Abstract
Yersinia pestis, the agent of plague, has arisen from a less virulent pathogen, Yersinia pseudotuberculosis, by a rapid evolutionary process. Although Y. pestis displays a large number of virulence phenotypes, it is not yet clear which of these phenotypes descended from Y. pseudotuberculosis and which were acquired independently. Y. pestis is known to replicate in macrophages, but there is no consensus in the literature on whether Y. pseudotuberculosis shares this property. We investigated whether the ability to replicate in macrophages is common to Y. pestis and Y. pseudotuberculosis or is a unique phenotype of Y. pestis. We also examined whether a chromosomal type III secretion system (TTSS) found in Y. pestis is present in Y. pseudotuberculosis and whether this system is important for replication of Yersinia in macrophages. A number of Y. pestis and Y. pseudotuberculosis strains of different biovars and serogroups, respectively, were tested for the ability to replicate in primary murine macrophages. Two Y. pestis strains (EV766 and KIM10+) and three Y. pseudotuberculosis strains (IP2790c, IP2515c, and IP2666c) were able to replicate in macrophages with similar efficiencies. Only one of six strains tested, the Y. pseudotuberculosis YPIII(p−) strain, was defective for intracellular replication. Thus, the ability to replicate in macrophages is conserved in Y. pestis and Y. pseudotuberculosis. Our results also indicate that a homologous TTSS is present on the chromosomes of Y. pestis and Y. pseudotuberculosis and that this secretion system is not required for replication of these bacteria in macrophages.
Yersinia pseudotuberculosis is a gram-negative bacterium that is transmitted by the fecal-oral route and typically causes self-limiting infections of the gut-associated lymphoid tissue (6). On rare occasions, Y. pseudotuberculosis can also cause fatal septicemia. Yersinia pestis is the agent of plague, an acute, often fatal infection of the lymphatic system that is transmitted by fleas or by aerosol (6). Although Y. pseudotuberculosis and Y. pestis utilize different modes of transmission and cause distinct diseases, both pathogens display a common tropism for lymphoid tissue (6). In addition, Y. pseudotuberculosis and Y. pestis are closely related at the genetic level. In fact, Y. pestis can be considered to be a subspecies of Y. pseudotuberculosis (1). Three biovars of Y. pestis have been recognized, Antiqua, Mediaevalis, and Orientalis, each of which is linked to a major pandemic (31). There are 21 different serologic variants of Y. pseudotuberculosis based on O-antigen typing (40). Sequence analysis of O-antigen gene clusters from many Y. pseudotuberculosis and Y. pestis strains has revealed that Y. pestis is most closely related to, and likely evolved from, serogroup O1b Y. pseudotuberculosis (40).
A number of virulence factors that are common to Y. pseudotuberculosis and Y. pestis have been identified. For example, both harbor a ∼70-kb virulence plasmid (known as pCD1 in Y. pestis and pYV in Y. pseudotuberculosis) that encodes a type III secretion system (TTSS) (11). Several antiphagocytic or antiinflammatory toxins (Yops) that are secreted by this system promote extracellular bacterial replication in vivo (11). Y. pestis carries two additional plasmids, pMT1 (∼100 kb) and pPCP1 (∼9.5 kb), that are not found in Y. pseudotuberculosis (20, 31). The presence of pMT1 and pPCP1 is thought to endow Y. pestis with increased virulence (pMT1 and pPCP1) and vector-borne transmissibility (pMT1) (20, 31). The complete DNA sequences of pCD1, pMT1, pPCP1, and Y. pestis CO92 (biovar Orientalis) and KIM (biovar Mediaevalis) chromosomes have recently been determined (13, 21, 24, 30, 33). Interestingly, Y. pestis encodes a TTSS that is distinct from the TTSS encoded on pCD1 or pYV (13, 30). The chromosomal TTSS in Y. pestis (referred to here as TTSS-2) is closely related to the TTSS encoded by Salmonella pathogenicity island 2 (SPI-2) of Salmonella enterica serovar Typhimurium that promotes replication of this bacterium in macrophages (19).
Bubonic plague typically initiates as a subcutaneous infection that is transmitted by fleas (6, 31). Pneumonic plague can occur when the bacteria are inhaled directly into the lungs. The bubonic and pneumonic forms of the disease have been studied by experimental introduction of Y. pestis into rodents or nonhuman primates. Bacterial multiplication initially occurs at the site of infection. Later, the bacteria enter the lymphatic system and spread to regional lymph nodes and then disseminate via the bloodstream to other organs such as spleen and liver. Following extensive replication in these organs, the bacteria enter the bloodstream, and the host dies of septicemic shock.
Y. pestis is considered to be a facultative intracellular pathogen (6, 31). It is believed that Y. pestis replicates within macrophages during the early stages of infection at peripheral host sites, while extracellular growth is predominant during the later stages of systemic infection. A number of studies have shown that Y. pestis can replicate in macrophages both in vivo and in vitro (7, 9, 15, 22, 41). Straley and Harmon (41) reported that wild-type Y. pestis KIM, or a KIM strain lacking pCD1 and pPCP1, could grow in mouse peritoneal macrophages. In addition, observations made by electron microscopy indicated that Y. pestis replicates in macrophage phagolysosomes, as nascent phagosomes containing bacteria fused with secondary lysosomes labeled with thorium dioxide (41). More recently, Oyston et al. (28) investigated the role of the response regulator PhoP for Y. pestis survival in macrophages. A phoP mutant of Y. pestis GB displayed a reduced ability to survive in J774A.1 macrophages and a reduced ability to kill mice by the subcutaneous route of infection (28). These results suggest that PhoP regulates one or more genes important for intracellular replication of Y. pestis in macrophages and that intracellular proliferation is important for disease when the infection is initiated from a peripheral location.
