Abstract
Type III secretion systems of enteric bacteria enable translocation of effector proteins into host cells. Secreted proteins of verotoxigenic Escherichia coli O157 strains include components of a translocation apparatus, EspA, -B, and -D, as well as “effectors” such as the translocated intimin receptor (Tir) and the mitochondrion-associated protein (Map). This research has investigated the regulation of LEE4 translocon proteins, in particular EspA. EspA filaments could not be detected on the bacterial cell surface when E. coli O157:H7 was cultured in M9 minimal medium but were expressed from only a proportion of the bacterial population when cultured in minimal essential medium modified with 25 mM HEPES. The highest proportions of EspA-filamented bacteria were detected in late exponential phase, after which filaments were lost rapidly from the bacterial cell surface. Our previous research had shown that human and bovine E. coli O157:H7 strains exhibit marked differences in EspD secretion levels. Here it is demonstrated that the proportion of the bacterial population expressing EspA filaments was associated with the level of EspD secretion. The ability of individual bacteria to express EspA filaments was not controlled at the level of LEE1-4 operon transcription, as demonstrated by using both β-galactosidase and green fluorescent protein (GFP) promoter fusions. All bacteria, whether expressing EspA filaments or not, showed equivalent levels of GFP expression when LEE1-4 translational fusions were used. Despite this, the LEE4-espADB mRNA was more abundant from populations with a high proportion of nonsecreting bacteria (low secretors) than from populations with a high proportion of secreting and therefore filamented bacteria (high secretors). This research demonstrates that while specific environmental conditions are required to induce LEE1-4 expression, a further checkpoint exists before EspA filaments are produced on the bacterial surface and secretion of effector proteins occurs. This checkpoint in E. coli O157:H7 translocon expression is controlled by a posttranscriptional mechanism acting on LEE4-espADB mRNA. The heterogeneity in EspA filamentation could arise from phase-variable expression of regulators that control this posttranscriptional mechanism.
A number of gram-negative enteric pathogens export effector proteins into host cells via a type III secretion system (TTSS). These include Salmonella spp., Yersinia spp., Shigella spp., and enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC, respectively) (10, 18, 23, 26). The effectors have numerous functions, including inhibition of phagocytosis (8, 20), invasion (19), cytotoxicity (17, 27, 35), and bacterial attachment (7, 33), mostly through effects on signal transduction (3, 17, 24, 31, 34, 48, 55). In general, these TTSSs comprise a basal apparatus, with proteins present in the inner and outer membranes, and a needle complex that allows injection of effector proteins through the host cell membrane (10, 26, 47, 53, 61). In EPEC and EHEC, EscF is considered to form a short needle that is elongated by a filament made up of EspA (61). The EspA filaments allow formation of a pore in the host cell membrane involving the secreted proteins EspD and EspB (36, 53, 54). The EspA filaments have been shown to be produced transiently during lesion formation, and their loss from the surface potentially allows the subsequent intimate interaction of the bacterial surface factor intimin and the translocated intimin receptor (Tir) (36).
EHEC O157 is considered to have originated from an EPEC O55 strain that acquired Shiga-like toxin-carrying phage (50). Both EHEC and EPEC strains carry the locus of enterocyte effacement (LEE), which encodes a TTSS (15, 38, 39). At least seven open reading frames are present, and the first three encode proteins required for regulation and assembly of the basal apparatus, LEE1-3 (6, 57). The next operon (LEE4) encodes the secreted proteins EspA, EspB, and EspD (14, 36, 46, 58, 59), as well as factors whose function is not clear, including SepL (37) and EspF (11, 41, 42). EPEC O127 (E2348/69) and EHEC O157:H7 (EDL933) differ in that the cloned EPEC LEE, but not the cloned EHEC O157 LEE, confers the ability to form attaching-and-effacing lesions on E. coli K-12 (16, 39). This difference may reflect the biology of the organisms, with EHEC O157 strains possibly behaving as commensals in ruminant hosts, whereas EPEC strains are overt animal and human pathogens. Among EHEC O157 strains (Shiga-like toxin and intimin positive), there is marked variation in the ability to secrete Tir and EspD into tissue culture medium (40). There was a correlation between high-secretor status, extent of attaching-and-effacing lesions, and strains of human disease origin. Cattle isolates were more likely to be low-level EspD secretors (40).
The aim of this study was to investigate the expression and secretion of the translocon proteins EspA and EspD to understand the heterogeneity in expression levels between isolates. Immunofluorescence microscopy was used to demonstrate that EspA filament expression was heterogeneous in all of the EHEC O157 strains tested. The proportion of the bacterial population expressing EspA filaments correlated with secreted EspD levels. High EspD secretors had a higher proportion of bacteria expressing EspA filaments than did low secretors. The molecular basis of this expression pattern lay not with the transcription of LEE4 or LEE1-3 but with a mechanism controlling the translation of the LEE4-espADB mRNA transcript.
MATERIALS AND METHODS
Minimal media.
Two defined media were used in this study, M9 minimal medium and minimal essential medium modified with 25 mM HEPES (MEM-HEPES). Glucose was added to the MEM-HEPES to give a final concentration of 0.2%. M9 minimal medium was prepared with a final glucose concentration of 0.2% (52). Antibiotics were included, when required, at the following concentrations: chloramphenicol (CAM), 12.5 μg/ml; kanamycin (KAN), 25 μg/ml.
Protein analysis.
Bacteria were cultured in MEM-HEPES to an optical density at 600 nm (OD600) of 0.8. Secreted proteins were extracted by trichloroacetic acid precipitation as described previously (40). For protein localization experiments, whole-cell fractions were prepared by centrifugation (20 min, 4,000 × g), two washes in 20 ml of phosphate-buffered saline (PBS), and suspension in 400 μl of protein A buffer (10 mM NaCl, 50 mM Tris-HCl [pH 7.6], 1 mM EDTA, 0.1 mM dithiothreitol). Membrane fractions were prepared by the method of Kabach (28), with final suspension in 400 μl of protein A buffer. Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Colloidal blue (Invitrogen, Groningen, The Netherlands). Western blotting for Esps was done with a polyclonal antibody for EspA (60) and a monoclonal antibody against EspD (40). Bacteria were stained for EspA filaments following fixation with 4% paraformaldehyde for 5 min. Aliquots of fixed bacteria were air dried onto slides and incubated with a 1/100 dilution of the EspA antibody (in PBS-0.1% bovine serum albumin [BSA])for 60 min at room temperature. After three washes with PBS-0.1% BSA, the samples were incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin antibody (1/500; Dako) for 30 min, and the slide was washed three times with PBS-0.1% BSA. The slide was then examined by fluorescence microscopy with appropriate filter sets, and the images were captured with Leica software.
Plasmid-based promoter-GFP fusion construction.
