phosphorylated product. Positions of molecular mass markers in each gel (in kilodaltons) are indicated on the left. In both gels, GST alone was used as a control. (B) MBP phosphorylation mediated by Pkn1 and PknD in the in vitro kinase reaction. The solid and open arrows represent phosphorylated Pkn1 and PknD as in panel A. Solid arrowheads represent phosphorylated MBP.
Expression of Pkn1 and PknD in C. trachomatis serovar L2.
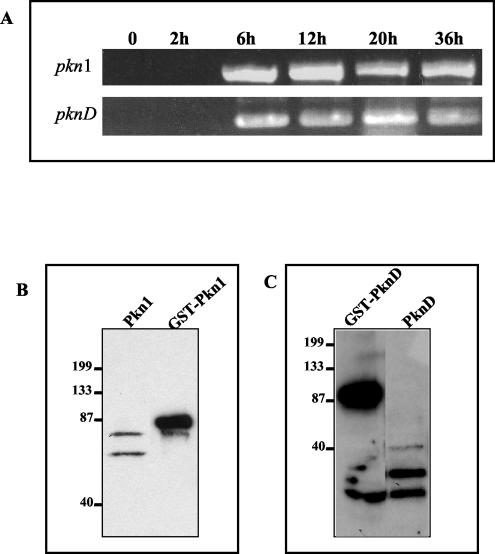
We extended our analysis to investigate expression of these kinase genes during the developmental cycle of Chlamydia. As described in Materials and Methods, transcription of kinase genes was analyzed by RT-PCR of total RNA extracted from infected L2 cells at 2, 6, 12, 20, and 36 h postinfection and normalized for the number of C. trachomatis genomes present as described in an earlier study (6). As shown in Fig. 4A, RNA transcripts for pkn1 and pknD were detected as early as 6 h postinfection. These results clearly indicate that both putative kinase genes present in C. trachomatis serovar L2 are transcriptionally active and are expressed during the developmental cycle.
FIG. 4.
Transcription analysis and protein expression of pkn1 and pknD in Chlamydia. (A) RT-PCR analysis using primers specific for Pkn1 and PknD was performed on total RNA samples isolated at different time intervals representing early (2 h), mid (6 to 12 h) and late (20 to 36 h) phases of the developmental cycle of Chlamydia. Uninfected L2 fibroblasts were used as control (0). (B and C) EBs were harvested after 40 h of infection and purified by density gradient centrifugation. Purified EBs were boiled in SDS-PAGE buffer, and samples were subjected to electrophoresis, transferred, and immunoblotted with anti-Pkn1 and anti-PknD antibodies. GST-Pkn1 and GST-PknD were used as positive controls. Positions of molecular mass markers in panels B and C are indicated on the left (molecular masses shown in kilodaltons).
Antipeptide antibodies were then used to detect expression of native chlamydial Pkn1 and PknD. We looked for the presence of Pkn1 and PknD proteins in density gradient centrifugation-purified EBs prepared from infected cultures harvested 40 h postinfection. Whole-cell lysates of uninfected L2 fibroblasts were used as controls. Anti-Pkn1 antibodies recognized two major protein bands, one at 70 and the other at approximately 65 kDa (Fig. 4B), representing either different phosphorylated forms or a degraded form of Pkn1. Western analysis of EB lysates revealed two major protein bands with molecular masses of 55 to 60 and 45 to 50 kDa that reacted with anti-PknD antibodies (Fig. 4C). No protein signal could be detected at 107 kDa, the expected molecular mass of PknD. The size of one of the protein bands was the same as that observed in GST-PknD preparations from E. coli (45 to 50 kDa), which indicated the presence of some putative cleavage site or processing site in PknD that generates the processed protein products in Chlamydia. These two protein bands might represent different phosphorylated forms of cleaved C terminus of native chlamydial-derived PknD. An antibody-peptide inhibition assay with anti-PknD proved the specificity of the lower-molecular-mass protein highlighted in Western analysis of EBs (data not shown). This result indicated that the lower-molecular-mass protein is a cleaved/processed PknD.
