Abstract
Kinetoplastid glycosomes contain a variety of metabolic activities, such as glycolysis, β-oxidation of fatty acids, lipid biosynthesis, and purine salvage. One advantage of sequestering metabolic activities is the avoidance of cellular oxidative damage by reactive oxygen species produced as a by-product of metabolism. Little is known about how glycosomes themselves withstand these toxic metabolites. We previously isolated an iron superoxide dismutase from Leishmania chagasi that is expressed at low levels in the early logarithmic promastigote stage and increases toward the stationary promastigote and amastigote stages. We have since identified a second highly homologous Lcfesodb gene that is expressed at high levels in the early logarithmic promastigote stage and decreases toward the stationary promastigote and amastigote stages. Localization studies using green fluorescent protein fusions have revealed that LcFeSODB1 and LcFeSODB2 are localized within the glycosomes by the last three amino acids of their carboxyl termini. To better understand the specific role that FeSODB plays in parasite growth and survival, a single-allele knockout of the Lcfesodb1 gene was generated. The parasites with these genes exhibited a significant reduction in growth when endogenous superoxide levels were increased with paraquat in culture. Furthermore, the FeSODB1-deficient parasites exhibited a significant reduction in survival within human macrophages. Our results suggest that LcFeSODB plays an important role in parasite growth and survival by protecting glycosomes from superoxide toxicity.
Despite the evolution of a complex mammalian immune response against foreign pathogens, Leishmania continues to plague humans, causing death and disease worldwide. This places an emphasis on elucidating the molecular mechanisms employed by Leishmania to establish a successful infection. Leishmania is an intracellular protozoan parasite that infects mammalian macrophages. These parasites possess a digenic life cycle, consisting of an extracellular promastigote form that multiplies and develops within the alimentary tract of the sand fly vector and an intracellular amastigote form that resides and multiplies within the phagosome of the mammalian host macrophage.
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are toxic metabolites that damage all living cells. The superoxide anion (O2˙−) is produced by the reduction of oxygen and is the fundamental oxidant among a battery of reactive intermediates, as it is involved in numerous reactions generating a plethora of increasingly toxic intermediates, including hydrogen peroxide (H2O2), hydroxyl radicals (˙OH), and peroxynitrite (ONOO−). Collectively, these prooxidants are toxic to DNA and can cause peroxidation of lipids and the inactivation of iron-sulfur enzymatic centers of dehydratases (15). Important reactions involving O2˙− are demonstrated in the following reactions: O2˙− + Fe3+ → Fe2+ + O2; 2 O2˙− + 2H+ → H2O2 + O2; O2˙− + H2O2 → ˙OH + −OH + O2; O2˙− + HOCl → ˙OH + O2 + Cl−; and O2˙− + ˙NO → ONOO−.
In transforming from promastigotes to amastigotes, Leishmania parasites undergo numerous biochemical changes in order to adapt to their new environment. Numerous genes have been shown to be differentially expressed in the promastigote and amastigote stages, and many morphological and metabolic changes have been documented. A particularly interesting organelle that houses many metabolic activities, such as glycolysis, β-oxidation of fatty acids, ether lipid biosynthesis, and purine salvage, is the glycosome. Glycosomes are found in Leishmania and other kinetoplastid parasites. One of the benefits of compartmentation of metabolic activities is the avoidance of cellular oxidative damage caused by ROS that are produced as a result of metabolism. Little is known about the antioxidant defenses of Leishmania that protect these organelles and the important metabolic enzymes contained in them from oxidative damage.
Superoxide dismutases (SODs) comprise a family of metalloenzymes containing iron (FeSOD), manganese (MnSOD), nickel (NiSOD), or copper and zinc (Cu/ZnSOD) at the active site (1, 35). SODs dismutate O2˙− into H2O2 and O2 before it exacerbates prooxidant damage by initiating the formation of even more toxic species, such as ˙OH and ONOO−. SODs have been shown to contribute to the pathogenicity of many parasites by protecting them from O2˙− toxicity (2, 3, 6, 16, 25, 28, 32, 34). We have previously isolated an FeSOD from Leishmania chagasi, Lcfesodb1, which is expressed at high levels in the stationary promastigote and amastigote stages of infection. We present here the isolation from L. chagasi of a second member of the FeSODB gene family (Lcfesodb2) that is expressed at high levels in the early logarithmic promastigote stage. We have shown that both LcFeSODB1 and LcFeSODB2 are localized to the glycosomes by the last three amino acids of their carboxyl termini, suggesting that these proteins may act to protect important glycosomal enzymes from O2˙− toxicity. In order to further characterize the role that Lcfesodb plays in parasite survival, we generated a single-allele knockout of the Lcfesodb1 gene using homologous-recombination technology. A single-allele knockout of Lcfesodb1 resulted in a decreased level of growth when the endogenous level of O2˙− within the parasites was increased with paraquat and a decreased level of survival within macrophage cells, suggesting that Lcfesodb1 is important for the growth and survival of L. chagasi. Such an antioxidant defense within Leishmania glycosomes had not been clearly defined previously.
MATERIALS AND METHODS
Materials.
All enzymes, [α-32P]dCTP, Hybond N+, Rapid-Hyb buffer, and protein molecular weight markers, were purchased from Amersham Biosciences. DNA and RNA molecular weight markers, agarose and parasite medium, medium components, and hygromycin B and Geneticin drugs were supplied by Life Technologies, Inc. All other chemicals were purchased from Sigma unless stated otherwise. Nucleic acid quantifications were performed using the Beckman DU 640 spectrophotometer.
Parasites.
L. chagasi (HMOM/BR/00/1669), Leishmania donovani (1S2D), and Leishmania major (Friedlin strain) parasites were used in this study. L. chagasi promastigote parasites were cultured at 26°C in hemoflagellate minimal essential medium consisting of minimal essential medium supplemented with sodium pyruvate (1.1×), essential (0.5×) and nonessential (1×) amino acids, glucose (0.15%), sodium bicarbonate (0.22% [wt/vol]), 10% fetal calf serum (inactivated at 56°C for 30 min), 10 μg of hemin/ml, and 35 mM HEPES sodium salt. The promastigotes were inoculated at 106/ml and harvested at log or stationary phase, defined by morphological and concentration criteria as previously described (36). Log-phase parasites were ovoid to cigar shaped and at a concentration of 4 × 106 to 8 × 106/ml. Stationary-phase parasites were needle shaped and at a concentration of 5 × 107 to 7 × 107/ml.
Infection of U937 cells.