In contrast to what is seen with Y. pestis, it is generally believed that Y. pseudotuberculosis is unable to survive in macrophages (12). Y. pseudotuberculosis typically initiates infection by penetrating M cells that overlie the Peyer's patches of the gut-associated lymphoid tissue. It is likely that Y. pseudotuberculosis encounters macrophages soon after passing through M cells (16). In fact, when rabbits were experimentally infected via ileal loops with a serogroup O1 Y. pseudotuberculosis strain, intact bacteria could be detected inside macrophages that are found in Peyer's patches (16). However, there are conflicting data in the literature concerning the ability of Y. pseudotuberculosis to replicate in macrophages. Mills and Finlay (27) have shown that YPIII, a serogroup O3 strain of Y. pseudotuberculosis, is unable to survive in mouse J774A.1 macrophages. In contrast, Tsukano et al. (42) reported that a serogroup O4b Y. pseudotuberculosis strain replicated in mouse peritoneal macrophages. These observations may indicate that the ability of Y. pseudotuberculosis to replicate in macrophages is a serogroup-specific property. Therefore, it is possible that Y. pestis inherited the ability to replicate in macrophages from a Y. pseudotuberculosis ancestor rather than acquiring this property independently.
We present evidence that the ability to replicate in macrophages is conserved between Y. pestis and Y. pseudotuberculosis. We tested a set of six bacterial strains representing two different biovars of Y. pestis (Mediaevalis and Orientalis) and three different serogroups of Y. pseudotuberculosis (O1, O2, and O3) for the ability to replicate in primary mouse macrophages. Two Y. pestis strains and three Y. pseudotuberculosis strains were essentially indistinguishable in terms of their ability to replicate in macrophages. One strain of Y. pseudotuberculosis, the YPIII strain (serogroup O3), was defective for intracellular replication. We speculate that YPIII has acquired a mutation during laboratory passage that results in an intracellular replication defect. In addition, although the chromosomal TTSS-2 appears to be conserved in Y. pestis and Y. pseudotuberculosis, we obtained evidence that this secretion system is not required for replication of these bacteria in primary murine macrophages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Y. pestis and Y. pseudotuberculosis strains used in this study are listed in Table 1. All strains used in this study were cured of the virulence plasmid pCD1 or pYV+. IP2790c, IP2515c, and IP2666c were obtained from Michel Simonet (University Lille 2) (38). YPIII(p−) (5) and EV766 (34) were obtained from Stanley Falkow (Stanford University). Y. pestis KIM10+ (32) was obtained from Kathleen McDonough (Wadsworth Center, New York State Department of Health). Y. pestis and Y. pseudotuberculosis strains were cultivated at 26°C on heart infusion (HI) (Difco) and Luria-Bertani (LB) agar plates, respectively. Cultures were grown in broth with aeration for 20 h at 26°C to prepare bacteria for infection assays. Y. pestis was grown in HI broth containing 1% glucose, 0.2% xylose, and 1 mM MgCl2 (41). Y. pseudotuberculosis was grown in LB broth. Bacterial cultures were washed once in phosphate-buffered saline (PBS) and then resuspended in PBS, and the number of viable bacteria in the inoculum was determined by spreading serial 10-fold dilutions of the inoculum on HI or LB agar plates. The plates were incubated for 2 or 3 days at 28°C to obtain input CFU. The Escherichia coli strains used, NovaBlue (Novagen), DH5α (36), and S17-1 (37) lysogenized with λpir (23), were grown in LB broth or on LB agar plates at 37°C. Bacterial growth medium was supplemented with ampicillin at 100 μg/ml, kanamycin at 25 μg/ml, tetracycline at 20 μg/ml, or streptomycin at 30 μg/ml when appropriate.
TABLE 1.
Yersinia strains
| Species and strain | Relevant characteristics | Reference or source |
|---|---|---|
| Y. pestis | ||
| KIM10+ | Biovar Medievalis, pCD1−, pPCP1− | 32 |
| KIM10+/GFP | KIM10+ containing p67GFP3.1, Apr | This work |
| EV766 | Biovar Orientalis, pCD1−, Δpgm | 34 |
| EV766/GFP | EV766 containing p67GFP3.1, Apr | This work |
| EV766.1/GFP | EV66/GFP y0514::kan, Apr, Kmr | This work |
| Y. pseudotuberculosis | ||
| IP2790c | Serogroup O1, pYV− | 38 |
| IP2790.1c | IP2790c y0514::kan, Kmr | This work |
| IP2790c/GFP | IP2790c containing p67GFP3.1, Apr | This work |
| IP2790.1c/GFP | IP2790c/GFP y0514::kan, Apr, Kmr | This work |
| IP2515c | Serogroup O2, pYV− | 38 |
| IP2515c/GFP | IP2515c containing p67GFP3.1, Apr | This work |
| IP2666c | Serogroup O3, pYV− | 38 |
| IP2666c/GFP | IP2666c containing p67GFP3.1, Apr | This work |
| YPIII(p−) | Serogroup O3, pYV− | 5 |
| YPIII(p−)/GFP | YPIII(p−) containing p67GFP3.1, Apr | This work |
PCR and DNA sequencing.