Immunoprecipitation and in vitro kinase assay with chlamydial-derived Pkn1 and PknD proteins.
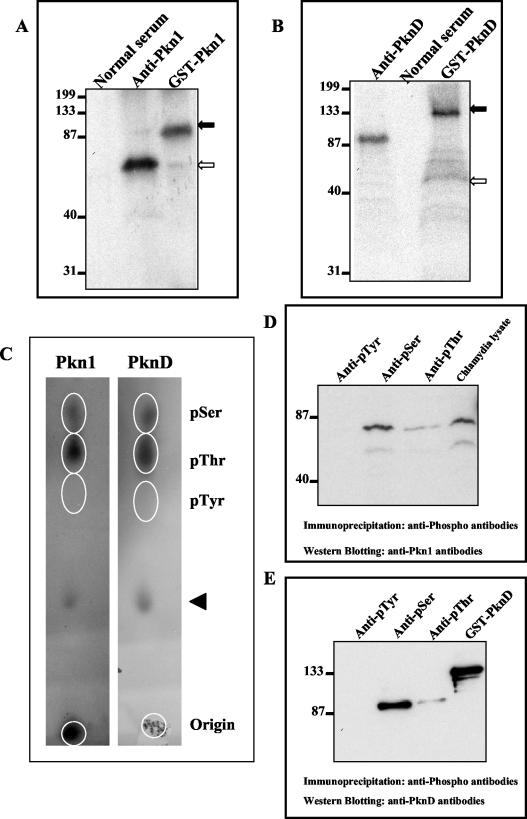
It was important to check the functional activity of the Ser/Thr kinases present in Chlamydia. Immunoprecipitations were carried out with anti-Pkn1 and anti-PknD antibodies to pull down both native chlamydial kinases from lysates of Chlamydia EBs. Immunoprecipitations were performed using protein G- and protein A-conjugated magnetic beads. Anti-Pkn1 and anti-PknD antibodies were cross-linked to beads, and chlamydial proteins were captured. Protein-captured beads were directly used for the in vitro kinase assay. A phosphorylated protein with a molecular mass of 70 kDa, representing Pkn1, could be detected after the kinase reaction with material immunoprecipitated with anti-Pkn1 antibody (Fig. 5A). The kinase assay with anti-PknD immunoprecipitates showed a phosphorylated protein with a molecular mass of 100 to 105 kDa (Fig. 5B). No phosphorylated products were observed in the kinase assay with material immunoprecipitated with normal rabbit serum. These results clearly suggest that both the kinases are present in EBs and are functionally active.
FIG. 5.
Phosphorylation of native chlamydial-derived Pkn1 and PknD and phosphoamino acid analysis. Chlamydia-derived Pkn1 and PknD kinase proteins were immunoprecipitated from chlamydial lysates using anti-Pkn1 and anti-PknD antibodies. Immunocomplex-bound beads were directly used for in vitro kinase assay using [γ-32P]ATP. Reactions were electrophoresed, transferred onto membranes, and subjected to autoradiography. Immunoprecipitation of chlamydial lysates by normal rabbit serum was used as a negative control. (A and B) The kinase assaywith purified GST-Pkn1 or GST-PknD was used as a positive control. Solid arrows represent phosphorylated GST-Pkn1 and GST-PknD, whereas open arrows represent phosphorylated Pkn1 (panel A) and PknD (panel B). Positions of molecular mass markers (in kilodaltons) in each panel are indicated on the left. (C) Phosphoamino acid analysis of GST-Pkn1 and GST-PknD by thin-layer chromatography. γ-32P-labeled Pkn1 and PknD proteins after an in vitro kinase reaction were electroblotted, excised, and acid hydrolyzed. Hydrolyzed labeled amino acids were spotted along with marker pSer, pThr, and pTyr amino acids on cellulose thin-layer chromatography plates and separated at high voltage. Marker phosphoamino acids were detected by ninhydrin staining, whereas labeled phosphoamino acids were detected by autoradiography. Spots of the labeled amino acids are encircled in each panel. The arrowhead represents unhydrolyzed proteins. (D and E) Immunoprecipitations from Chlamydia-infected cell lysates were performed using anti-pSer, anti-pThr, or anti-pTyr antibodies. Phosphoimmunocomplexes were separated on SDS-PAGE and transferred to membranes. Western analysis of blots was performed using anti-Pkn1 and anti-PknD antibodies. Panel D represents immunoblotting with anti-Pkn1 antibodies. Five percent input chlamydial lysate was used as a positive control. Panel E represents immunoblotting with anti-PknD antibodies, and GST-PknD was used as a positive control.