Differentiation of the human macrophage cell line U937 (American Type Culture Collection) into macrophages and infection with stationary-phase Leishmania promastigotes were performed using standard methods as previously described (31). U937 cells were seeded at a concentration of 2.5 × 105/cm2 in eight-chamber slides and differentiated into adherent macrophages by treatment with 7.5 ng of phorbol myristate acetate (Sigma) per ml of RPMI 1640 with 10% fetal calf serum, 2 mM glutamine, and 50 μg of Gentamicin (RPMI)/ml with 5% CO2 at 37°C for 72 h. Adherent cells were washed three to five times with warm RPMI medium, followed by incubation with L. chagasi parasites at a parasite-to-U937-cell ratio of 10:1 for 6 h. Nonengulfed parasites were washed away three to five times with warm RPMI. Warm fresh RPMI medium was then added to the flask containing the infected macrophages, and the flask was incubated with 5% CO2 at 37°C. The level of infection was determined 6, 18, and 42 h postinfection by optical microscopy following Diff Quick staining of cell preparations (22). Values are expressed either as the percentage of macrophages infected or as the total number of amastigotes per 100 macrophages.
Sequencing of iron-SOD genes.
Two converging primers, Forward Primer 1 (5′-CTGCACCACTCGAAGCACCA-3′) and Reverse Primer 2 (5′-CAGGTAGTACGCGTGCTCCCA-3′), based on conserved amino acid sequence of FeSODs (24), were synthesized based on Leishmania codon usage.
The 426-bp product obtained by amplifying L. chagasi genomic DNA was subsequently labeled with [α-32P]dCTP (Amersham Biosciences) and used as a probe to screen a λZAPII L. chagasi (MHOM/BR/82/BA-2, CI) sheared genomic library, kindly provided by S. Reed (Seattle Biomedical Research Institute, Seattle, Wash.). A total of ∼105 PFU were plated and lifted in duplicate. Potential positive clones were treated by secondary screening to recover isolated positive clones. Isolated clones were amplified using standard protocols, and DNA was extracted using a Lambda Maxi kit (Qiagen). Sequencing of the purified recombinant λ DNA was performed at the University of Calgary DNA Sequencing Facility, Calgary, Alberta, Canada.
Southern and Northern blots.
Genomic DNA was isolated from logarithmic promastigotes by lysing parasites in lysis buffer (10 mM Tris-Cl, pH 8.3, 50 mM EDTA, 1% sodium dodecyl sulfate [SDS]). RNase A (100 μg/ml) was added to the suspension and incubated overnight at 37°C, followed by the addition of 100 μg of proteinase K/ml and incubation at 42°C overnight. DNA was extracted by performing a phenol-chloroform treatment followed by ethanol precipitation. Genomic DNA was digested with various restriction enzymes and separated on a 1% agarose gel. The DNA was transferred to Hybond N+ membranes via capillary action and fixed with long-wave UV light for 5 min. The blots were blocked with Rapid-Hyb buffer and incubated at 68°C with [α-32P]dCTP random-primed labeled DNA probes for 2 h, after which the blots were washed successively with decreasing salt concentrations. Total RNA was isolated from promastigotes using the acid guanidinium isothiocyanate method (4). Total RNAs were separated in a 1.2% formaldehyde-containing agarose gel, transferred onto Hybond N+ membranes, and baked at 80°C for 2 h. The blots were blocked with Rapid-Hyb buffer and incubated at 62°C with [α-32P]dCTP random-primed labeled DNA probes for 2 h and washed in decreasing salt concentrations. The RNA blots were hybridized with [α-32P]dCTP-labeled α-tubulin as a loading control.
Generation of Lcfesodb1 gene knockouts.
The generation of knockout constructs was based on the pX63-HYG and pX-NEO vectors (kindly provided by S. M. Beverley), which consist of a pSP6-T3 (Life Technologies Inc.) backbone and a cassette comprising a gene encoding resistance to hygromycin B (hyg) or Geneticin (neo) and flanking dihydrofolate reductase-thymidylate synthase (DHFR) sequences that are required for proper gene expression (5). To disrupt the Lcfesodb1 locus, the target Lcfesodb1 DNA sequences were cloned into pX63-HYG and pX63-NEO such that the hyg expression cassette (2,837 bp) and neo expression cassette (2,687 bp) were flanked at both ends by Lcfesodb1 DNA. This strategy permitted subsequent excision of the entire insert, excluding vector DNA, for transfection into the parasites to promote homologous recombination with chromosomal Lcfesodb1 sequence. The 5′ Lcfesodb1 homologous-region insert was PCR amplified, yielding a 466-bp fragment (nucleotides 80 to 538) that was cloned into the HindIII/XhoI sites in both vectors. The unique 3′ Lcfesodb1 homologous-region insert was PCR amplified, yielding a 1,456-bp (nucleotides 888 to 2217) fragment that was cloned into the SmaI/BglII sites of both vectors, resulting in the pX63HYGFeSODB1KO and pX63NEOFeSODB1KO vectors. The nucleotide numbers refer to the Lcfesodb1 sequence (GenBank accession no. AF003963). The vectors were digested with HindIII and BglII, and the knockout fragment containing the selectable gene marker was purified. The procedures used in the transfection of L. chagasi promastigotes have been described previously (14, 23). Wild-type parasites were transfected with 5 μg of the linearized knockout constructs by electroporation, and transformants were selected at 50 μg of hygromycin B/ml. For gene complementation studies, the entire coding region of the Lcfesodb1 gene was cloned into the BamHI sites of the pX-Neo expression vector to generate the pX-FeSODB1 vector. Plasmids were sequenced for the correct orientation. Single-allele knockout parasites were transfected with 40 μg of the pX-FeSODB1 plasmid by electroporation and selected at 1 mg of Geneticin/ml. It is known that continuous subpassage of parasites may result in a decrease in virulence; therefore, the Δsodb1hyg and wild-type strains were electroporated and subpassaged in parallel the same number of times under the same culture conditions.
Western blot analysis.