The oligonucleotides 5′-GGGAAAAACTGTGCCATAAAAGCG-3′ (y0514.1.1) and 5′-TTTGTCCAACCTGTAGCGTGG-3′ (y0514.1.2) were purchased from Life Technologies and were used to amplify Y. pestis and Y. pseudotuberculosis DNA sequences by PCR using an Eppendorf Mastercycler. The reactions were performed with whole bacteria as a source of template in 100 μl of 1× PCR buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.3]) containing 0.5 U of Taq DNA polymerase (Roche), 0.25 μM concentrations of each primer, and 160 μM concentrations of deoxynucleoside triphosphate. The reactions were cycled under the following parameters: 1 cycle at 94°C for 2 min, 25 cycles to amplify the target DNA (30 s at 94°C, 30 s at 50°C, and 1 min kb−1 at 72°C), and 1 cycle at 72°C for 5 min. The PCR products were inserted into the EcoRV site of pETBlue-2 by using commercially available reagents (Perfectly Blunt cloning kit; Novagen). Plasmid DNA was isolated from NovaBlue transformants and sequenced using an ABI 3100 automated genetic analyzer and an ABI PRISM BigDye Terminator cycle sequencing kit (Applied Biosystems). The resulting sequences were used to perform BLASTN searches of the Y. pestis KIM (13) or CO92 (30) or Y. pseudotuberculosis IP32953 (http://bbrp.llnl.gov/bbrp/html/microbe.html) genomic databases.
Strain constructions.
Inducible expression of green fluorescent protein (GFP) in Y. pestis and Y. pseudotuberculosis was achieved as follows. The gfp3mut3.1 gene was isolated from pGFPmut3.1 (Clontech) and inserted between the HindIII and XmaI sites of pMMB67EH (17). The resulting plasmid, p67GFP3.1, in which gfp3mut3.1 expression is under the control of the tac promoter, was constructed using DH5α as a host. p67GFP3.1 was introduced into the Yersinia strains listed in Table 1 either by electroporation (10) or by conjugation as previously described (4). Bacteria containing p67GFP3.1 express GFP after 1 h of induction with 300 μM IPTG (isopropyl-β-d-thiogalactopyranoside). GFP expression is undetectable in the absence of IPTG induction.
The open reading frame (ORF) encoding a putative secretin protein of TTSS-2 (ORF y0514 in KIM) (13) was inactivated by allelic recombination in EV766 and IP2790c as follows. The ORF y0514 PCR product (a product amplified from the Y. pestis EV766 chromosome by using PCR and primers y0514.1.1 and y0514.1.2 as described above) was inserted into pETBlue-2 (see above), and the resulting plasmid was designated pETBlue-2.1. Plasmid pETBlue-2.1 was digested with EcoRV, which cuts once near the middle of ORF y0514. A kan gene, encoding resistance to kanamycin (Kmr), was isolated from pBSL86 (2) by digestion of this plasmid with PstI. The kan gene was treated with Klenow (Roche) to create blunt ends and inserted at the EcoRV site of pETBlue-2.1 to disrupt the y0514 ORF. The resulting plasmid, pETBlue-2.2, was digested with NotI and XbaI. The mutated ORF y0514 containing kan was isolated and inserted between the NotI and XbaI restriction sites in pSB890 (29), which encodes resistance to tetracycline (Tetr). The resulting plasmid (pSB890.1) was introduced by electroporation into S17-1λpir, and transformants were selected on LB plates containing tetracycline and kanamycin. pSB890.1 was conjugated from S17-1λpir into IP2790c or EV766/GFP. Transconjugants were selected on M63 minimal agar plates (26) or Yersinia selective agar plates (Oxoid) containing kanamycin. Kmr transconjugants were grown for several generations in the absence of kanamycin or tetracycline and then plated on LB agar plates lacking NaCl and supplemented with 5% sucrose to select against the sacB gene carried on pSB890. Sucrose-resistant colonies were tested for a Kmr and Tets phenotype. The replacement of the wild-type ORF y0514 sequence with the mutated sequence was verified by Southern blot analysis using a DIG DNA labeling and detection kit (Roche) and the ORF y0514 PCR product as a probe.
Bone marrow macrophage isolation and culture conditions.
Bone marrow-derived macrophages were isolated from C57BL/6 female mice (Taconic Laboratories) as previously described (8). Briefly, the bone marrow cells isolated from two femurs were suspended in culture medium (see below) and seeded into 100-mm-diameter petri dishes (Nunc) at 4 × 106 cells/plate. After 5 days of incubation at 37°C in an atmosphere of 5% CO2, the macrophages were collected and used for infection assays or were subcultured for up to two additional passages. All tissue culture reagents were purchased from Life Technologies unless otherwise specified. The culture medium was Dulbecco's modified Eagle medium with high glucose (4,500 μg/ml) supplemented with 20% heat-inactivated fetal bovine serum (HyClone), 30% L-cell-conditioned medium, 2 mM l-glutamine, and 1 mM sodium pyruvate. The medium used for infection assays (infection medium) was the same as described above except that the concentrations of fetal bovine serum and L-cell-conditioned medium were reduced to 10 and 15%, respectively.
Macrophage infections.