Pkn1 and PknD are autophosphorylated on Ser and Thr amino acid residues.
To demonstrate that E. coli-expressed Pkn1 and PknD are autophosphorylated at the Ser and/or Thr residues, phosphoamino acid analysis of autophosphorylated GST-Pkn1 and GST-PknD was performed using thin-layer chromatography. As shown in Fig. 5C, the γ-32P-labeled phosphoamino acids for Pkn1 and PknD migrated with the marker pSer and pThr residues in thin-layer chromatography. No signal could be detected at the pTyr position. This result indicates that Pkn1 and PknD are autophosphorylated at Ser and Thr residues but not at Tyr. To demonstrate that native Pkn1 and PknD from Chlamydia are also autophosphorylated at Ser/Thr residues, specific mouse monoclonal antibodies against pSer, pThr, and pTyr were used for immunoprecipitation of native Chlamydia-derived phosphorylated proteins. Western blotting of phosphoimmunocomplexes with specific anti-Pkn1 and anti-PknD antibodies recognized phosphorylated Pkn1 and PknD. As shown in Fig. 5D, two different phosphorylated forms of Pkn1 (represented by 70- and 65-kDa bands) were detected in immunoprecipitation with anti-pSer and anti-pThr antibodies. Western analysis of phosphoimmunocomplexes with anti-PknD antibodies showed a protein band of ∼100 to 105 kDa with anti-pSer and anti-pThr antibodies (Fig. 5E). No signal could be detected with anti-pTyr immunoprecipitates. These results further confirm that native Chlamydia-derived Pkn1 and PknD proteins are functionally active and are phosphorylated at Ser/Thr residues as predicted.
In vivo and in vitro interaction of Pkn1 and PknD.
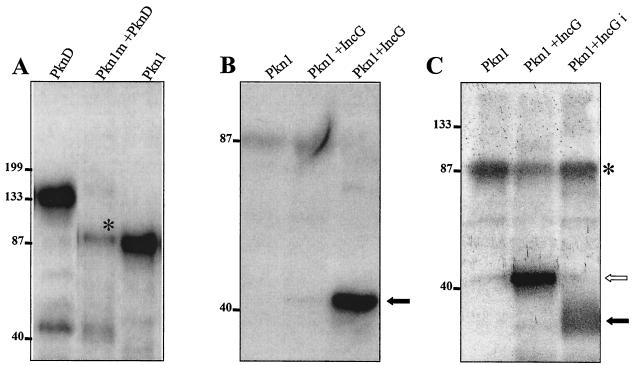
Screening of a C. trachomatis serovar L2 library with Pkn1 and PknD as bait in the Bacteriomatch two-hybrid system revealed interacting partner proteins like polymorphic membrane proteins, PlsX (fatty acid-phospholipid synthesis protein), IhfA (integration host factor), and several other hypothetical proteins. Multiple clones were obtained for each interacting partner. Interestingly, when Pkn1 was used as bait to screen the chlamydial library, two independent interacting clones were identified as PknD. When PknD was used as bait, three interacting clones were identified as Pkn1 (up to 375 μg of carbenicillin resistance per ml). To prove the interaction of Pkn1 and PknD with each other, a mutation of pkn1 was made in kinase subdomain II, where the codon for a conserved lysine residue at position 42 was replaced with an alanine codon by in vitro mutagenesis. In eukaryotic protein kinases, this residue is highly conserved and is essential for the ATP binding activity of the proteins. Mutant Pkn1 was expressed in E. coli as a GST fusion protein (GST-Pkn1 [K→A]) and purified. In in vitro kinase assays, it had no autophosphorylation activity (data not shown). GST-Pkn1 [K→A] was used in an in vitro kinase assay with PknD to examine whether Pkn1 could act as substrate for PknD. As shown in Fig. 6A, a phosphorylated Pkn1 could be detected along with the autophosphorylated PknD, which indicates that PknD is able to interact with and phosphorylate the mutant GST-Pkn1 [K→A]. Efforts to coimmunoprecipitate these kinases from chlamydial lysates using anti-Pkn1 and anti-PknD antibodies were unsuccessful. Nevertheless, the results of the in vitro kinase assay clearly support the interactions predicted by the bacterial two-hybrid system and are strong evidence for functional interactions of these kinases.