In order to allow single-cell levels of promoter activity to be determined, the enhanced green fluorescent protein (GFP) gene (egfp), but without a promoter, was cloned into pACYC184. Primers GFP 5′ and GFP 3′ (Table 1) were used to amplify the gfp gene from peGFP (extended-half-life GFP; Clontech). These primers incorporate BamHI (GFP 5′) and BglII (GFP 3′) sites at their termini, allowing cloning into the BamHI site of pACYC184. All of the restriction enzymes used were from New England Biolabs. The resultant plasmid (pAJR70) has single BamHI and KpnI sites 5′ of the gfp gene, allowing promoters of interest to be cloned in frame with the reporter gene, creating translational fusions. Promoters for LEE1-4 were amplified from ZAP193 and cloned into pAJR70 to create pAJR71-75 (Table 2). These fusions provide a readout that is a combination of both transcription and translation initiation activities. Any transcriptional regulation acting on the promoter can be detected by using these fusions. As they also contain the wild-type ribosome binding site, they provide a more realistic readout of gene expression than a standard transcriptional fusion that uses a nonnative ribosome binding site. These fusions, while described as translational, are unlikely to provide a readout of other posttranscriptional control mechanisms, as the mRNA structure of the fusions will differ completely from that of the wild-type transcript.
TABLE 1.
Oligonucleotide primers used in this study
| Primers | Sequencea |
|---|---|
| lacI 5′ | gggggagctcCGTTATTTCTTGATGTCTCTGAC |
| lacI 3′ | ggggatccGCCTGGGGTGCCTAATGAGTGAG |
| lacA 5′ | ccggatccAATGACCGAAAGAATAAGAG |
| lacA 3′ | aaaactgcagATGTCTTTTGTGACGATACT |
| lacZ 5′ | ccggatccggtaccATGACCATGATTACGGATTCAC |
| lacZ 3′ | ccagatctCCTTACGCGAAATACGGGC |
| pSepL 5′ | cgggatccGATTGAGGCCTTGTTCAAGG |
| pSepL 3′ | cgggtaccTTCAATACCATTAGCCATTGG |
| lacI up 5′ | CGCGGTATGGCATGATAGCG |
| lacA down 3′ | CATGCCGGATGCGGCTAATG |
| lac mid 5′ | GTGACGTCTGCTTGCTGCAT |
| lac mid 3′ | CAGCAGGATATCCTGCACC |
| espA short 5′ | ccggatccCGATTGTCGAAGATAAAC |
| espA short 3′ | cggtgaccTGCATTTGATGTATCCAT |
| pgi probe 5′ | ACATCAATCCAACGCAGACC |
| pgi probe 3′ | TTCATCTTCTCCAGCACCGC |
| espA probe 5′ | ACATCCGTTGTTAATGTGAGTGCG |
| espA probe 3′ | GCAATTTTGGCATCCACAAG |
| sepL probe 5′ | ATCAAAACCCCGCATCTGTT |
| sepL probe 3′ | TTTCCTTGCGCTACCTTTGC |
| espD probe 5′ | GCTTAACGTAAATAACGATACCCTG |
| espD probe 3′ | CTCACCACTAATACCAAACAAT |
| escC probe 5′ | ATTCGCTAGATGCAGATTTTATCGG |
| escC probe 3′ | GTTACTTGATATTCAGGATGGC |
| lee1 5′ | ccggatccCTGTAACTCGAATTAAGTAGAG |
| lee1 3′ | ccggtaccGTATGGACTTGTTGTATGTGAATT |
| lee2 5′ | cgggatccGCGAACGCGCTCAATAATCTG |
| lee2 3′ | cgggtaccTGCTGCTTCCATTGATCTTTC |
| lee3 5′ | cgggatccCTGCTCGTCTCCGAGCATGCC |
| lee3 3′ | cgggtaccAACTAAAAGATTCATCTGCAGGCT |
| GFP 5′ | ccggatccggggtaccATGGTGAGCAAGGGCGAGGAGC |
| GFP 3′ | cgggtaccAACTAAAAGATTCATCTGCAGGCT |
The lowercase portions of some sequences are nonmatching sequence incorporating a restriction enzyme site.
TABLE 2.
Plasmids used in the study
| Plasmid(s) | Description |
|---|---|
| pIB307 | pMAK705-based vector for allelic exchange; temperature-sensitive replicon (4) |
| pDG005 | Low-copy-number vector containing MG1655 lacZYA region |
| pDG028 | Low-copy-number vector containing sac-kan cassette (21) |
| pACYC184 | Low-copy-number cloning vector |
| pAJR26 | lacIA flanking regions from ZAP193 cloned into pIB307 |
| pAJR27 | lacIA flanking regions from ZAP1 cloned into pIB307 |
| pAJR33 | pAJR26 with sac-kan cassette from pDG28 cloned between lacIA fragments |
| pAJR34 | pAJR27 with sac-kan cassette from pDG28 cloned between lacIA fragments |
| pAJR39 | pAJR26 with promoterless lacZ gene cloned between lacIA fragments |
| pAJR43 to 48 | pAJR39 with sepL promoter region from described ZAP strain cloned in frame of lacZ (pAJR43, ZAP1; pAJR44, ZAP21; pAJR45, ZAP41; pAJR46, ZAP46; pAJR47, ZAP58; pAJR48, ZAP193) |
| pAJR55 to 60 | pAJR39 with espA promoter region from described ZAP strain cloned in frame of lacZ (pAJR55, ZAP1; pAJR56, ZAP21; pAJR57, ZAP41; pAJR58, ZAP46; pAJR59, ZAP58; pAJR60, ZAP193) |
| peGFP | Clontech commercial vector |
| pAJR70 | pACYC184 cut with BamHI, gfp gene cloned BamHI/BglII |
| pAJR71 | pAJR70 cut with BamHI/KpnI; LEE1 promoter cloned in frame 5′ to gfp |
| pAJR72 | pAJR70 cut with BamHI/KpnI; LEE2 promoter cloned in frame 5′ to gfp |
| pAJR73 | pAJR70 cut with BamHI/KpnI; LEE3 promoter cloned in frame 5′ to gfp |
| pAJR74 | pAJR70 cut with BamHI/KpnI; LEE4 promoter cloned in frame 5′ to gfp |
| pAJR100 | pACYC184 cut with BamHI; lacZ gene cloned BamHI/Bgl/II |
| pAJR101 | pAJR100 cut with BamHI/KpnI; LEE1 promoter cloned in frame 5′ to lacZ |
| pAJR102 | pAJR100 cut with BamHI/KpnI; LEE2 promoter cloned in frame 5′ to lacZ |
| pAJR103 | pAJR100 cut with BamHI/KpnI; LEE3 promoter cloned in frame 5′ to lacZ |
| pJTPr-2 | pCB267 with LEE4 (sepL) promoter cloned 5′ to lacZ |
Quantification of GFP fusion levels.