Twenty-four hours before infection, 2 × 105 macrophages were seeded into the wells of a 24-well tissue culture plate in 1 ml of infection medium. Bacteria were grown as described above and used to infect macrophages at a multiplicity of infection (MOI) of 5. After addition of bacteria, the plate was centrifuged for 4 min at 50 × g to facilitate bacterial contact with macrophages. The macrophages were washed twice with PBS after 20 min of incubation, and fresh infection medium containing 8 μg of gentamicin/ml was added to each well. This is referred to as the 0-h time point. After 1 h of incubation, the wells were washed twice with PBS, and fresh infection medium containing 2 μg (for Y. pestis infections) or 4.5 μg (for Y. pseudotuberculosis infections) of gentamicin/ml was added to each well. The infected macrophages were incubated for an additional 1 to 25 h.
CFU assay.
At various times postinfection, the infected macrophages were washed three times with PBS and lysed by incubation in 0.5 ml of 0.1% Triton X-100 in PBS. After incubation for 10 min at 37°C, the lysates were removed, the wells were rinsed with 0.5 ml of HI broth, and the lysates and HI rinses from each well were pooled into a 2-ml microcentrifuge tube. The samples were sonicated for three pulses of 1 s each using a Microson XL200 ultrasonic cell disruptor equipped with a microprobe P-1 (Misonix) to disperse the bacteria. Serial 10-fold dilutions of the samples were spread on HI or LB agar plates, the plates were incubated at 28°C for 2 or 3 days, and output CFU were enumerated.
Immunofluorescence microscopy.
Macrophages were seeded into tissue culture plates and infected with bacteria as described above except that the wells contained 12-mm-diameter glass coverslips that had been washed with acetone and heated to 180°C for 4 h to ensure that they were lipopolysaccharide free. At different times postinfection, IPTG was added (300 μM final concentration) to the incubation medium to induce GFP expression in intracellular bacteria. After 1 h of incubation, the cells were washed twice with PBS and fixed for 20 min in 2.5% paraformaldehyde at room temperature. All subsequent steps were done at room temperature. The fixed cells were incubated in 0.5% Triton X-100 in PBS for 1 min to permeabilize membranes and then in 3% bovine serum albumin in PBS for 10 min to reduce nonspecific antibody binding. Bacteria were immunolabelled by incubation in rabbit anti-Yersinia antiserum SB349 (3) diluted 1:1,000 in 3% bovine serum albumin for 30 min. After washing three times with PBS, the fixed cells were incubated with goat anti-rabbit secondary antibody conjugated to lissamine rhodamine under conditions recommended by the supplier (Molecular Probes). After washing, the coverslips containing fixed macrophages were inverted onto a drop of mounting medium (3) on a glass microscope slide. The slides were examined by epifluorescence microscopy using a Zeiss Axioplan2 microscope equipped with a 40× objective. Images were captured using a Spot camera (Diagnostic Instruments, Inc.) and processed using Adobe Photoshop 5.5.
RESULTS
Survival and replication of Y. pestis in macrophages.
We began our study by confirming that Y. pestis cured of pCD1 and pPCP1 can survive and replicate in primary murine macrophages (41). For this purpose, we isolated macrophages from C57BL/6 mice and infected them with KIM10+, a biovar Mediaevalis strain that lacks pCD1 and pPCP1 (Table 1). We experimented with different incubation media that have been used to selectively inhibit replication of extracellular Y. pestis (28, 41). We found that a standard tissue culture medium supplemented with 2 μg of gentamicin/ml (28) was effective at selectively inhibiting replication of extracellular Y. pestis in our assays (data not shown). This medium was therefore used in all subsequent experiments.
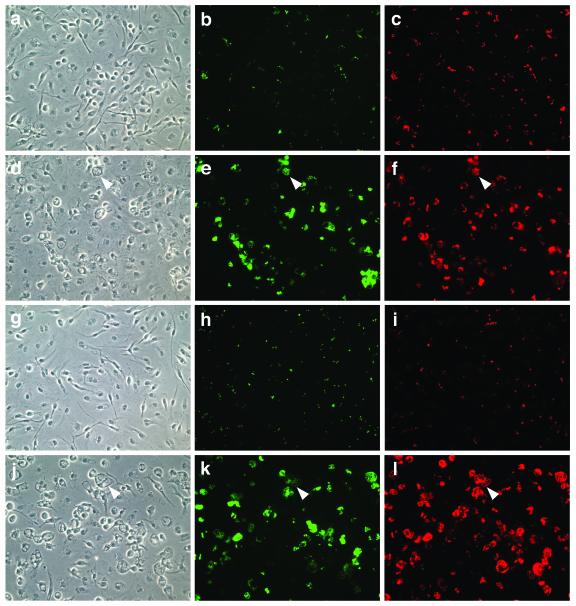
We initially used fluorescence microscopy to assay for survival and replication of Y. pestis in macrophages. A plasmid encoding GFP under the control of an IPTG-inducible promoter was introduced into the KIM10+ strain. The resulting strain, KIM10+/GFP, was grown at 26°C and used to infect C57BL/6 macrophages growing on coverslips at an MOI of 5. After a brief incubation to allow phagocytosis, gentamicin was added to the cultures at a low concentration to kill extracellular bacteria (Materials and Methods). The cultures were incubated for various times, and 1 h prior to fixation, IPTG was added to the cultures to induce GFP expression within intracellular bacteria. The fixed cells were incubated with anti-Yersinia antibody and a secondary antibody conjugated with lissamine rhodamine to label bacteria. The coverslips were examined by phase or epifluorescence microscopy using appropriate filters to detect viable bacteria and total bacteria. No increase in the number of intracellular KIM10+/GFP was observed during the first 4 h of infection (data not shown). However, most of the bacteria that could be detected at 4 h were GFP positive and therefore viable (Fig. 1a to c). An increase in the number of GFP-positive intracellular bacteria was observed at 8 h postinfection (data not shown), and by 25 h postinfection, many of the macrophages contained large numbers of GFP-positive bacteria (Fig. 1d to f). Similar results were obtained when the assay was carried out with the Y. pestis strain EV766/GFP (biovar Orientalis) (Table 1) (data not shown).