FIG. 6.
Phosphorylation of interacting partners of Pkn1. (A) E. coli-expressed and purified mutant GST-Pkn1 [K→A] and GST-PknD were used in an in vitro kinase assay. Both kinases were eluted from glutathione Sepharose beads by reduced glutathione, and the eluted proteins were used in the kinase assay. A band of Pkn1 marked with * indicates the phosphorylation of mutant Pkn1 by PknD. A faint band of lower-molecular-mass phosphorylated PknD is visible in the same lane. An autophosphorylated GST-Pkn1 bound to glutathione beads is also shown in the panel as a positive control. (B) Phosphorylation of E. coli-expressed and glutathione-eluted GST-IncG with the GST-Pkn1. A very faint phosphorylation product could be seen in the lane when reduced glutathione-eluted Pkn1 and IncG were used in the kinase reaction, whereas a solid arrow represents a strong phosphorylated product when Pkn1 bound to GST beads was used with the glutathione-eluted IncG. (C) Pkn1-mediated phosphorylation of immunoprecipitated IncG from C. trachomatis total lysates in an in vitro kinase reaction. The open arrow represents phosphorylated GST-IncG, whereas the solid arrow represents IncGi (immunoprecipitated with anti-IncG antibody from chlamydial lysates). *, the residual autophosphorylating activity of Pkn1.
Interaction of Pkn1 with IncG.
It has previously been shown that two inclusion membrane proteins (IncA and IncG) of Chlamydia are phosphorylated during the infection process (28, 29). In our Bacteriomatch two-hybrid screening, we did not isolate any clone encoding the Incs that interacted with either of the chlamydial kinases. Since these proteins could still be interacting partners with chlamydial kinases, IncA and IncG (ORFs CT119 and CT118, respectively) of C. trachomatis serovar L2 were cloned in pTRG of the Bacteriomatch two-hybrid system and tested for interactions with the chlamydial kinases as bait. The IncA target interacted even with the empty bait vector, so IncA was not pursued further. Interaction of IncG with Pkn1 was indicated by an increase in carbenicillin resistance (up to 350 μg of carbenicillin per ml). To further verify the interaction of Pkn1 with IncG, IncG was expressed as a GST fusion protein in E. coli and used as a substrate in an in vitro kinase reaction with Pkn1. As shown in Fig. 6B, IncG was phosphorylated in the kinase reaction with Pkn1. These results clearly suggest that Pkn1 interacts with IncG. Attempts were also made to determine if native IncG from Chlamydia could be phosphorylated with Pkn1 in the in vitro kinase reaction. IncG was immunoprecipitated from total chlamydial EB lysates by using rabbit polyclonal anti-IncG antibodies (kindly provided by Ted Hackstadt). Immunoprecipitated IncG bound to protein A-coated Sepharose beads was directly used in the kinase assay with glutathione-eluted GST-Pkn1. Phosphorylated IncG could be seen in the kinase assay with Pkn1 (Fig. 6C), thus indicating that the native chlamydial IncG could be phosphorylated with chlamydial kinase Pkn1.