ZAP193 was transformed with pAJR70-74 (Table 2) under standard conditions. The resultant transformants were grown overnight in M9 minimal medium or MEM-HEPES containing CAM and then diluted 1:50 into fresh, MEM-HEPES-CAM or M9 minimal medium-CAM on the next morning. Typically, 15 ml was cultured in Erlenmeyer flasks shaken at 200 rpm, 37°C. The optical density of the cultures was monitored by determination of the OD600. At an OD600 of 0.8, a 20-μl aliquot was removed and smeared onto a glass slide. This was heat fixed (65°C, 15 min), passed briefly through a Bunsen flame, and then chemically fixed by flooding of the slide with 4% paraformaldehyde. After 20 min, the slide was washed three times with PBS and a coverslip was applied with DAKO fluorescent mounting medium. Images were acquired on a Zeiss LSM 510 confocal system with single images of a projected focus stack consisting of four optical sections with a depth of 0.7 μm. The total GFP produced by the population was determined by analyzing 100-μl aliquots of culture with a fluorescent plate reader (Fluorostar Optima; BMG). Promoterless plasmid pAJR70 in ZAP193 acted as a control for background fluorescence, which, at the appropriate OD, was subtracted from the other values.
Creation of plasmids suitable for allelic exchange.
In order to create plasmids that would facilitate chromosomal exchange, flanking regions of the lac operon were PCR amplified and cloned into a temperature-sensitive plasmid (pIB307; Table 2). Primer pairs lacI 5′-lacI 3′ and lacA 5′-lacA 3′ were used to amplify strain-specific lac sequences from E. coli O157:H7, ZAP193, and ZAP1. These products were cleaned with a Qiagen PCR purification kit, digested with BamHI, recleaned, and then ligated under standard conditions. One microliter of this ligation reaction mixture was then used as a template for PCR with primers lacI 5′ and lacA 3′ to produce 1.4-kb lacIA fragments specific for each strain. These fragments were digested with SacI and PstI and then cloned into the same sites in pIB307 (Table 2) to produce pAJR26 (ZAP193) and pAJR27 (ZAP1). To produce plasmids that would allow the chromosomal lacIA intergenic region to be replaced with a selectable marker and a counterselection gene, a sac-kan cassette was cloned into the BamHI sites of pAJR26 and pAJR27. The sac-kan cassette from pDG28 (Table 2) was excised with BamHI, cleaned with the Qiagen PCR cleanup kit, and ligated into pAJR26 and pAJR27 under standard conditions. The resultant plasmids (pAJR33 and pAJR34) therefore had strain-specific lacIA regions flanking a sac-kan cassette.
Creation of lac deletion strains.
The method of Hamilton et al. (25) was used for the allelic exchange of pAJR33 and pAJR34 into ZAP1 and ZAP193, respectively. Plasmid pAJR33 was also used for allelic exchange of the lacZY genes of ZAP41. Briefly, the strain to be manipulated was electroporated with the plasmid containing the appropriate flanking regions and plated at 30°C on Luria-Bertani broth (LB)-CAM plates. Ten transformants were then inoculated into prewarmed LB-KAN at 42°C. After 48 h of growth in log phase at 42°C, a large number of cointegrates were obtained, as determined by PCR with primers lacI′ up and lacA′ down (Table 1). The 42°C culture was then used to inoculate LB containing KAN (12.5 μg/ml) at 30°C, and log-phase growth was carried out for 48 h at 30°C. Serial dilutions were then plated onto LB-KAN (S-Gal) plates (Sigma-Aldrich, Dorset, United Kingdom) at 30°C. S-Gal plates allowed lac mutant colonies to be easily identified. Colonies were then replica plated on LB-KAN, LB-CAM, and LB plates at 30°C to determine if gene replacements had been obtained. Colonies that appeared to have undergone allelic exchange were analyzed by PCR with primer pairs lac mid 5′-lac mid 3′ and lacI up 5′-lacA down 3′ and plated on MacConkey agar indicator plates (Oxoid, Basingstoke, United Kingdom) to confirm their lactose fermentation status. Loss of the original plasmids was confirmed by PCR with pIB307-specific primers. ZAP1 with the lac region deleted but containing the sac kan cassette was termed ZAP1001, the ZAP41 strain with lac deleted was termed ZAP1041, and the ZAP193 strain with lac deleted was termed ZAP1193 (Table 3).
TABLE 3.
Strains used in the study
| Strain | Details | Source, reference |
|---|---|---|
| ZAP1 | O157:H7 Redhouse Dairy human-outbreak strain phage type 2 | Our stocks, 51 |
| ZAP21 | Bovine diarrhea strain STEC 413/89-1 | Our stocks, 13 |
| ZAP41 | O157:H7 bovine strain | Our stocks, 40 |
| ZAP46 | O157:H7 bovine strain | Our stocks, 40 |
| ZAP58 | O157:H7 human disease outbreak strain | Our stocks, 40 |
| ZAP193 | NCTC 12900; O157 stx Nalr | Our stocks |
| ZAP198 | O157 stx Nalr | Our stocks, 49 |
| ZAP1001 | ZAP1 sac-kan ΔlacZY | This study |
| ZAP1041 | ZAP41 sac-kan ΔlacZY | This study |
| ZAP1193 | ZAP193 sac-kan ΔlacZY | This study |
| ZAP1193-sep1-5 | ZAP1193 with sepL promoter regions from ZAP strains cloned in frame with lacZ and exchanged in place of the sac-kan cassette (sep-1, ZAP1; sep-2, ZAP21; sep-3, ZAP41; sep-4, ZAP46; sep-5, ZAP58) | This study |
| ZAP1193-espA1-6 | ZAP1193 with espA promoter regions from ZAP strains cloned in frame with lacZ and exchanged in place of the sac-kan cassette (espA1, ZAP1; espA2, ZAP21; espA3, ZAP41; espA4, ZAP46; espA5, ZAP58, espA6, ZAP193) | This study |
Construction of plasmids to facilitate single-copy promoter fusions.
To allow the activities of different promoters to be compared on the chromosome, plasmids suitable for allelic exchange containing a promoterless lacZ gene were constructed. Primers lacZ 5′ and lacZ 3′ (Table 1) were used to amplify the lacZ gene from pDG005 (Table 2). These primers incorporate BamHI sites at their 5′ ends and BglII sites at their 3′ termini, allowing cloning into pAJR26. The resultant plasmid (pAJR39) contains single BamHI and KpnI sites 5′ of the lacZ gene, allowing promoters of interest with their initial coding sequence to be cloned in frame with the reporter gene. In addition, the promoterless lacZ gene was cloned into the BamHI site of pACYC184 with the same primers for construction of low-copy-number plasmid-based promoter fusions.
Creation of single-copy reporter strains in E. coli O157.
The reporter plasmids derived from pAJR39 and pAJR40 were used to create single-copy chromosomal fusions in place of the sac-kan cassette of ZAP1193. Putative promoter regions for sepL and espA were amplified for five EHEC strains with appropriate primers (Table 1). The promoters were cloned into an allelic-exchange vector (pAJR39) to create a series of plasmids containing promoter-lacZ fusions as detailed in Table 2 (pAJR43 to -48 and pAJR55 to -60). Individual plasmids containing the sepL::lacZ or espA::lacZ fusion from the different EHEC strains were then exchanged into the chromosome of ZAP1193 in place of the sac-kan cassette. To achieve this, the plasmids were electroporated into the target strain and transformants were selected on LB-CAM plates at 30°C. Ten transformants were then inoculated into LB-CAM broth at 42°C and subcultured four times over a 48-h period. Serial dilutions of these cultures were plated on LB-CAM plates at 42°C, and individual colonies were screened for primary integrates with appropriate primer pairs. Primary integrates were then inoculated into LB at 30°C for 48 h of logarithmic growth, and serial dilutions were plated onto LB-sucrose (6%, wt/vol; no NaCl) agar (4). Individual colonies were replica plated on LB, LB-CAM, and LB-KAN at 30°C to identify those that had undergone successful allelic exchange. Selected promoters were also cloned into the pACYC184-based lacZ fusion vector (Table 2).