FIG. 1.
Analysis of Y. pestis and Y. pseudotuberculosis replication in macrophages by microscopy. C57BL/6 macrophages were infected with KIM10+/GFP (a to f) or IP2790c/GFP (g to l) at an MOI of 5. The infected cells were incubated for 4 h (a to c and g to i) or 25 h (d to f and j to l) and then fixed, and bacteria were labeled with a rabbit anti-Yersinia antibody (red). One hour before fixation, IPTG was added to the cells to induce GFP expression in viable bacteria (green). The samples were visualized by phase (left panels) or epifluorescence (middle and right panels) microscopy. Representative images were captured using a digital camera. Macrophages containing large numbers of intracellular bacteria displayed an enlarged and vacuolated morphology under phase-contrast microscopy (arrowheads).
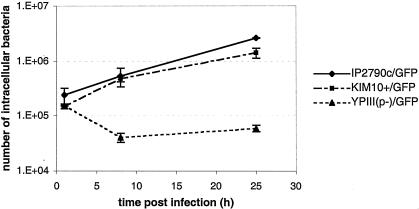
We next performed a CFU assay (Materials and Methods) to measure the number of viable bacteria that could be recovered from C57BL/6 macrophages at different times postinfection. Surprisingly, the number of CFU of KIM10+/GFP or EV766/GFP that could be recovered from the macrophages remained relatively constant from 1 to 25 h postinfection (data not shown). To determine whether our inability to detect an increase in CFU over time was due to bacterial aggregation, macrophages infected with KIM10+/GFP in the presence of IPTG were lysed and samples of the lysates were examined by epifluorescence microscopy. Large aggregates of GFP-positive bacteria were observed in the lysates, even after vigorous vortexing of the samples (data not shown). We found that these aggregates could be disrupted by briefly sonicating the lysates (42), and when sonicated lysates were used for the CFU assay, the number of viable intracellular bacteria increased with time (Fig. 2). Eight hours after infection, the number of intracellular KIM10+/GFP increased on average by threefold, and after 25 h, it increased by ninefold (Fig. 2). Therefore, our results show that Y. pestis cured of pCD1 and pPCP1 can survive and replicate in primary murine macrophages.
FIG. 2.
Kinetics of Y. pestis and Y. pseudotuberculosis replication in macrophages. C57BL/6 macrophages were infected with IP2790c/GFP (♦), KIM10+/GFP (▪), or YPIII(p−)/GFP (▴). Intracellular bacteria were enumerated at the indicated times postinfection by a CFU assay. The values represent averages ± standard deviations (error bars) of triplicate samples for IP2790c and KIM10+ and of duplicate samples for YPIII(p−).
Survival and replication of serogroup O1 Y. pseudotuberculosis in macrophages.
We next analyzed two plasmid-cured strains of Y. pseudotuberculosis for the ability to survive and replicate in macrophages. The strains tested were IP2790c, a serogroup O1 isolate, and YPIII(p−), a serogroup O3 isolate. YPIII, the parental strain of YPIII(p−), has been shown to be unable to replicate in J774A.1 macrophages (27). The GFP expression plasmid was introduced into IP2790c and YPIII(p−), and the resulting strains (Table 1) were used to infect C57BL/6 macrophages as described above. Interestingly, the microscopic assay revealed that IP2790c/GFP survived and replicated in macrophages as well as did KIM10+/GFP (Fig. 1g to l), while YPIII(p−) displayed a clear defect in intracellular growth (data not shown). The number of viable intracellular IP2790c/GFP increased on average by twofold 8 h after infection and by 12-fold 25 h after infection (Fig. 2). In contrast, the number of viable intracellular YPIII(p−)/GFP decreased by fourfold in the first 8 h of infection and then remained relatively constant between 8 and 25 h postinfection (Fig. 2). Thus, the YPIII strain is defective for intracellular survival (27), while the IP2790c strain shares with Y. pestis the ability to survive and replicate in macrophages.
The ability to replicate in macrophages is conserved between Y. pestis and Y. pseudotuberculosis.
To determine whether the ability to replicate in macrophages is specific to serogroup O1 strains of Y. pseudotuberculosis or is a more general property of the species, we assayed for this phenotype in additional strains of different serogroups. The strains tested were the serogroup O2 strain IP2515c/GFP and the serogroup O3 strain IP2666c/GFP (Table 1). YPIII(p−)/GFP was analyzed in parallel as a negative control. CFU assays performed on C57BL/6 macrophages infected with these strains showed that IP2515c/GFP and IP2666c/GFP were able to survive and replicate in macrophages (Table 2). In contrast, YPIII(p−)/GFP showed a decrease in viability over time (Table 2). We also assayed intracellular replication of IP2515c/GFP and IP2666c/GFP by fluorescence microscopy. The numbers of GFP-positive intracellular IP2515c/GFP and IP2666c/GFP increased over time as expected (data not shown). In summary, these data indicate that the ability to survive and replicate in macrophages is conserved between Y. pseudotuberculosis and Y. pestis. Our results also suggest that the YPIII strain may have a genetic difference from other Y. pseudotuberculosis strains that renders it defective for survival in macrophages.