DISCUSSION
Analysis of 35 prokaryotic genomes sequenced so far indicates that the presence of eukaryote-type protein kinases and phosphatases in prokaryotes is more common than previously thought (16). Genome sequencing of Chlamydiae suggested the existence of three putative Ser/Thr kinases and one PP2C-type protein phosphatase (31). In this study, we established that Pkn1 and PknD, predicted to be putative kinases, are indeed functional protein kinases expressed by C. trachomatis serovar L2 and that they interact with several chlamydial proteins.
Analysis of the amino acid sequence of PknD from C. trachomatis serovar L2 showed that it has a characteristic protein kinase signature, including all 12 conserved domains of eukaryotic Ser/Thr kinases. A GST-PknD fusion protein expressed in E. coli as well as PknD isolated from chlamydial EBs showed autophosphorylating activity, thus confirming that it is a functional kinase. Phosphoamino acid analysis and Western blot analysis with anti-pSer and anti-pThr antibodies confirmed that PknD is a Ser/Thr kinase. In E. coli, as well as in Chlamydia, PknD undergoes protein processing generating a cleaved C-terminal product of ∼45 to 50 kDa. The cleaved product reacts with antibodies raised against a C-terminal PknD peptide. The N-terminal kinase domain present in full-length PknD is phosphorylated, as is the cleaved C-terminal product. Western analysis of purified EB lysates with anti-PknD peptide antibodies did not detect full-length PknD protein at its expected size of ∼107 kDa (see Fig. 4C). However, kinase assays using material immunoprecipitated from chlamydial lysates with anti-PknD antibodies showed a phosphorylated protein at the expected size of full-length PknD (see Fig. 5B). These results suggest that the amount of full-length PknD present in Chlamydia is probably too low to be detectable by Western blot, but functionally, PknD is a very active kinase (detectable by kinase assay).
Sequence analysis of Pkn1 showed the presence of all the subdomains with conserved motifs except subdomain VIb, the catalytic domain. All known eukaryotic and prokaryotic kinases contain this catalytic domain with the highly conserved motif, H-R-D-L-K-x-x-N. The Asp residue in this motif is considered to be involved in the catalytic reaction, accepting the proton from the attacking hydroxyl of the substrates. The lysine residue in this loop helps to facilitate the phosphotransfer reaction by stabilizing the negative charge generated on the substrate-hydroxyl group during the phosphotransfer reaction. Though E. coli-derived GST-Pkn1 as well as chlamydial-derived Pkn1 showed autophosphorylating activity, the absence of this catalytic domain in chlamydial Pkn1 raises the intriguing possibility that this kinase differs in binding of the substrate and the catalytic reaction.