L. chagasi lysates were prepared by harvesting log- and stationary-phase parasites as described previously (20). Briefly, the parasites were washed three times with phosphate-buffered saline (PBS; 1×), resuspended in hypotonic buffer (5 mM Tris-Cl [pH 7.8], 0.1 mM EDTA [pH 8.0], 5 mM phenylmethylsufonyl fluoride, 100,000 IU of aprotinin, 20 μg of leupeptin/μl) and freeze-thawed in liquid nitrogen. The equivalent of 5 × 106 parasites/lane were boiled for 10 min and separated on a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane by semidry blotting. Western blot analysis was performed using anti-LcFeSODB1 antiserum and an enhanced-chemiluminescence kit (Amersham Biosciences). Anti-LcFeSODB1 polyclonal antiserum was obtained from the University of Calgary Animal Research Facility. His-tagged LcFeSODB1 protein was excised from a Coomassie blue-stained SDS-polyacrylamide gel electrophoresis (PAGE) gel, macerated in the presence of liquid nitrogen, resuspended in Freund's complete adjuvant, and passed several times through a 21.5-gauge needle before inoculation into New Zealand White rabbits. Further inoculations were done at 4-week intervals using gel-purified protein resuspended in Freund's incomplete adjuvant. After four inoculations, the rabbits were bled and the specificity of the antiserum was tested. Monoclonal antibody (E7) to β-tubulin was purchased from the Developmental Studies Hybridoma Bank, University of Iowa.
SOD cytochrome c activity assay.
The SOD activity in L. chagasi parasite lysates was determined by the reduction of ferricytochrome c according to standard methods (19). Briefly, reaction mixtures contained 0.05 M potassium phosphate buffer at pH 7.8, 0.02 mM EDTA, 0.01 mM ferricytochrome c, 0.05 mM xanthine, and 1.6 × 10−2 U of xanthine oxidase/ml. Absorbance at 550 nm was monitored at 25°C, and the rate of reduction of ferricytochrome c was calculated. Under these conditions, the amount of SOD required to inhibit the rate of reduction of ferricytochrome c by 50% is defined as 1 U of activity.
Subcellular localization of LcFeSODB.
To determine the subcellular localization of LcFeSODB1 and LcFeSODB2 within L. chagasi, various LcFeSODB fusions were constructed with green fluorescent protein (GFP). The gene fusions were expressed in the parasites in the pXNEO expression vector. Fusion constructs were generated by PCR amplifying the coding region of GFP with Forward Primer 3 (5′ GTCGGATCCATGGTGAGCAAGGGCGAGG 3′) and Reverse Primer 4 (5′ CCGGAATTCGTACTTGTACAGCTCGTCC 3′) containing a mutated GFP stop codon (TAC) to allow fusion with a downstream gene. The entire coding regions of LcFeSODB1 and LcFeSODB2 that were to be fused to GFP were PCR amplified using Forward Primer 5 (5′ CCGGAATTCATGCCGTTCGCTGTTCAGCCG 3′) (for both LcFeSODB1 and LcFeSODB2) and Reverse Primer 6 (5′ CCGTCTAGATTAAAGCTGGCTAGAGGCGAAATCCC 3′) (for LcFeSODB1) and Reverse Primer 7 (5′ CCGTCTAGATTACAGATCACTGTTGACGTAGTGGG 3′) (for LcFeSODB2). Amplification of the LcFeSODB coding regions without their respective SQL and SDL amino acids was done using Forward Primer 5 and Reverse Primer 8 (5′ CCGTCTAGATTAAGAGGCGAAATCCCAGTC 3′) (for LcFeSODB1 without SQL) and Reverse Primer 9 (5′ CCGTCTAGATTAGTTGACGTAGTGGGAGCC 3′) (for LcFeSODB2 without SDL). Amplification of GFP with the amino acids SQL or SDL added to the carboxyl terminus was done using Forward Primer 3 and Reverse Primer 10 (5′CCGTCTAGATTAAAGCTGGCTGTACTTGTACAGCTCGTCCATGCCG 3′) (for GFP with SQL) and Reverse Primer 11 (5′ CCGTCTAGATTACAGATCACTGTACTTGTACAGCTCGTCCATGCCG 3′) (for GFP with SDL). The underlined sequences identify the restriction sites used and the start codon, stop codon, or matched stop codon. The GFP and LcFeSODB PCR products were digested with EcoRI and ligated together. The ligated products were PCR amplified using Forward Primer 3 and the corresponding reverse primers and cloned into the pXNEO BamHI/XbaI restriction sites. The newly constructed vectors were transfected into L. chagasi parasites by electroporation and selected with Geneticin to a final concentration of 50 μg/ml.
Approximately 5 × 106 mid-log-phase parasites were washed with 1× PBS and applied to poly-l-lysine-coated coverslips for 15 min. The parasites were washed with PBS and fixed with 4% paraformaldehyde-0.01% electron microscopy-grade gluteraldehyde in PBS (pH 7.0) at 24°C for 30 min and washed twice with PBS. The parasites were then incubated in 50 mM glycine (in 1× PBS) for 15 min and then washed with PBS. The coverslips were blocked, and the cells were permeabilized with 2% goat serum-0.1% Triton X-100 for 30 min. Primary antibody was diluted (1:200) in 2% goat serum, centrifuged at 18,000 × g for 10 min, and applied to the coverslips for 1 h. The coverslips were washed six times with PBS (5 min per wash). Secondary antibody, Alexa Fluor 594 (Molecular Probes), was diluted (1:500) in 2% goat serum, centrifuged for 10 min, and applied to the coverslips for 1 h in the dark. The coverslips were then washed six times with PBS (5 min per wash). Twenty microliters of 50% glycerol was applied to a glass slide, and the coverslips (parasite side down) were placed on top and sealed with nail polish. The parasites were photographed with a fluorescent microscope.
In vitro drug assays.
Treatment of parasites with paraquat involved seeding the stationary-phase parasites at a density of 106/ml in hemoflagellate minimal essential medium as previously described (23). The parasites were permitted to recover for 24 h prior to the addition of freshly prepared paraquat at a concentration that inhibited wild-type parasite growth by 50% under the conditions used (500 μM). After 6 days of exposure to paraquat, parasite viability was measured microscopically according to flagellar motility by assessing 200 promastigotes as motile (viable) or nonmotile (nonviable), as previously described (36). Experiments with <95% parasite viability before treatment with paraquat were discounted.
GenBank accession numbers.
The coding regions of the fesodb genes were amplified from L. donovani (IS2D) and L. major (Friedlin strain) genomic DNAs using primers based on L. chagasi Lcfesodb1 and Lcfesodb2 sequences (GenBank accession numbers AF003963 and AF312581, respectively). The sequences of the genes isolated from L. donovani and L. major have been deposited in GenBank with the following accession numbers: L. donovani LddFeSODB1, AF312585, and LddFeSODB2, AF312582; L. major LmFeSODB1, AF312586, and LmFeSODB2, AF312583.