TABLE 2.
Replication of different Y. pseudotuberculosis strains in macrophages
| Strain | No. (105) of viable bacteria recovered from macrophages at the indicated hour postinfectionb
|
Increase (fold)a | |
|---|---|---|---|
| 1 | 25 | ||
| IP2790c/GFP | 3.10 ± 0.6 | 16 ± 8 | 5.16 |
| IP2790.1c/GFP | 3.05 ± 0.55 | 14.5 ± 9.5 | 4.75 |
| IP2515c/GFP | 1.40 ± 0.62 | 7.05 ± 3.05 | 5.05 |
| IP2666c/GFP | 0.84 ± 0.08 | 12.2 ± 3.65 | 14.46 |
| YPIII(p−)/GFP | 1.39 ± 0.21 | 0.55 ± 0.13 | 0.42 |
The n-fold increase was calculated by dividing the average number at 25 h by the average number at 1 h.
The numbers represent the averages ± standard deviations from two independent experiments.
Analysis of the chromosomal TTSS-2 in Y. pestis and Y. pseudotuberculosis.
The observation that plasmid-cured strains of Y. pestis and Y. pseudotuberculosis can survive and replicate in macrophages suggests that the genes required for this phenotype are encoded on the chromosome and are common to both species. The TTSS-2 found in Y. pestis (13, 30) is similar in gene content and sequence to the TTSS present in Salmonella serovar Typhimurium SPI-2. Since the SPI-2 TTSS is required for intracellular replication of serovar Typhimurium (19), it seemed logical to investigate whether TTSS-2 is present in any strains of Y. pseudotuberculosis. To answer this question, we performed a BLASTN search of the incomplete Y. pseudotuberculosis IP32953 (serogroup O1) genome database (http://bbrp.llnl.gov/bbrp/html/microbe.html) by using as queries a number of TTSS-2 sequences obtained from the KIM genome (13). For example, as queries we used sequences that are predicted to encode a secretin (ORF y0514) and an AraC family transcription factor (ORF y0517) in TTSS-2. The results showed that IP32953 contains sequences that are homologous to TTSS-2. We next designed oligonucleotides complementary to sequences that encode the secretin (y0514) in TTSS-2. Using these oligonucleotides as primers in PCRs, we were able to amplify products of the predicted sizes from EV766, IP2790c, YPIII(p−), IP2515c, and IP2666c genomic DNA (data not shown). Therefore, it is likely that TTSS-2 is present in Y. pseudotuberculosis as well as Y. pestis.
The observation that TTSS-2 is present in Y. pestis and Y. pseudotuberculosis strains raised the possibility that this secretion system might be a common factor that is important for survival and replication of these bacteria in macrophages. To investigate this possibility, we inactivated TTSS-2 in Y. pseudotuberculosis IP2790c and Y. pestis EV766/GFP by insertion of a kan gene into the secretin ORF (y0514) (Materials and Methods). The GFP expression plasmid was then introduced into IP2790.1c. C57BL/6 macrophages were infected with IP2790.1c/GFP or EV766.1/GFP (Table 1), and intracellular replication was assayed as before by microscopy. The strains with kan insertions in ORF y0514 replicated in macrophages as well as did the parental controls (data not shown). A CFU assay performed with IP2790.1c/GFP showed that inactivation of ORF y0514 did not compromise bacterial replication in C57BL/6 macrophages (Table 2). Therefore, a functional TTSS-2 does not appear to be required for intracellular replication of Y. pestis or Y. pseudotuberculosis in macrophages.
DISCUSSION
Y. pestis and Y. pseudotuberculosis are closely related at the genetic level. It has been hypothesized that Y. pestis is a clone of Y. pseudotuberculosis serogroup O1b that evolved 1,500 to 20,000 years ago (1, 40). The goal of this work was to determine whether the ability of Y. pestis to replicate in macrophages was acquired from a Y. pseudotuberculosis ancestor or was acquired independently by Y. pestis during its rapid evolution.
Six Yersinia strains cured of the virulence plasmid were tested for the ability to survive and replicate in primary murine macrophages. We found that two Y. pestis strains, EV766 (biovar Orientalis) and KIM10+ (biovar Mediaevalis), could survive and replicate in bone marrow-derived macrophages, as expected (28, 41). Although we did not test a biovar Antiqua strain for this phenotype, it is likely that all biovars of Y. pestis are able to survive and replicate in macrophages. Four Y. pseudotuberculosis strains were tested for survival and replication in macrophages. The IP2790c (serogroup O1), IP2515c (serogroup O2), and IP2666c (serogroup O3) strains were phenotypically very similar to Y. pestis in terms of their ability to survive and replicate in macrophages. These findings confirm that Y. pestis and Y. pseudotuberculosis strains cured of the virulence plasmid can replicate in macrophages (41, 42). The serogroup O3 strain YPIII(p−), derived from YPIII, had a unique phenotype. The majority of internalized YPIII(p−) did not survive, which is consistent with the results obtained with the parental YPIII strain analyzed by the Finlay laboratory (27). We speculate that the YPIII strain used in our study and that of Mills and Finlay (27) acquired a genetic mutation during laboratory passage that decreases intracellular fitness. Experiments are under way in our laboratory to try to clarify this issue.