Measurement of β-galactosidase activity.
Assays for β-galactosidase were performed as described in reference 44, with the following modifications. One hundred microliters of bacterial culture was removed and permeabilized by addition of 900 μl of Z buffer (44), 75 μl of CHCl3, and 37 μl of 0.1% SDS and vortexing for 10 s. The assay tubes were then incubated at 25°C for 5 min before addition of 200 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg ml−1). The reaction was stopped by addition of 500 ml of 1 M NaCO3 to the reaction mixture. To remove cell debris, the tubes were centrifuged for 4 min (10,000 × g; Beckman). The resulting supernatant was removed, the absorbances at 420 and 550 nm were determined, and Miller units were calculated as previously described (43).
Northern analysis.
Bacterial cultures (250 ml) were grown in MEM-HEPES to an OD600 of 0.8. Total RNA was extracted with a Qiagen RNeasy midi-prep kit or Ambion Totally RNA kits in accordance with the manufacturer's instructions. RNA was quantified both spectrophotometrically (Cecil CE2021; Aurius) and by running 2-μl aliquots of total extracted RNA on nondenaturing Tris-borate-EDTA gels with denaturing buffer (New England Biolabs). Electrophoresis of RNA on 1.0% (wt/vol) agarose formaldehyde-morpholinepropanesulfonic acid gels was performed as previously described (52). After electrophoresis, the RNA was blotted onto Hybond-N nylon membranes (Amersham-Pharmacia Biotech) by downward transfer (9) and UV cross-linked. RNA dot blot assays were performed by hybridization of a range of RNA concentrations (10 to 0.1 ng) directly onto a Hybond-N nylon membrane and UV cross-linking. Prehybridization and hybridization for all blots were performed at 42°C with Ultrahyb buffer (Ambion Inc.). Samples were probed with single-stranded DNA probes specific for either espA, sepL, espD, or escC (Table 1) or pgi (phosphoglucose isomerase) as a control (Table 1). Probes were labeled with [32P]dCTP and Ready-To-Go DNA labeling beads (Amersham-Pharmacia Biotech). The 32P signal was recorded on BioMax MR film (Kodak, Rochester, N.Y.) and also detected with a PhosphorImager (Bio-Rad GS-525; Bio-Rad), which allowed further quantification with Multi-analyst software (Bio-Rad).
RESULTS
Medium-dependent secretion of EspA and -D is associated with LEE1-4 expression levels.
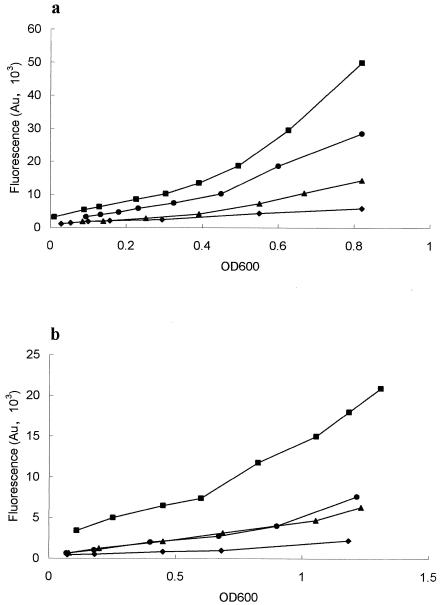
It is known that MEM-HEPES is permissive for type III secretion of EspA, -B, and -D by EPEC and EHEC (12, 14, 32, 40). By contrast, no secretion of Esps was detected when six different strains of EHEC O157 were cultured in M9 minimal medium (data not shown). The basis for this was investigated with LEE1, LEE2, LEE3, and LEE4 translational fusions to egfp (pAJR71-74; Table 2) transformed into Shiga-like-toxin-negative strain E. coli O157:H7 NCTC 12900 (ZAP193, Table 3). Expression from these promoters was measured in MEM-HEPES and M9 minimal medium (Fig. 1). Expression from all four operons was increased in MEM-HEPES and was associated with the formation of EspA filaments (Fig. 2a to d) and secretion of EspD and EspA (data not shown) in MEM but not in M9 minimal medium. For example, LEE1 expression was approximately fourfold higher in MEM-HEPES (28,500 U) than in M9 minimal medium (7,500 U) at the same optical density (OD600 = 0.6). The difference was more pronounced for LEE3 expression, which was approximately 10-fold higher in MEM-HEPES (9,500 U) than in M9 minimal medium (940 U) at the same optical density (OD600 = 0.6).
FIG. 1.
Expression of LEE1-4 operons in MEM-HEPES (a) and M9 minimal medium (b). LEE1-4 promoter-egfp fusions were constructed as described in Materials and Methods and transformed into ZAP193 (Table 3). Fluorescence was assayed throughout the growth curve as described in Materials and Methods and expressed as relative fluorescence units (Au). Symbols: ▪, LEE1; •, LEE2; ▴, LEE3; ♦, LEE4.
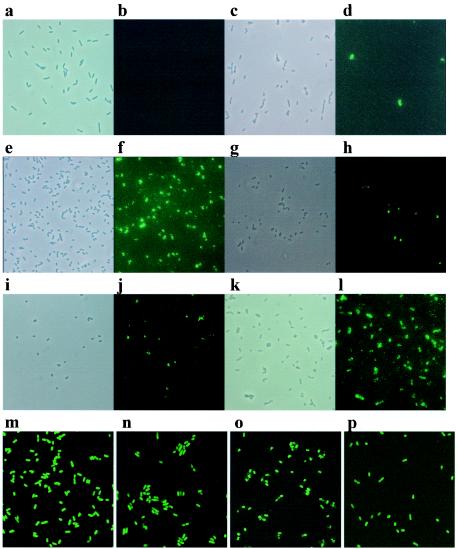
FIG. 2.
Micrographs showing EspA and LEE1-4 expression from E. coli O157:H7. Bacteria were visualized by phase-contrast microscopy (a, c, e, g, i, and k). EspA filaments were visualized by indirect immunofluorescence (b, d, f, h, and j) as described in Materials and Methods. (a and b) ZAP193 cultured in M9 minimal medium. (c and d) ZAP193 cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES to an OD600 of 0.4. (e and f) ZAP193 cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES to an OD600 of 0.8. (g and h) ZAP41 cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES to an OD600 of 0.8. (i and j) ZAP58 cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES to an OD600 of 0.8. (k and l) ZAP193 cultured in MEM-HEPES overnight. Images m to p are of enhanced GFP fluorescence captured by confocal microscopy as described in Materials and Methods. All of the bacteria visualized were fluorescent; therefore, phase images are not shown. m, LEE1::egfp in ZAP193; n, LEE2::egfp in ZAP193; o, LEE3::egfp in ZAP193; p, LEE4::egfp in ZAP193.