As mentioned earlier, annotation of the C. trachomatis serovar D genome predicted the presence of three ORFs encoding putative eukaryote-like Ser/Thr kinases. In this study, we characterized the two ORFs encoding Pkn1 and PknD and proved that they are functional Ser/Thr kinases of Chlamydia. We also cloned and sequenced the C. trachomatis serovar L2 homolog of the third ORF encoding Pkn5 that was predicted to be a eukaryote-like Ser/Thr kinase in the original serovar D genome annotation. However, autophosphorylation activity could not be detected with the GST-Pkn5 fusion protein. Moreover, GST-Pkn5 was not phosphorylated either in the presence of GST-Pkn1, GST-PknD, both GST-Pkn1 and GST-PknD, purified chlamydial lysates, or by a eukaryotic cell lysate (L2 fibroblast extract). Further, GST-Pkn5 was not able to phosphorylate MBP. These negative results call into question whether Pkn5 is a functional kinase. Detailed analysis of the Pkn5 N-terminus conserved region that encodes the eukaryote-like kinase subdomains showed the presence of all the subdomains except subdomains I (the absence of Gly residues) and XI (lack of the conserved Arg residue). The residues in these two subdomains have very important structural roles. Subdomain I has a consensus motif, G-x-G-x-x-G-x-V, that stabilizes the negative charges of α and β phosphates of ATP during phosphorylation. In eukaryotic and prokaryotic Ser/Thr kinases, the first two Gly residues are highly conserved. Replacement of the first Gly residue of the murine cAMP-dependent protein kinase reduces catalytic activity 200-fold, and substitution of the second Gly residue reduces catalytic activity by more than 1,000-fold (12). The absence of the Arg residue in subdomain XI is highly detrimental to activity, as Arg plays an important role in stabilizing the peptide-protein substrate binding lobe. Hence, we speculate that Pkn5 has no or very poor autophosphorylating activity due to the lack of Gly residues in subdomain I and the lack of conserved Arg in subdomain XI. Aminoglycoside 3′ phosphotransferases are the evolutionary descendants of eukaryotic-like protein kinases, with whom they share catalytically essential subdomains II through VII and IX (3, 13). However, even though these antibiotic kinases bind and utilize ATP in a manner similar to their protein-specific cousins, they do so without using the subdomain I motif. RT-PCR analysis of pkn5 at different developmental stages indicated that pkn5 is in fact a transcriptionally active gene in Chlamydia. We suggest that chlamydial Pkn5 is not a protein kinase but could be an aminoglycoside 3′ phosphotransferase, phosphorylating nonprotein substrates such as antibiotics in order to neutralize their toxic properties (11, 19).
Further sequence analysis of Pkn1 and PknD indicated the presence of potential transmembrane domains at the N terminus of Pkn1 (amino acids 207 to 224) and PknD (amino acids 663 to 681). These kinases may act as transmembrane kinases and might serve as receptors for external environmental signals to regulate cellular functions as is known for transmembrane kinases from Streptomyces and Myxococcus (25, 14) species. A comparative analysis for coexistence of histidine kinases and Ser/Thr kinases in prokaryotes showed that bacteria containing Ser/Thr kinases display mostly developmental morphogenesis, like Myxococcus xanthus, or pathogenicity, like Mycobacterium tuberculosis, implying that there may be underlying molecular reasons as to why some bacteria use Ser/Thr kinases in addition to His kinases (14). In the Ser/Thr kinase signaling cascade, phosphorylation of Ser or Thr residues is very stable, and the signal is amplified exponentially by phosphorylation of downstream effector molecules. This type of signaling might be required by prokaryotes for some cellular events involving sustained expression. Detection of RNA transcripts at the early mid-phase of the developmental cycle as well as the detection of protein expression in EBs suggest that they might be activated by specific signaling requirements that may occur at differentiation. So, it is reasonable to speculate that the two functional Ser/Thr kinases present in Chlamydia that we described may play some essential role during intracellular growth of Chlamydia via interacting with host-signaling pathways. The presence of one putative PP2C-type protein phosphatase (CT259) in C. trachomatis further argues for a functional kinase-based signaling cascade in Chlamydia for interaction with host signaling pathways.
One of the major goals in the study of these Ser/Thr protein kinases is to identify their cellular targets, as there could be many different targets in signaling pathways (21). Screening of the chlamydial library to search for probable targets or interacting partners revealed interaction of the two kinases with each other and with several other proteins. Further analysis demonstrated that the two kinases do interact with each other in vitro in the kinase assay. Signal transduction cascades in eukaryotes involve interactions of different kinases with each other. Thus, we can speculate that the interaction of Pkn1 and PknD may have functional significance in Chlamydia.