The chromosomal TTSS (TTSS-2) encoded by Y. pestis (13, 30) is homologous to the TTSS encoded by SPI-2 of serovar Typhimurium that plays a key role in intracellular replication (19). To determine whether TTSS-2 might be important for intracellular proliferation of Y. pestis and Y. pseudotuberculosis, we investigated whether TTSS-2 is encoded in the genome of different Y. pseudotuberculosis strains. BLASTN searches of the IP32953 baseline assembly (http://bbrp.llnl.gov/bbrp/html/y.pseudo.htm) with TTSS-2 sequences from Y. pestis KIM revealed that TTSS-2 is encoded by this serogroup O1 strain. Furthermore, we were able to amplify by PCR TTSS-2 sequences from all Y. pseudotuberculosis strains examined in this study. Therefore, it is likely that Y. pestis inherited TTSS-2 from its Y. pseudotuberculosis ancestor. We inactivated TTSS-2 in Y. pestis EV766 and Y. pseudotuberculosis IP2790c by the insertion of a kan resistance cassette into the ORF predicted to encode a secretin (ORF y0514). Surprisingly, the resulting secretin mutants replicated in macrophages as well as the parental strains did. Therefore, TTSS-2 does not appear to be required for replication of Y. pestis or Y. pseudotuberculosis in primary murine macrophages.
Oyston et al. (28) have shown that Y. pestis phoP mutants are defective for survival in macrophages. As in the case of serovar Typhimurium (14, 18), PhoP-regulated gene expression may allow Y. pestis to resist destruction in macrophages. It is likely that PhoP plays an important role in replication of Y. pseudotuberculosis in macrophages as well. The genes regulated by PhoP that contribute to intracellular replication of Y. pestis (or Y. pseudotuberculosis) have not been identified. Although Y. pestis and Y. pseudotuberculosis may share some similarities in survival strategies with serovar Typhimurium, there are likely to be important differences as well. A property of intracellular Y. pestis and Y. pseudotuberculosis that appears to be unique is the tendency of these bacteria to form tight aggregates. It is not clear what causes this aggregation phenotype, but it could be due to the expression of a surface structure that is required for survival in the phagosome. The pH 6 antigen is a fimbrial adhesin that is expressed by Y. pestis and Y. pseudotuberculosis when these bacteria are grown at 37°C and pH 6 (25). The pH 6 antigen is expressed by Y. pestis in macrophage phagosomes (25), but to our knowledge, it has never been reported that pH 6 antigen mediates tight aggregation of Y. pestis. Moreover, the pH 6 antigen does not appear to be required for intracellular survival of Y. pestis (25).
Several studies have reported that Y. pestis replicates in macrophages in vivo (7, 9, 15, 22). For example, Finegold (15) used transmission electron microscopy to study the course of pneumonic plague in an experimentally infected nonhuman primate and observed intact Y. pestis inside alveolar macrophages. The ability of Y. pestis to proliferate in macrophages is likely to be important in the early stages of plague pathogenesis for several reasons. This ability would allow the bacterium to avoid destruction upon entering the host, as the bacterium will exist at ambient temperatures prior to entry and will be efficiently phagocytosed by resident macrophages. This ability could also provide a replicative niche in which the bacterium can become conditioned to growth at 37°C, while protected from neutrophils that are recruited to the site of infection. Finally, this ability may provide the bacterium with a vehicle for transport from the initial site of infection to local lymph nodes. The fact that the 50% lethal dose of a Y. pestis phoP mutant is increased 75-fold when mice are challenged subcutaneously is consistent with the idea that intracellular replication is important at early stages of infection from peripheral routes (28). It would be interesting to determine whether phoP mutants are also attenuated for virulence in the aerosol or intravenous route of infection. It is also clear that extracellular growth of Y. pestis is essential for virulence during the systemic phase of the infection process. This is highlighted by the fact that the antibiotic streptomycin, which does not efficiently enter mammalian cells, is an effective treatment for plague.
Does intracellular replication play an equally important role in the pathogenesis of Y. pseudotuberculosis infections? It is possible that, like Y. pestis, Y. pseudotuberculosis replicates transiently in macrophages at early stages of the infection process. Fujimura et al. (16) studied the early stages of invasion of a Y. pseudotuberculosis serogroup O1 strain into rabbit Peyer's patches and observed intact bacteria within intraepithelial macrophages. Macrophages could serve as transient sites for bacterial multiplication in Peyer's patches or mesenteric lymph nodes and as vehicles for dissemination of Y. pseudotuberculosis to deeper tissues. An interesting fact that is often overlooked is that the name “pseudotuberculosis” derives from the observation that Y. pseudotuberculosis infections of mesenteric lymph nodes can resemble, at the histopathological level, infections caused by the intracellular pathogen Mycobacterium tuberculosis. Furthermore, Y. pseudotuberculosis lacking pYV can multiply by as much as 1 or 2 logs in the lymphatic organs of mice for a period of several days after infection (39). However, as in the case with Y. pestis, sustained replication of Y. pseudotuberculosis in lymphatic organs does require pYV, which promotes extracellular replication (39).
Given the role of pYV in promoting extracellular replication, it is important to consider how the presence of this plasmid would impact Yersinia replication in macrophages. Y. pseudotuberculosis strains that contain pYV and that are cultured at 37°C to upregulate expression of the TTSS can partially resist phagocytosis by macrophages (35). However, approximately 50% of the bacteria that come into contact with macrophages under these conditions are internalized (35). Therefore, the ability to avoid intracellular killing could provide a significant survival advantage to those wild-type bacteria that are phagocytosed by macrophages. In preliminary experiments, we infected C57BL/6 macrophages with isogenic pYV-negative and pYV-positive isolates of IP2790 grown at 26°C, conditions which downregulate expression of the TTSS. We found that under these conditions, the pYV-positive strain was able to replicate as well as the pYV-negative strain over a period of 24 h (data not shown). Therefore, the presence of the virulence plasmid does not appear to decrease the ability of Y. pseudotuberculosis to replicate in macrophages.