EspA filaments are expressed by a subpopulation of bacteria.
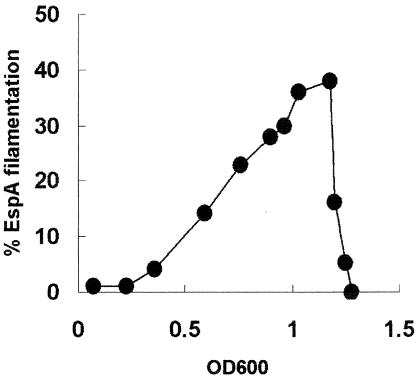
Initial characterization of nine wild-type E. coli O157 strains cultured in MEM-HEPES revealed that EspA filaments were detectable only on a proportion of the bacteria. The proportion depended on the strain and exact culture conditions (Fig. 2 and 3) but varied from 1 to 95%. In order to study this variation, it was necessary to have a consistent EspA phenotype to start any analysis. This was achieved by culturing strains overnight in M9 minimal medium, which resulted in lower levels of LEE1-4 expression (Fig. 1b) and no detectable EspA filamentation (Fig. 2a and b). Strains were then subcultured (to an OD600 of 0.05) into MEM-HEPES, which induced LEE1-4 operon expression (Fig. 1a) and stimulated EspA filament expression and protein secretion (Fig. 2c to f). As shown in Fig. 2c to f for ZAP193, only a proportion of the bacterial population expressed EspA filaments and this proportion increased throughout the exponential phase of growth but then dropped off rapidly as the bacteria entered stationary phase (Fig. 3). This pattern was reproducible and observed with different strains. Western blot analysis confirmed that the polyclonal anti-EspA antibody recognizes a 25-kDa protein, the expected size of EspA (Fig. 4a).
FIG. 3.
EspA filament expression during the growth of E. coli O157:H7. ZAP193 was cultured overnight in M9 minimal medium and subcultured into MEM-HEPES to an OD600 of 0.05. Bacteria were labeled to detect EspA filaments as described in Materials and Methods. The results of a typical experiment are shown. Experiments were repeated three times for both ZAP193 and ZAP198, and both strains gave the same pattern of expression.
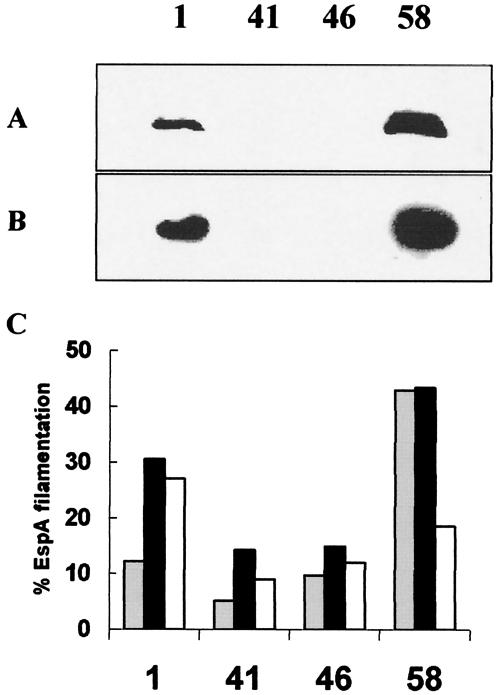
FIG. 4.
Analysis of EspA and EspD expression by E. coli O157 strains. Western blot analysis of EspA (A) and EspD (B) in bacterial supernatants from four wild-type E. coli O157 strains: ZAP1, ZAP58 (both high secretors), ZAP41, and ZAP46 (both low secretors). Bacteria were cultured overnight in M9 minimal medium and subcultured into MEM-HEPES. The supernatants were obtained at an OD600 of 0.8, and proteins were separated and detected as described in Materials and Methods. (C) Proportions of bacteria expressing EspA filaments were determined by immunofluorescence microscopy as described in Materials and Methods. Samples were prepared at OD600s of 0.4 (░⃞); 0.8 (▪); and 1.2 (□).
High and low levels of EspD secretion by wild-type E. coli O157:H7 strains correlates with the proportion of bacteria that express EspA filaments.
Our previous work has shown that E. coli O157:H7 strains isolated from both human disease outbreaks and cattle show a marked heterogeneity in their level of EspD secretion into MEM-HEPES (40). To test whether this heterogeneity was linked to the proportion of the population expressing EspA filaments, two high-secretion and two low-secretion strains (ZAP1,-41, -46, and -58; Table 3) were cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES as described before. The bacteria were then sampled at different points in the growth curve and the proportion of bacteria expressing EspA filaments was determined, as well as the level of EspA and -D in the supernatants. High-secretion strains ZAP1 and ZAP58 had easily detectable levels of EspA and EspD in their supernatants at an OD600 of 0.8, whereas the low-secretion strains had barely detectable levels by comparison (Fig. 4a and b). Increased exposure of the blots did permit detection of EspA and -D from strains ZAP41 and -46 (data not shown). The high-secretion strains had a higher proportion of bacteria producing EspA filaments at the three optical densities examined (Fig. 4c). When strains were cultured overnight in MEM-HEPES (rather than M9 minimal medium) and subcultured into MEM-HEPES, the proportion of EspA+ bacteria increased, with high-secretion strains giving populations containing 70 to 95% EspA+ bacteria, whereas low secretors remained under 30% (Fig. 2k and l, Fig. 4c and data not shown). These growth conditions produced the largest disparity in EspA filamentation between high- and low-secretor strains and were used to analyze differences in mRNA transcript levels (shown later in this report).
Production of EspA filaments by individual bacteria does not correlate with LEE1-4 expression.
The LEE1, -2, -3, and -4::egfp fusions on pACYC184 in E. coli O157:H7 (ZAP193) were used to determine if the variable expression of EspA filaments resulted from differences in expression from the LEE1-4 promoters in individual cells. Bacteria were again cultured overnight in M9 minimal medium and then subcultured into MEM-HEPES. While the proportion of bacteria expressing EspA filaments was again variable, GFP levels in each cell were very similar, as shown by fluorescence and confocal microscopy at all of the optical densities examined (Fig. 2m to p). In addition, the LEE1-4 promoters from E. coli O157:H7 (ZAP1) were cloned in front of lacZ and transformed into ZAP1 and ZAP41 derivatives with the lacZY region deleted (Table 3). The levels of β-galactosidase from each promoter fusion were equivalent in the two strain backgrounds when the bacteria were cultured in MEM-HEPES (Table 4). Previous work with primer extension has revealed at least two putative promoters for espADB (LEE4) (2, 43), so single-copy, chromosomally integrated lacZ constructs to both regions were made in ZAP1193 (Δlac, Materials and Methods). This allowed putative promoters from different strains to be analyzed in an E. coli O157:H7 (Δlac) high-secretor background. No activity from espA promoters was detected under permissive secretion conditions, in contrast to the sepL promoter regions, which gave measurable and equivalent activity for the regions cloned from five E. coli EHEC strains (Table 5). Sequence analysis demonstrated that all five regions were identical over a 220-bp region proximal to the sepL AUG start codon (data not shown).