Recent studies in Chlamydia have shown that a group of proteins known as Incs are localized to the inclusion membrane (2, 28). One Inc protein, IncA of C. psittaci, is exposed at the surface of the inclusion and is phosphorylated in host cells during the infection process. Recently, it has been shown that IncG of C. trachomatis interacts with a mammalian host protein, 14-3-3β, in the yeast two-hybrid system (29). 14-3-3 proteins are a family of dimeric α-helical pSer/Thr binding proteins. Labeling with 32P orthophosphate showed that IncG is phosphorylated in Chlamydia-infected cells. The bacterial two-hybrid analysis showed interaction of IncG and Pkn1. Therefore, we examined whether chlamydial kinases could phosphorylate IncG in vitro in the kinase assay. Phosphorylation of E. coli expressed IncG as well as immunoprecipitated IncG from Chlamydia with Pkn1 in the kinase assay confirmed that a chlamydial kinase is in fact responsible for the phosphorylation of IncG. At present, the functional significance of IncG phosphorylation is not known, but it seems possible that phosphorylation of some chlamydial proteins might be responsible for specific interactions with host proteins in mediating the signaling required for maintenance of the infection process.
In summary, this study reports the first demonstration of functional Ser/Thr kinases in C. trachomatis. While mutagenesis or screening of mutants would be the most direct means for identifying the functional significance of these Ser/Thr kinases in chlamydial development, the absence of a genetic system requires us to use alternate, less direct approaches. Further functional characterization of the identified substrates and targets for the individual kinases might provide intriguing insights into the fundamental question as to the significance of the existence of these eukaryote-like enzymes in Chlamydia and add to our understanding of chlamydial pathogenicity.
Acknowledgments
We thank Harlan D. Caldwell for providing C. trachomatis serovar L2, Ted Hackstadt for providing rabbit polyclonal antisera against IncG, and Denny B. Buxton and Robert S. Adelstein (NHLBI, National Institutes of Health) for providing advice and facilities to perform the phosphoamino acid analysis. We also thank Michael N. Flora and the Uniformed Services University of the Health Sciences Biomedical Instrumentation Center for oligonucleotide synthesis and DNA sequencing. We also thank Priscilla Wyrick for helping us get started in Chlamydia research and for her continuing support.
This work was supported by grant AI 44033 from the National Institute of Allergy and Infectious Diseases.
The opinions or assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
Editor: J. T. Barbieri
REFERENCES
- 1.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, S. 1987. Phosphotransferase sequence homology. Nature 329:21. [DOI] [PubMed] [Google Scholar]
- 4.Buxton, D. B., and R. S. Adelstein. 2000. Calcium-dependent threonine phosphorylation of nonmuscle myosin in stimulated RBL-2H3 mast cells J. Biol. Chem. 275:34772-34775. [DOI] [PubMed] [Google Scholar]
- 5.Cozzone, A. J. 1993. ATP-dependent protein kinases in bacteria. J. Cell. Biochem. 51:7-13. [DOI] [PubMed] [Google Scholar]
- 6.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048-1060. [DOI] [PubMed] [Google Scholar]
- 7.Galyov, E. E., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1993. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 361:730-732. [DOI] [PubMed] [Google Scholar]
- 8.Han, G., and C. C. Zhang. 2001. On the origin of Ser/Thr kinases in a prokaryote. FEMS Microbiol. Lett. 200:79-84. [DOI] [PubMed] [Google Scholar]
- 9.Hanks, S., and A. M. Quinn. 1991. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200:38-62. [DOI] [PubMed] [Google Scholar]
- 10.Hanlon, W. A., M. Inouye, and S. Inouye. 1997. Pkn9, a Ser/Thr protein kinase involved in the development of Myxococcus xanthus. Mol. Microbiol. 23:459-471. [DOI] [PubMed] [Google Scholar]
- 11.Heinzel, P., O. Werbitzky, J. Distler, and W. Piepersberg. 1988. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3′-phosphotransferase. Relationships between antibiotic and protein kinases. Arch. Microbiol. 150:184-192. [DOI] [PubMed] [Google Scholar]
- 12.Hemmer, W., M. McGlone, I. Tsigelny, and S. S. Taylor. 1997. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 272:16946-16954. [DOI] [PubMed] [Google Scholar]