In summary, our data show that the ability to replicate in macrophages is common to Y. pestis and Y. pseudotuberculosis. It is therefore highly probable that Y. pestis inherited this property from its Y. pseudotuberculosis ancestor. The ability to replicate in macrophages is unlikely to be responsible for the increased virulence of Y. pestis. However, the fact that this phenotype has been conserved during evolution suggests that the importance of intracellular replication to the pathogenesis of Y. pestis and Y. pseudotuberculosis may be underestimated.
Acknowledgments
We thank Michel Simonet and Stanley Falkow for providing the Yersinia strains used in this study. We also thank members of the Bliska laboratory for helpful comments on the manuscript.
This work was supported by Public Health Research grant RO1 AI48507 (J.B.B.).
Editor: B. B. Finlay
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52, 54, 56. [PubMed]
- 3.Black, D. S., and J. B. Bliska. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 37:515-527. [DOI] [PubMed] [Google Scholar]
- 4.Bliska, J. B., and D. S. Black. 1995. Inhibition of the Fc receptor-mediated oxidative burst in macrophages by the Yersinia pseudotuberculosis tyrosine phosphatase. Infect. Immun. 63:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolin, I., L. Norlander, and H. Wolf-Watz. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of fleaborne plague. J. Immunol. 85:348-363. [PubMed] [Google Scholar]
- 8.Celada, A., P. W. Gray, E. Rinderknecht, and R. D. Schreiber. 1984. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 160:55-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charnetzky, W. T., and W. W. Shuford. 1985. Survival and growth of Yersinia pestis within macrophages and an effect of the loss of the 47-megadalton plasmid on growth in macrophages. Infect. Immun. 47:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conchas, R. F., and E. Carniel. 1990. Highly efficient electroporation of Yersinia. Gene 87:133-137. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R. 1998. The Yersinia deadly kiss. J. Bacteriol. 180:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stanier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J. Infect. Dis. 179(Suppl. 2):S326-S330. [DOI] [PubMed] [Google Scholar]
- 15.Finegold, M. J. 1969. Pneumonic plague in monkeys: an electron microscopic study. Am. J. Pathol. 54:167-185. [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura, Y., T. Kihara, and H. Mine. 1992. Membranous cells as a portal of Yersinia pseudotuberculosis entry into rabbit ileum. J. Clin. Electron Microsc. 25:35-45. [Google Scholar]
- 17.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 18.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 21.Hu, P., J. Elliott, P. McCready, E. Skowronski, J. Garnes, A. Kobayashi, R. R. Brubaker, and E. Garcia. 1998. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen, W. A., and M. J. Surgalla. 1968. Plague bacillus: survival within host phagocytes. Science 163:950-952. [DOI] [PubMed] [Google Scholar]
- 23.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Transcomplementation-dependent replication of a low molecular weight origin from plasmid RK6. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 24.Lindler, L. E., G. V. Plano, V. Burland, G. F. Mayhew, and F. R. Blattner. 1998. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 66:5731-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindler, L. E., and B. Tall. 1993. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol. Microbiol. 8:311-324. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Mills, S. D., and B. B. Finlay. 1998. Isolation and characterization of Salmonella typhimurium and Yersinia pseudotuberculosis-containing phagosomes from infected mouse macrophages: Y. pseudotuberculosis traffics to terminal lysosomes where they are degraded. Eur. J. Cell Biol. 77:35-47. [DOI] [PubMed] [Google Scholar]
- 28.Oyston, P. C. F., N. Dorrell, K. Williams, S.-R. Li, M. Green, R. W. Titball, and B. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, L. E., S. Hobbie, J. E. Galan, and J. B. Bliska. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNFα production and the downregulation of the MAP kinases p38 and JNK. Mol. Microbiol. 27:953-965. [DOI] [PubMed] [Google Scholar]
- 30.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 31.Perry, R. D., and J. D. Fetherstone. 1997. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry, R. D., S. C. Straley, J. D. Fetherston, D. J. Rose, J. Gregor, and F. R. Blattner. 1998. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 66:4611-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portnoy, D. A., and S. Falkow. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosqvist, R., I. Bolin, and H. Wolf-Watz. 1988. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect. Immun. 56:2139-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 38.Simonet, M., and S. Falkow. 1992. Invasin expression in Yersinia pseudotuberculosis. Infect. Immun. 60:4414-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonet, M., S. Richard, and P. Berche. 1990. Electron microscopic evidence for in vivo extracellular localization of Yersinia pseudotuberculosis harboring the pYV plasmid. Infect. Immun. 58:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37:316-330. [DOI] [PubMed] [Google Scholar]
- 41.Straley, S. C., and P. A. Harmon. 1984. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect. Immun. 45:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsukano, H., F. Kura, S. Inoue, S. Sato, H. Izumiya, T. Yasuda, and H. Watanabe. 1999. Yersinia pseudotuberculosis blocks the phagosomal acidification of B10.A mouse macrophages through the inhibition of vacuolar H+-ATPase activity. Microb. Pathog. 27:253-263. [DOI] [PubMed] [Google Scholar]