TABLE 4.
Miller assay results for plasmid-based LEE1-4 promoter fusions to β-galactosidase in high- and low-secretor E. coli O157 EspD backgrounds
| Plasmid | Avg no. of Miller units ± SD (n = 3) |
|
|---|---|---|
| ZAP1001a (high secretor) | ZAP1041a (low secretor) | |
| pAJR101 (LEE1::lacZ) | 7,429 ± 122 | 7,659 ± 187 |
| pAJR102 (LEE2::lacZ) | 2,364 ± 79 | 1,984 ± 97 |
| pAJR103 (LEE3::lacZ) | 113 ± 11 | 144 ± 45 |
| pJTPr-2 (LEE4::lacZ) | 353 ± 57 | 351 ± 42 |
Strains were grown in MEM-HEPES to an OD600 of 0.6, and Miller assays were carried out as described in Materials and Methods.
TABLE 5.
Miller assay results obtained with chromosomal sepL′ and espA′ translational fusions to lacZ in EHEC O157 ZAP1193
| Promoter source | Avg no. of Miller units (± SD, n = 3) for indicated chromosomally integrated translational β-galactosidase fusions in ZAP1193a |
|
|---|---|---|
| sepL::lacZ | espA::lacZ | |
| ZAP1 | 50 ± 2 | 3 ± 2 |
| ZAP21 | 50 ± 3 | 4 ± 2 |
| ZAP41 | 49 ± 1 | 2 ± 1 |
| ZAP46 | 49 ± 2 | 2 ± 1 |
| ZAP58 | 47 ± 2 | 4 ± 1 |
| ZAP193 | NDb | 2 ± 1 |
| No promoter | 2 ± 1 | 2 ± 1 |
Strains were grown in MEM-HEPES to an OD600 of 0.6, and Miller assays were carried out as described in Materials and Methods.
ND, not determined for this construct.
Translocon protein secretion is inversely related to levels of the espADB mRNA transcript.
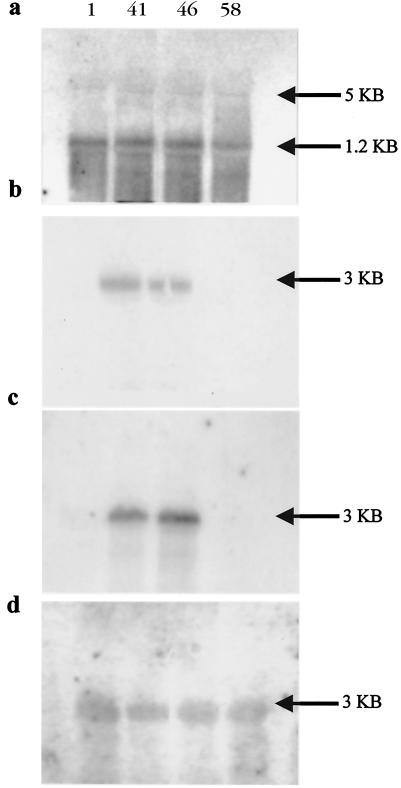
The levels of LEE4 mRNA in two high-secretor and two low-secretor strains were investigated. In order to maximize the secretion differences between the strains, overnight MEM-HEPES cultures were used to inoculate MEM-HEPES for exponential growth and samples were taken at an OD600 of 0.8. For the high-secretion strains, this produced populations that had more than 80% of the bacteria expressing EspA filaments, compared to less than 30% for the low-secretor strains. Northern analysis was carried out with probes to sepL, espA, and espD mRNAs. As shown in Fig. 5a, all of the strains produced equivalent levels of a 1- to 1.5-kb transcript when a sepL probe was used. In addition, a faint band of approximately 5 kb was detected. This concurs with previous work suggesting that initially a 5.6-kb transcript is produced from the sepL promoter, which is then processed to give separate species, including the 2.8-kb espADB and 1.5-kb sepL mRNAs (22, 43). Both low-secretion strains (ZAP41 and ZAP46) were found to contain an mRNA transcript of approximately 3 kb (Fig. 5b and c) when either an espA or an espD probe was used. In contrast, the same transcript was less abundant in high-secretion strains (Fig. 5b and c). As controls, the RNA extracts were probed for LEE2 (escC) and phosphoglucose isomerase (pgi) transcripts. The strains were found to contain very similar levels of both of these transcripts (Fig. 5d and data not shown).
FIG. 5.
Northern analyses of LEE operon expression. Detection of transcript mRNA with an sepL probe (a) in high- and low-secretor strains: ZAP1 and -58 (high secretors, Table 3) and ZAP41 and -46 (low secretors, Table 3). The top arrow indicates a transcript of approximately 5 kb, and the lower arrow indicates a transcript of approximately 1.2 kb. Detection of transcript mRNA with an espA probe (b) and an espD probe (c). The arrow indicates a transcript of approximately 3 kb. Detection of transcript mRNA with a LEE2 (escC) probe (d) in high- and low-secretor strains. The arrow indicates a transcript of approximately 3 kb. All strains (Table 3) were grown in MEM-HEPES to an OD600 of 0.6, and the total RNA was extracted as described in Materials and Methods. The probes used are defined in Table 2.
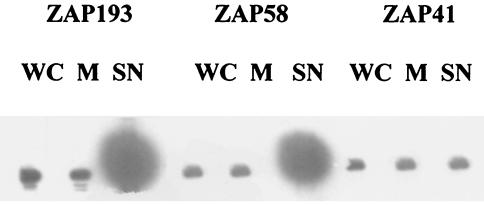
The abundance of the espADB mRNA in the two low-secretor strains prompted the question of whether this mRNA is translated and the protein is maintained in the cell as a pool rather than being secreted. Western blots were carried out on the high- and low-secretor strains for EspD present in whole cells, membranes, and supernatants (Fig. 6). There were equivalent and relatively low levels of EspD detectable in whole cells of both phenotypes, and most of this protein was membrane associated.
FIG. 6.
EspD localization in E. coli O157:H7. ZAP1, -41, -193, and -58 were cultured overnight in MEM-HEPES, subcultured into MEM-HEPES, and grown to an OD600 of 0.8. Supernatant (SN), whole-cell (WC), and membrane (M) proteins were prepared as described in Materials and Methods. Western blotting for EspD was carried out as described in Materials and Methods.
DISCUSSION
The aim of this study was to investigate the molecular basis of the variation in secreted EspD levels demonstrated by wild-type E. coli O157:H7 strains (40). Previous work had shown that the majority of cattle isolates secrete significantly lower levels of EspD than do many human outbreak strains. This research therefore focused on the expression of the LEE4 translocon proteins, EspA and EspD, in both high- and low-secretion strains.