- 13.Hon, W. C., G. A. McKay, P. R. Thompson, R. M. Sweet, D. S. Yang, G. D. Wright, and A. M. Berghuis. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89:887-889. [DOI] [PubMed] [Google Scholar]
- 14.Inouye, S., R. Jain, T. Ueki, H. Nariya, C. Y. Xu, M. Y. Hsu, B. A. Fernandez-Luque, J. Munoz-Dorado, E. Farez-Vidal, and M. Inouye. 2000. A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microbiol. Comp. Genomics 5:103-120. [DOI] [PubMed] [Google Scholar]
- 15.Janda, L., P. Tichy, J. Spizek, and M. Petricek. 1996. A deduced Thermomonospora curvata protein containing serine/threonine protein kinase and WD-repeat domains. J. Bacteriol. 178:1487-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennelly, P. J. 2001. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Koehler, J. E., R. R. Burgess, N. E. Thompson, and R. S. Stephens. 1990. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. J. Biol. Chem. 265:13206-13214. [PubMed] [Google Scholar]
- 18.Koul, A., A. Choidas, A. K. Tyagi, K. Drlica, Y. Singh, and A. Ullrich. 2001. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology 147:2307-2314. [DOI] [PubMed] [Google Scholar]
- 19.Martin, P., E. Jullein, and P. Courvalin. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 2:615-625. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto, A., S. K. Hong, H. Ishizuka, S. Horinouchi, and T. Beppu. 1999. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene 146:47-56. [DOI] [PubMed] [Google Scholar]
- 21.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 22.Motley, S. T., and S. Lory. 1999. Functional characterization of a serine/threonine protein kinase of Pseudomonas aeruginosa. Infect. Immun. 67:5386-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay, S., V. Kapatral, W. Xu, and A. M. Chakrabarty. 1999. Characterization of a Hank's type serine/threonine kinase and serine/threonine phosphoprotein phosphatase in Pseudomonas aeruginosa. J. Bacteriol. 181:6615-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadvornik, R., Vomastek, T., Janecek, J., Technikova, Z., and P. Branny. 1999. Pkg2, a novel transmembrane protein Ser/Thr kinase of Streptomyces granaticolor. J. Bacteriol. 181:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 27.Peirs, P., L. De Wit, M. Braibant, K. Huygen, and J. Content. 1997. A serine/threonine protein kinase from Mycobacterium tuberculosis. Eur. J. Biochem. 244:604-612. [DOI] [PubMed] [Google Scholar]
- 28.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 29.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 30.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 31.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 32.Udo, H., M. Inouye, and S. Inouye. 1997. Biochemical characterization of Pkn2, a protein Ser/Thr kinase from Myxococcus xanthus, a gram-negative developmental bacterium. FEBS Lett. 400:188-192. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J., C. Li, H. Yang, A. Mushegian, and S. Jin. 1998. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J. Bacteriol. 180:6764-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstock, H., D. Dean, and G. Bolan. 1994. Chlamydia trachomatis infections. Infect. Dis. Clin. North. Am. 8:797-819. [PubMed] [Google Scholar]
- 35.Yarden, Y., and A. Ullrich. 1988. Growth factor receptor tyrosine kinases Annu. Rev. Biochem. 57:443-478. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, C. C. 1993. A gene encoding a protein related to eukaryotic protein kinases from the filamentous heterocystous cyanobacterium Anabaena PCC 7120. Proc. Natl. Acad. Sci. USA 90:11840-11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, C. C. 1996. Bacterial signalling involving eukaryotic-type protein kinases. Mol. Microbiol. 20:9-15. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, C. C., A. Friry, and L. Peng. 1998. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 180:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, C. C., and L. Libs. 1998. Cloning and characterization of the pknD gene encoding an eukaryotic-type protein kinase in the cyanobacterium Anabaena sp. PCC7120. Mol. Gen. Genet. 258:26-33. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, W., M. Inouye, and S. Inouye. 1996. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryotic-like Ser/Thr protein kinases. Mol. Microbiol. 20:435-447. [DOI] [PubMed] [Google Scholar]