RESULTS
Isolation and characterization of the Lcfesodb gene cluster.
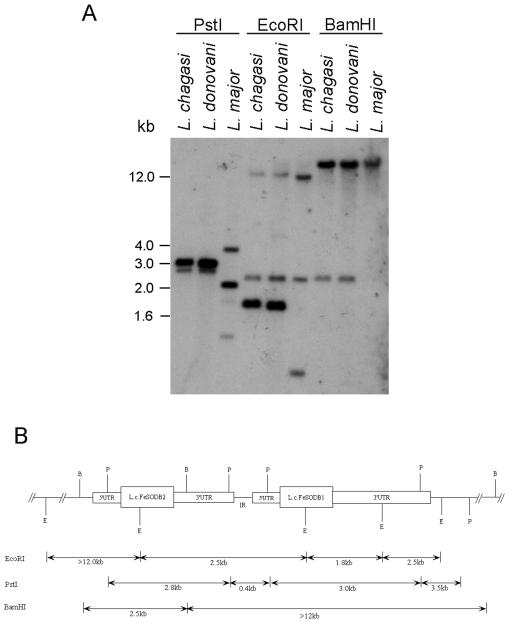
One Lcfesodb gene (now called Lcfesodb1) was previously isolated (23). The isolation of the second fesodb gene (now called Lcfesodb2) is presented herein. A λZAPII L. chagasi genomic DNA library was screened, and a positive clone containing Lcfesodb sequence was digested with EcoRI, PstI, and BamHI (Fig. 1A). Extensive sequencing and Southern blot analysis of the insert revealed that the Lcfesodb2 gene is situated in tandem upstream of the Lcfesodb1 gene (Fig. 1B). The chromosomal localization of the Lcfesodb1 and Lcfesodb2 genes determined by pulsed-field gel electrophoresis revealed that they colocalize to the same high-molecular-weight chromosome, which supports their tandem genomic organization (data not shown). Similar results were obtained for the fesodb1 and fesodb2 genes of L. donovani (data not shown).
FIG. 1.
Southern blot analysis and restriction map of the Lcfesodb cluster. (A) Total genomic DNAs from L. chagasi, L. donovani, and L. major (500 ng) were isolated; restriction digested with PstI, EcoRI, and BamHI; resolved on a 1% agarose gel; and transferred to a nitrocellulose membrane. The membrane was hybridized with a radiolabeled Lcfesodb1 coding-region probe. (B) Restriction map of the L. chagasi Lcfesodb cluster determined by Southern blot hybridization, library screening, and sequence analysis of genomic DNA. The tentative positions of the 3′ UTR of Lcfesodb1 and the intergenic region (IR) are based on the known 5′ UTR Lcfesodb1 sequence. The sequence located upstream of the 5′ UTR of Lcfesodb1 and downstream of the Lcfesodb2 stop codon is representative of the 3′ UTR of Lcfesodb2 and the IR. The restriction fragment lengths shown below the schematic correspond to the hybridizing bands observed with several probes used for Southern blot analysis. P, PstI; E, EcoRI; B, BamHI.
FeSODB2 consists of 209 amino acids with a predicted molecular mass of 23.2 kDa, whereas LcFeSODB1 consists of 196 amino acids with a predicted molecular mass of 21.7 kDa. The amino acid sequences of LcFeSODB1 and LcFeSODB2 are 88% identical and differ primarily by the presence of a 13-amino-acid extension found at the carboxyl terminus of LcFeSODB2 (Fig. 2). The last three amino acids of LcFeSODB1 (SQL) and LcFeSODB2 (SDL) resemble the typical glycosomal localization signal sequence, SKL, suggesting that these proteins may be localized to the glycosomes. We have also amplified fesodb1 and fesodb2 genes from L. donovani (LddFeSODB1 and LddFeSODB2) and L. major (LmFeSODB1 and LmFeSODB2) genomic DNAs using primers flanking the coding region sequences of Lcfesodb1 and Lcfesodb2. Sequence comparisons of the fesodb1 and fesodb2 genes in the different species revealed >92% identity, suggesting a high degree of conservation of fesodb genes in Leishmania (Fig. 2).
FIG. 2.
Amino acid sequence comparison of L. chagasi, L. donovani, and L. major FeSODB coding regions. Sequence alignment of FeSODB1 and FeSODB2 reveals a higher degree of divergence in the carboxyl termini, where FeSODB2 contains a 13-amino-acid extension that is not present in FeSODB1. The solid boxes indicate amino acid differences. The arrows indicate residues involved in iron binding at the active site. To maintain alignment, dashes represent missing amino acids.
Lcfesodb1 and Lcfesodb2 genes are differentially expressed.
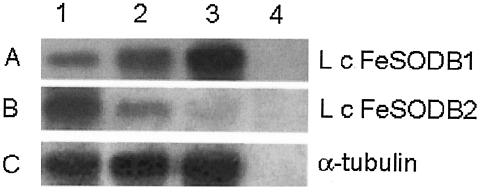
To study the expression pattern of Lcfesodb1 and Lcfesodb2 as the parasites progress through their life cycle, total RNAs were isolated from parasites at early logarithmic phase, stationary phase, and an amastigote-like stage (derived from infected U937 cells). The amount of RNA loaded in each lane was normalized using a probe complementary to the constitutively expressed α-tubulin gene of Leishmania (Fig. 3C). Northern blot hybridization using the specific 3′ untranslated region (UTR) of Lcfesodb1 revealed a single band of 3.8 kb that increased in intensity ∼1.7-fold from the early logarithmic to the stationary phase and ∼2.1-fold from the early logarithmic to the amastigote stage (Fig. 3A). In contrast, Northern blot hybridization using the specific 3′ UTR of Lcfesodb2 revealed a single band of 2.4 kb that decreased in intensity ∼2.4-fold from the early logarithmic to the stationary phase and ∼3.1-fold from the early logarithmic to the amastigote stage (Fig. 3B). These results demonstrate that the Lcfesodb genes are differentially expressed: Lcfesodb1 is expressed at low levels in the early logarithmic promastigote stage and at high levels in the stationary promastigote and amastigote stages, whereas Lcfesodb2 is expressed at high levels in the early logarithmic promastigote stage and at low levels in the stationary promastigote and amastigote stages. Similar results were observed for Ldfesodb1 and Ldfesodb2 in L. donovani (data not shown).
FIG. 3.