EspA filaments were visualized on the surface of the bacteria by indirect immunofluorescence assay with an anti-EspA polyclonal antibody. Initial experiments examined a number of wild-type strains and demonstrated that only a proportion of the bacteria expressed EspA filaments and that strains originally characterized as high secretors had the populations with the highest proportions of EspA-positive bacteria. Such inherent heterogeneity makes it difficult to compare expression levels between strains. In this case, it was decided to start strains from an equal point, overnight growth in M9 minimal medium, which fails to induce secretion of the translocon proteins (Fig. 2), and study the transition to their expression in MEM-HEPES. Growth in MEM-HEPES increases LEE1-4 expression, resulting in secretion of EspA and -D (Fig. 1a). With this comparison, high secretors expressed filaments on a higher proportion of the population than did low-secretor strains (Fig. 2). These differences were supported by Western blot analysis of EspD and EspA levels in the bacterial supernatants (Fig. 4a and b). Attempts were made to quantitate the levels of filament-positive and -negative bacteria by flow cytometry, but the preparation steps resulted in the loss of EspA filaments, which appear extremely fragile in comparison to other surface appendages, such as fimbriae and flagella. The pattern of EspA expression was analyzed in more detail with two high-secretor Shiga-like toxin-negative E. coli O157 strains (ZAP193 and -198, Table 3). The highest proportions of EspA+ bacteria were shown to be present in late exponential phase, with filaments lost rapidly from these positive cells once the cultures entered stationary phase. Growth phase-dependent expression of translocon proteins has been observed for E. coli O157:H7, linked to quorum sensing (29, 56).
The molecular basis of this heterogeneity was then investigated. As with many phase-variable systems, we attempted to demonstrate variation at the transcriptional level with GFP and lacZ fusions to LEE4 both on low-copy-number vectors and by single-copy fusions exchanged into the chromosome. Previous reports have demonstrated that a promoter in front of sepL drives expression of LEE4 and espADB in EPEC but that a promoter in front of espA drives LEE4 expression in EHEC O157 (2, 22, 43). Our data reported here (and unpublished), which were obtained with promoter regions amplified from five E. coli O157 strains, show no evidence of any expression from a promoter in front of espA, with expression only detectable from sepL. None of these constructs displayed any phase-variable expression, either at the single-cell level (GFP fusions) or at the colony level (lacZ fusions). In addition to LEE4, analysis of LEE1-3 GFP fusions in ZAP193 also demonstrated consistent expression at the single-cell level under conditions that produce heterogeneous expression of EspA filaments (Fig. 2i to k). Taken together, the data indicate that the heterogeneity in EspA filament production, and therefore high- and low-level secretion, is controlled posttranscriptionally.
LEE4 mRNA levels were analyzed by Northern blotting with probes for sepL, espA, and espD. Expression from a promoter in front of sepL would produce an initial transcript length of approximately 5.6 kb (if expression occurs to the end of espF), as shown in two previous studies with EPEC (22, 43). Equivalent levels of sepL-containing transcripts were detected in both high- and low-secretor strains, confirming the fusion data that indicated no variation in expression from this promoter between the strains. The detection of two sepL transcripts indicates some processing of the initial 5.6-kb message. This processing of the transcript may explain the +1 transcriptional start site mapped for espA by Beltrametti et al. (2), as this could instead be an mRNA cleavage site. Probes directed against espA and espD both revealed a transcript of approximately 3 kb which will be the mRNA transcript for espA, espD, and espB. The intriguing result, obtained with both probes, was that this transcript was more abundant in the low-secretor strains than in the high-secretor strains; i.e., there is an inverse relationship between secreted protein (EspA and EspD) levels and detection of the associated espADB mRNA transcript. Control probes for LEE2 and phosphoglucose isomerase transcripts displayed little variation between the high- and low-secretor strains. Taken together with the fusion data, the data show that LEE4 is transcribed at the same level in both high- and low-secretor strains but translation and secretion of Esps is then restricted in the majority of bacteria in a low-secretor population. This was confirmed by an analysis of EspD levels in whole cells that showed that the abundant transcript was not being translated to give an intracellular pool of Esps in low-secretor strains and any EspD appeared to be associated with the cell membrane (Fig. 6c). The checkpoint in secretion therefore appears to lie in translation of the transcript. The reason for the much lower detectable levels of LEE4 and espADB mRNAs in the high-secretor strains is unknown, but this was repeatedly demonstrated with both espA and espD probes. One possibility is that secretion and translation are coupled at the type III secretion system, as suggested for the related Salmonella enterica serovar Typhimurium flagellar export system (30). If the mRNA is engaged within a type III secretion complex during secretion of EspADB, it may be more difficult to extract. Differences in the half-lives of the transcripts may also explain the findings and are being investigated. The concept of a checkpoint in Yop secretion at the posttranscriptional level has been discussed by Anderson and Schneewind for Yersinia sp. (1).
One conclusion from the data presented is that EspA secretion is phase variable in E. coli O157 and controlled at the posttranscriptional level, although the mechanism responsible for this variation remains to be demonstrated. The extent of this variation differs between strains and accounts for the observed differences in protein secretion levels (40). The population can be divided into bacteria that have EspA filaments and are actively secreting proteins and those that have no filaments and presumably are not secreting. Expression levels of the LEE1-4 operons are equivalent in both populations; the difference between them is that in the secreting subpopulation, the espADB transcript is being translated, leading to translocon production, whereas in the other subpopulation, translation is checked. High-secretor strains contain a higher proportion of cells that are actively producing the translocon proteins, whereas in the low-secretor strains, this proportion of cells is much lower. Phase variation of the TTSS translocon may keep expression limited in vivo, reducing the likelihood of an immune response to these antigens. In addition, the variation may allow other adhesins to function on the bacterial cell surface without interference from EspA filaments. Such adhesins may drive initial binding of EHEC O157, which could then lead to production of the translocon apparatus. A posttranscriptional checkpoint would have the translocon apparatus primed for immediate expression in response to a suitable stimulus. Recent work suggests that flagellar phase variation in S. enterica is controlled by a posttranscriptional mechanism in which a control protein is proposed to bind to the 5′ untranslated region of the flagellar transcript, inhibiting ribosome binding and translation (5). For E. coli O157:H7, current work is attempting to identify factors that interact with the LEE4 mRNA transcript, including both proteins and small RNAs.
The production of EspA filaments therefore requires the right environmental conditions to lead to expression of LEE1-4, but then a further restriction is placed on whether of not the LEE4 espADB mRNA transcript is translated. To date, we have no evidence that this heterogeneous expression occurs in EPEC O127 strain E2348/69, suggesting that it may not be subject to the same posttranscriptional restriction. This difference could account for the finding that the cloned EPEC LEE in E. coli K-12 does lead to Esp secretion and attaching-and-effacing lesion formation, whereas the cloned EHEC O157 LEE does not (16, 39). We therefore speculate that a phase-variable factor, possibly not encoded on the LEE, enables translation of the espABD message in E. coli O157:H7. This further control of Esp production may be important for the success of the bacterium in ruminants, especially cattle. EPEC in humans and animals causes an acute and generalized infection of the gastrointestinal tract to which there is a strong adaptive immune response. This may, in part, result from less restricted Esp expression. By contrast, E. coli O157:H7 colonizes the terminal rectum in cattle (45), with no apparent immune response (our unpublished data), and can persist at this site for many weeks. The low-secretion phenotype displayed by the majority of bovine E. coli O157:H7 strains may be necessary to keep the expression of the type III secreted factors restricted until required at the specific colonization site.