Northern blot analysis. Ten micrograms of total RNAs from early log-phase L. chagasi parasites (lane 1), stationary-phase L. chagasi parasites (lane 2), U937 cells infected with L. chagasi parasites (lane 3), and uninfected U937 cells (lane 4) was extracted and resolved on a 1.2% agarose gel under denaturing conditions, blotted, and probed with the 3′ UTR of Lcfesodb1 (SA, 7.5 × 108 cpm/μg) (A), the 3′ UTR of Lcfesodb2 (specific activity, 5.3 × 108 cpm/μg) (B), or α-tubulin from L. chagasi (loading control) (specific activity, 3.5 × 108 cpm/μg) (C). No hybridizing bands of any size were observed with the α-tubulin probe and the RNA from the uninfected U937 cells (C, lane 4). Exposures: 26 h at −80°C (A and B) and 2 h at room temperature (C).
Replacement of the Lcfesodb1 gene by homologous recombination.
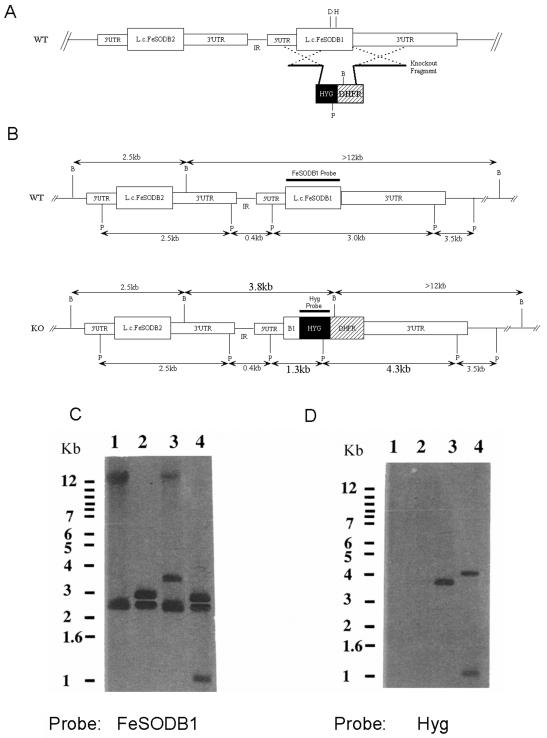
The high expression of Lcfesodb1 in the stationary promastigote- and amastigote-like stages suggests that the LcFeSODB1 protein may play an important role in intracellular survival. To help define the role of Lcfesodb1 in parasite survival, we attempted to create a homozygous Lcfesodb1 gene deletion in L. chagasi using sequential rounds of gene replacement to replace both alleles of the endogenous Lcfesodb1 gene with HYG and NEO drug-resistant genes. In the first round of transfection, the knockout fragment containing a HYG/DHFR resistance cassette flanked by the 5′ and unique 3′ homologous Lcfesodb1 regions (pX63HygFeSODB1) was transfected into wild-type L. chagasi promastigotes (Fig. 4A). The predicted homologous-recombination event was designed to knock out the aspartate-161 and histidine-165 amino acid residues near the carboxyl terminus of LcFeSODB1, which are essential in coordinating the iron atom in the active site of FeSOD enzymes for activity (Fig. 4A) (24). We were aware that this knockout strategy could replace one Lcfesodb1 allele alone or both Lcfesodb1 and Lcfesodb2 alleles, because Lcfesodb2 is situated upstream of Lcfesodb1 and shows similarity to the 5′ UTR and the 5′ half of the coding region of Lcfesodb1.
FIG. 4.
Schematic representation and Southern blot analysis of Lcfesodb1 knockout parasites. (A) Schematic of homologous-recombination strategy employed to knock out the 360-bp region of Lcfesodb1 that contains the active-site aspartic acid-161 (D) and histidine-165 (H) residues, which act as ligands to coordinate the iron. The transfected knockout fragment consists of a 466-bp fragment, homologous to the 5′ UTR and coding region of Lcfesodb1, and a 1,465-bp fragment, homologous to the 3′ UTR of Lcfesodb1, that flank the HYG/DHFR cassette. The homologous-recombination event between the Lcfesodb1 allele and the knockout fragment DNA is represented by two dashed crosses. IR, intergenic region. (B) Schematic depicting the endogenous wild-type (WT) allele of the Lcfesodb cluster and the disrupted allele following homologous recombination. The expected BamHI (B) and PstI (P) restriction fragment sizes are given. (C and D) Southern blot analyses of L. chagasi Δsodb1hyg and wild-type genomic DNAs using Lcfesodb1-specific (C) and hygromycin (hyg)-specific (D) probes. Lanes 1, wild-type DNA digested with BamHI; lanes 2, wild-type DNA digested with PstI; lanes 3, Δsodb1hyg DNA digested with BamHI; lanes 4, Δsodb1hyg DNA digested with PstI. Molecular size marker positions are shown to the left of each blot.
Genomic DNAs from a hygromycin B-resistant mutant (Δsodb1hyg) and wild-type parasites were isolated, digested with BamHI and PstI, and analyzed using Southern blotting to confirm proper integration of the knockout fragment into only one Lcfesodb1 allele (Fig. 4C and D). Digestion of the wild-type genomic DNA with BamHI resulted in two bands (>12 and 2.5 kb), whereas the knockout resulted in three bands (>12, 3.8, and 2.5 kb) when hybridized with the Lcfesodb1 coding region probe (Fig. 4C, lanes 1 and 3). The 2.5-kb band was present in both the wild-type and knockout parasites at the same intensity and was determined to contain the Lcfesodb2 gene by using Lcfesodb2-specific probes (data not shown). The fact that the bands were of equal intensity supports the idea that Lcfesodb2 was not disrupted by a homologous-recombination event. The >12-kb band was present in both the wild-type and knockout parasites; however, the band in the knockout parasites was approximately half the intensity of the wild-type band, as would be seen if one allele of Lcfesodb1 was knocked out. The 3.8-kb hybridizing band in the Δsodb1hyg digest, absent in the digested wild-type DNA, is a result of the predicted integration of the HYG/DHFR cassette into the Lcfesodb1 gene, which introduces a BamHI site that is not present in this location in the wild-type allele (Fig. 4B and C, lane 3). Hybridization of these digests with a hyg-specific probe resulted in a single band of 3.8 kb corresponding to a fragment containing hyg gene sequences, as expected (Fig. 4B and C, lane 3).
Hybridization of PstI-digested Δsodb1hyg genomic DNA with the Lcfesodb1 coding region-specific probe also revealed the predicted pattern that would result from accurate integration into only the Lcfesodb1 gene locus (Fig. 4B). The 2.8- and 3.0-kb bands appearing in both the wild-type and Δsodb1hyg digests (Fig. 4C, lanes 2 and 4) represent fragments containing the uninterrupted Lcfesodb2 and Lcfesodb1 alleles, respectively (Fig. 4B). Hybridization with probes specific to each 3′ UTR verified their identities (data not shown). The additional 1.3-kb band appearing only in the Δsodb1hyg digested DNA (Fig. 4C, lane 4) represents the disrupted Lcfesodb1 allele fragment, because the HYG/DHFR cassette contains an internal PstI site (Fig. 4B). Hybridization of this blot with a hyg-specific probe showed the expected absence of hybridizing bands in the wild-type DNA and the presence of 4.3- and 1.3-kb bands containing hyg gene sequences (Fig. 4B and C, lane 4).
In further support of the finding that one allele of Lcfesodb1 had been disrupted, Northern blot analysis using a 3′ UTR-specific probe from Lcfesodb1 revealed a significant decrease in the intensity of the Lcfesodb1 transcript in the Δsodb1hyg parasites compared to the wild-type parasites (data not shown). Northern blot analysis using a 3′ UTR-specific probe from Lcfesodb2 did not reveal a difference in intensity of the Lcfesodb2 transcript in the Δsodb1hyg parasites compared to the wild type, suggesting that the Lcfesodb2 allele was not disrupted (data not shown). Taken together, these results confirm that one Lcfesodb1 allele was successfully disrupted by the hygromycin cassette and that the Lcfesodb2 gene was not affected by the homologous-recombination event. Repeated attempts to knock out the second Lcfesodb1 allele with the neo, sat, or GFP gene were unsuccessful. It is possible that both alleles of the Lcfesodb1 gene are essential for parasite survival; however, further tests would need to be done to come to this conclusion.
Reduced SOD activity within Δsodb1hyg parasites is due to the lack of one Lcfesodb1 allele.
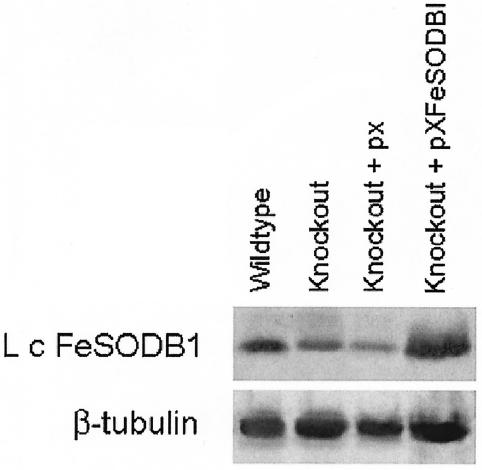
Western blot and densitometry analyses of wild-type and Δsodb1hyg promastigote lysate revealed an ∼1.5-fold decrease in LcFeSODB1 protein in the Δsodb1hyg parasites compared to the wild-type parasites (Fig. 5). To determine if there is a decrease in SOD activity in the Δsodb1hyg parasites compared to the wild type, a cytochrome c assay was performed on parasite lysate. The results revealed a significant (P < 0.025) decrease in SOD activity in the Δsodb1hyg parasites (3.6 ×10−6 ± 3.6 ×10−6 U/mg) compared to the wild type (12.7 ×10−6 ± 1.4 ×10−6 U/mg) (The activity is the average ± standard error of five independent trials.)
FIG. 5.
Western blot analyses of wild-type and Δsodb1hyg parasites. Lysates from stationary-phase wild-type, Δsodb1hyg (Knockout), Δsodb1hyg+pX (Knockout + px), and Δsodb1hyg+pX-FeSODB1 (Knockout + pXFeSODB1) parasites were separated on an SDS-12% PAGE gel and subjected to Western blotting using LcFeSODB1 antiserum (top). The same blots were stripped and incubated with monoclonal β-tubulin antibody to serve as a loading control (bottom).
To determine whether the decrease in SOD activity observed in the Δsodb1hyg parasites was in fact due to the absence of one Lcfesodb1 allele, the Δsodb1hyg parasites were rescued by transfection with a plasmid containing the Lcfesodb1 gene (pX-FeSODB1). Western blot analysis of the Δsodb1hyg parasites overexpressing LcFeSODB1 showed an ∼1.6-fold increase in LcFeSODB1 protein compared to Δsodb1hyg parasites transfected with the pX vector alone (Fig. 5). Cytochrome c analysis of lysates from Δsodb1hyg+pX-FeSODB1 parasites showed that the SOD activity in the Δsodb1hyg+pX-FeSODB1 parasites (12.4 × 10−6 ± 2.7 ×10−6 U/mg) increased to a level similar to that of the wild type (12.7 ×10−6 ± 1.4 ×10−6 U/mg) and considerably higher than that of lysates from Δsodb1hyg+pX parasites (5.9 × 10−6 ± 1.6 × 10−6 U/mg). Taken together, these complementation results show that the decreased level of SOD activity in the Δsodb1hyg parasites can be attributed to the absence of one Lcfesodb1 allele.
Cellular localization of LcFeSODB1 and LcFeSODB2.
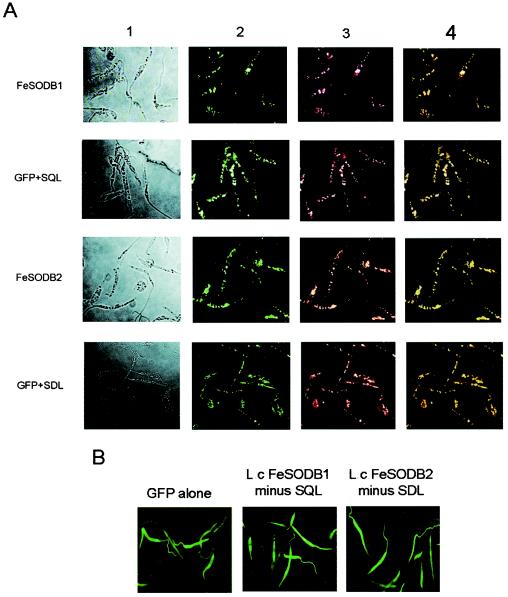
To gain a better understanding of the roles that LcFeSODB1 and LcFeSODB2 play in parasite survival, we studied the cellular localization of these proteins within L. chagasi. The last three amino acids of LcFeSODB1 (SQL) and LcFeSODB2 (SDL) conspicuously resemble the glycosomal targeting signal sequence, SKL. Previous mutational analysis of the SKL glycosomal targeting signal in Trypanosoma brucei showed that the signal is highly degenerate (30).