Acknowledgments
We are grateful to Trinad Chakraborty for providing the EspD monoclonal antibody and plasmid pJTPr-2. Help with obtaining and analyzing the confocal images was provided by Linda Sharp. We also thank J. McCluskey for advice regarding Northern analyses and Nicola Holden and Megan Porter for critical appraisal of the manuscript.
This work was funded as part of a DEFRA Veterinary Fellowship research program to D.L.G. and D.G.E.S.
Editor: V. J. DiRita
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 2.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. S., A. Marie-Cardine, B. Schraven, and J. B. Bliska. 2000. The Yersinia tyrosine phosphatase YopH targets a novel adhesin-regulated signalling complex in macrophages. Cell. Microbiol. 2:401-414. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive psc101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 5.Bonifield, H. R., and K. T. Hughes. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 185:3567-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 7.Celli, J., W. Y. Deng, and B. B. Finlay. 2000. Enteropathogenic Escherichia coli (EPEC) attachment to epithelial cells: exploiting the host cell cytoskeleton from the outside. Cell. Microbiol. 2:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Celli, J., M. Olivier, and B. B. Finlay. 2001. Enteropathogenic Escherichia coli mediates antiphagocytosis through the inhibition of PI 3-kinase-dependent pathways. EMBO J. 20:1245-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 11.Crane, J. K., B. P. McNamara, and M. S. Donnenberg. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell. Microbiol. 3:197-211. [DOI] [PubMed] [Google Scholar]
- 12.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djafari, S., F. Ebel, C. Deibel, S. Kramer, M. Hudel, and T. Chakraborty. 1997. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol. Microbiol. 25:771-784. [DOI] [PubMed] [Google Scholar]
- 14.Ebel, F., C. Deibel, A. U. Kresse, C. A. Guzman, and T. Chakraborty. 1996. Temperature- and medium-dependent secretion of proteins by Shiga toxin-producing Escherichia coli. Infect. Immun. 64:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, S. J., J. Yu, and J. B. Kaper. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebel, A., H. Ilchmann, M. Aelpfelbacher, K. Ehrbar, W. Machleidt, and W. D. Hardt. 2001. SopE and SopE2 from Salmonella typhimurium activate different sets of Rho GTPases of the host cell. J. Biol. Chem. 276:34035-34040. [DOI] [PubMed] [Google Scholar]
- 18.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 19.Galan, J. E., and D. Zhou. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc. Natl. Acad. Sci. USA 97:8754-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallois, A., J. R. Klein, L. A. H. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 166:5741-5748. [DOI] [PubMed] [Google Scholar]
- 21.Gally, D. L., T. J. Rucker, and I. C. Blomfield. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, M. D., M. Johnson, J. C. D. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 23.Goosney, D. L., S. Gruenheid, and B. B. Finlay. 2000. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16:173-189. [DOI] [PubMed] [Google Scholar]
- 24.Goosney, D. L., R. DeVinney, and B. B. Finlay. 2001. Recruitment of cytoskeletal and signaling proteins to enteropathogenic and enterohemorrhagic Escherichia coli pedestals. Infect. Immun. 69:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck, J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ide, T., S. Laarmann, L. Greune, H. Schillers, H. Oberleithner, and M. A. Schmidt. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 28.Kabach, H. R. 1971. Bacterial membranes. Methods Enzymol. 22:99-120. [Google Scholar]
- 29.Kanamaru, K., I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 30.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 31.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 32.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 65:2606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 34.Kenny, B., and B. B. Finlay. 1997. Intimin-dependent binding of enteropathogenic Escherichia coli to host cells triggers novel signaling events, including tyrosine phosphorylation of phospholipase C-γ1. Infect. Immun. 65:2528-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 36.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kresse, A. U., F. Beltrametti, A. Muller, F. Ebel, and C. A. Guzman. 2000. Characterization of sepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 182:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 40.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. E. Hoey, C. Currie, T. Chakraborty, D. G. E. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 42.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 44.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neves, B. C., S. Knutton, L. R. Trabulsi, V. Sperandio, J. B. Kaper, G. Dougan, and G. Frankel. 1998. Molecular and ultrastructural characterisation of EspA from different enteropathogenic Escherichia coli serotypes. FEMS Microbiol. Lett. 169:73-80. [DOI] [PubMed] [Google Scholar]
- 47.Nikolaus, T., J. Deiwick, C. Rappl, R. A. Freeman, W. Schröder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nougayrede, J. P., M. Boury, C. Tasca, O. Marches, A. Milon, E. Oswald, and J. De Rycke. 2001. Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect. Immun. 69:6785-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostroff, S. M., P. M. Griffin, R. V. Tauxe, L. D. Shipman, K. D. Greene, J. G. Wells, J. H. Lewis, P. A. Blake, and J. M. Kobayashi. 1990. A statewide outbreak of Escherichia coli O157:H7 infections in Washington State. Am. J. Epidemiol. 132:239-247. [DOI] [PubMed] [Google Scholar]
- 50.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 51.Roe, A. J., C. Currie, D. G. E. Smith, and D. L. Gally. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145-152. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw, R. K., S. Daniell, F. Ebel, G. Frankel, and S. Knutton. 2001. EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol. 3:213-222. [DOI] [PubMed] [Google Scholar]
- 55.Simonovic, I., M. Arpin, A. Koutsouris, H. J. Falk-Krzesinski, and G. Hecht. 2001. Enteropathogenic Escherichia coli activates ezrin, which participates in disruption of tight junction barrier function. Infect. Immun. 69:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sperandio, V., C. Y. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, K. A., C. B. O'Connell, P. W. Luther, and M. S. Donnenberg. 1998. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect. Immun. 66:5501-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wainwright, L. A., and J. B. Kaper. 1998. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol. Microbiol. 27:1247-1260. [DOI] [PubMed] [Google Scholar]
- 60.Wales, A. D., F. A. Clifton-Hadley, A. L. Cookson, M. P. Dibb-Fuller, C. M. Hayes, R. M. La Ragione, J. M. Roe, K. A. Sprigings, G. R. Pearson, and M. J. Woodward. 2001. Preliminary observations on E. coli O157:H7 in sheep. Epidemiology of verocytotoxigenic E. coli, p. 105-113. In G. Duffy, P. Garvey, J. Coia, W. Wasteson, and D. McDowell (ed.), Verocytotoxigenic E. coli in Europe, 5; concerted action CT98-3935. Teagasc, Dublin, Ireland.
- 61.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3:753-762. [DOI] [PubMed] [Google Scholar]