We created GFP-LcFeSODB1 and GFP-LcFeSODB2 fusion protein gene constructs and overexpressed them in L. chagasi parasites (selected at 50 μg of Geneticin/ml) to determine their localization. Fluorescence microscopy showed a distinct punctate pattern of fluorescence, suggesting compartmentation of the proteins in an organelle (Fig. 6A). Colocalization of the GFP fusion proteins with the glycosomal proteins HGPRT (29) (Fig. 6, columns 3 and 4) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (10, 11) (data not shown) confirmed that the GFP fusion proteins are localized to the glycosomes. To test whether the proposed targeting signal sequences SQL and SDL were capable of targeting the cytosolic GFP protein to the glycosome, we individually fused the SQL and SDL sequences to the carboxyl terminus of GFP. The fluorescence patterns resembled the punctate pattern seen with the LcFeSODB1 and LcFeSODB2 fusion proteins and were colocalized with the glycosomal marker proteins HGPRT (Fig. 6A) and GAPDH (data not shown). GFP-LcFeSODB1 and GFP-LcFeSODB2 fusion proteins lacking the SQL and SDL signal sequences exhibited GFP fluorescence similar to that in parasites expressing GFP alone (Fig. 6B). These parasites exhibited fluorescence throughout the organism, including the flagella, characteristic of a cytoplasmic localization pattern. We have thought about generating monoclonal antibodies to LcFeSODB1 and LcFeSODB2 proteins to further complement our localization studies; however, because the amino acid sequences of these proteins are virtually identical, it is highly likely that we would not be able to differentiate between the LcFeSODB1 and LcFeSODB2 proteins in immunolocalization or fractionation studies. Therefore, we have used specific GFP fusion protein constructs and immunolocalization with control glycosomal proteins to demonstrate that the LcFeSODB1 and LcFeSODB2 proteins are localized to the glycosomes, and we have used this technology to identify the specific amino acids involved in targeting these proteins to the glycosomes.
FIG. 6.
Cellular localization of LcFeSODB1 and LcFeSODB2. (A) Fluorescence patterns of L. chagasi parasites expressing GFP fusion proteins with LcFeSODB1 (FeSODB1), SQL alone (GFP+SQL), LcFeSODB2 (FeSODB2), or SDL alone (GFP+SDL). Column 1, parasites observed under bright-field illumination; column 2, GFP fluorescence; column 3, Alexa Fluor 594 fluorescence; column 4, merged images of columns 2 and 3. (B) Fluorescence patterns of L. chagasi parasites expressing GFP alone and GFP fusion proteins with LcFeSODB1 lacking the last three amino acids (SQL) and LcFeSODB2 lacking the last three amino acids (SDL).
Δsodb1hyg parasites exhibit decreased survival upon exposure to paraquat and within human macrophages.
In order to determine whether the single-allele knockout of Lcfesodb1 affects parasite viability compared to the wild type, Δsodb1hyg parasite growth was monitored with and without hygromycin B. No significant differences were observed in the growth curves or in the gross morphology of the Δsodb1hyg parasites compared to the wild type (data not shown). To determine the effect that knocking out one allele of Lcfesodb1 has on the ability of L. chagasi to survive an increased level of endogenous O2˙−, the ability of the parasites to survive in culture in the presence of paraquat was examined. Paraquat is a redox-cycling drug that penetrates the cytosol to significantly increase endogenous O2˙− production. After 6 days of exposure to paraquat, the percent survival of the Δsodb1hyg parasites (46.2% ± 9.7%) was significantly (P < 0.02) lower than that of the wild-type parasites (76.9% ± 6.3%). The deficiency in survival of the Δsodb1hyg parasites was rescued to a level slightly higher than that of the wild type after transfection with pX-FeSODB1 (86.6% ± 1.6%) and considerably higher than that of Δsodb1hyg transfected with pX (52.7% ± 2.3%). (The data represent the averages ± standard errors of four independent trials.)
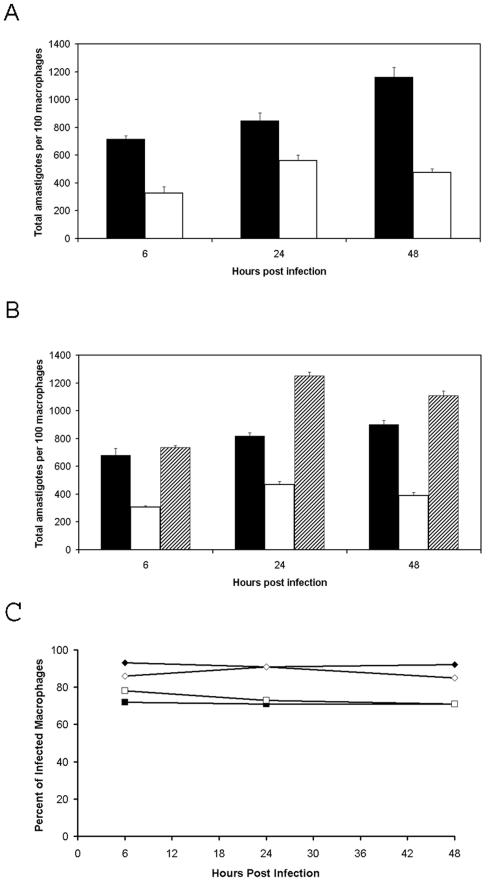
To study the effect of knocking out one allele of the Lcfesodb1 gene on the ability of the parasites to survive intracellularly, we performed an infection study using U937 cells. Differentiated U937 cells were infected with stationary-phase wild-type and Δsodb1hyg parasites and incubated for various times. Remarkably, after 48 h of infection, the total number of Δsodb1hyg amastigotes per 100 macrophages decreased to less than half that of the wild-type amastigotes (475 and 1,162, respectively) (Fig. 7A). The reduced infection level of the Δsodb1hyg parasites was rescued by overexpressing LcFeSODB1 in these parasites, which exhibited a slightly increased infection level after 48 h compared to the control wild-type parasites transfected with the pX vector alone (1,110 and 900, respectively) (Fig. 7B). This could imply that the parasites overexpressing FeSODB1 exhibit enhanced survival and replication compared to the control wild-type parasites. These numbers were significantly (P < 0.001) higher than the infection level of Δsodb1hyg amastigotes transfected with the pX vector alone (390). The percentages of infected macrophages in the wild-type and Δsodb1hyg parasites and in the Δsodb1hyg parasites transfected with pX and the Δsodb1hyg parasites transfected with pX-FeSODB1 were similar at 6, 24, and 48 h postinfection (Fig. 7C). These results suggest that LcFeSODB1 is important for the survival of L. chagasi under conditions of increased endogenous O2˙− and for survival within macrophages.
FIG. 7.
Survival of L.chagasi wild-type and Δsodb1hyg parasites within U937 cells. (A and B) Differentiated U937 cells were incubated with stationary-phase wild-type (solid bars) or Δsodb1hyg (open bars) promastigotes (A) and with wild-type promastigotes transfected with the pX vector (solid bars), Δsodb1hyg promastigotes transfected with the pX vector (open bars), or Δsodb1hyg promastigotes transfected with the pX-FeSODB1 vector (shaded bars) (B) for 6 h as described in Materials and Methods. Nonengulfed parasites were washed away, and the infected U937 cells were incubated for 6, 24, and 48 h, after which the infected macrophages were stained with Diff Quick and examined by optical microscopy to determine the level of infection, which was expressed as the total number of amastigotes per 100 infected macrophages. (C) Percentages of infected macrophages for wild-type parasites (solid diamonds), Δsodb1hyg parasites (open diamonds), Δsodb1hyg parasites transfected with pX (solid squares), and Δsodb1hyg parasites transfected with pX-FeSODB1 (open squares). The averages plus standard errors of four independent experiments are shown. *, P < 0.001.
DISCUSSION
With the emergence of multidrug-resistant Leishmania parasites, the toxicity of current drug therapy and the increased tendency for patient relapse have strengthened the drive for the development of better drugs and an effective vaccine. Understanding the mechanisms that allow Leishmania to survive intracellularly is important for designing such treatment strategies and for drug development. The production of O2˙− has been recognized as an important mechanism involved in the killing of Leishmania and many other pathogens (8, 13, 21, 26, 33). The ability of SODs to dismutate O2˙− and our previous findings that L. chagasi possesses FeSODs which are not found in humans attracted our interest to these proteins as potential targets for therapeutics. We have previously hypothesized that LcFeSODB may protect the parasites from macrophage-derived O2˙− toxicity; however, the evidence presented here has caused us to reevaluate our previous hypothesis. Interestingly, we have isolated a second fesodb gene (Lcfesodb2) that is highly homologous to Lcfesodb1 and whose expression is high in the early logarithmic promastigote stage and decreases toward the stationary promastigote and amastigote stages. This contrasts with the expression of Lcfesodb1, which is low in the early logarithmic stage and increases toward the stationary and amastigote stages. We demonstrate that LcFeSODB1 and LcFeSODB2 are both localized to the glycosomes by the final three amino acids located at the ends of their carboxyl termini. Furthermore, we show that a single-allele knockout of the Lcfesodb1 gene results in a significant decrease in survival of the parasites under conditions of increased endogenous O2˙− levels and within macrophages.
In this study, we present a distinct member of the L. chagasi fesodb gene cluster, Lcfesodb2, which was determined to be located in tandem with and upstream of the Lcfesodb1 gene. Southern blot hybridization patterns of L. chagasi and L. donovani genomic DNAs appear to be very similar, which suggests that the two organisms have maintained similar genomic arrangements of the fesodb genes throughout their evolution. Although the Southern blot hybridization pattern of the fesodb genes in L. major differs from those in L. chagasi and L. donovani (Fig. 1A), sequence analysis of isolated fesodb1 and fesodb2 genes from L. chagasi, L. donovani, and L. major demonstrates a high degree of conservation, with >90% identity in amino acid sequence (Fig. 2). This suggests a low rate of evolution for the individual fesodb genes, which in turn may reflect an important role of FeSODB in oxidative-stress defense. Although we have not performed biochemical or gene knockout studies of FeSODB in L. donovani or L. major, their high amino acid sequence identity and the presence of identical glycosomal targeting signal sequences leads us to hypothesize that the FeSODB genes may have similar functions in those species.
Northern blot analysis revealed differential expression of the Lcfesodb1 and Lcfesodb2 genes in L. chagasi, where the expression of Lcfesodb1 increases from the promastigote to the amastigote stage and Lcfesodb2 expression decreases from the promastigote to the amastigote stage. The high level of expression of Lcfesodb2 in the early logarithmic promastigote stage suggests that the gene may be important for survival within the midgut of the sandfly, and the high level of expression of Lcfesodb1 in the stationary promastigote and amastigote stages suggests that it may be important for survival within macrophages. The possession of SODs in both the promastigote and amastigote stages is clearly beneficial to the parasites. Research indicates that the midgut environment of the sandfly is potentially lethal for the developing parasite and that expression of stage- and species-specific molecules promotes parasite survival and growth during this stage (27). It is very likely that between blood meals, the parasites endure conditions of overcrowding and starvation within the sandfly. Such conditions have been shown to result in increased oxidative stress on other cells, leading to the induction of antioxidant enzymes, such as catalase (18). As the parasites enter the mammalian macrophage, they are exposed to an entirely different environment, which is much harsher than the environment in the gut of the sandfly. Among other changes, heat shock and an increased acidic and oxidative environment undoubtedly impose many metabolic stresses on the parasites that lead to increased intracellular oxidative pressures. The possession of SODs, such as LcFeSODB1 and LcFeSODB2, would therefore be beneficial for parasite survival in controlling endogenous superoxide levels. What is not clear at the moment is why Leishmania possesses two highly homologous FeSODs, both targeted to the glycosomes and with completely different expression profiles. Perhaps Lcfesodb2 predated Lcfesodb1 in evolution, and over time the parasites acquired an FeSODB that is up-regulated toward the amastigote stage and as a result acquired an enhanced ability to survive intracellularly.
To advance our understanding of the specific role that LcFeSODB1 plays in survival, we used GFP fusion proteins to determine the localization of LcFeSODB within the parasites. The results show that LcFeSODB1 and LcFeSODB2 are both localized to the glycosomes and hence are not likely to be available to react directly with external O2˙− generated from macrophages, since O2˙− is a charged molecule that does not diffuse through lipid membranes (12, 17). This suggests that the proteins may have evolved to protect the parasites against O2˙− formed in the glycosomes as a result of changes in metabolism that may occur in the parasites in response to oxidative stress, starvation, and/or overcrowding within the sandfly gut or within the phagolysosomes of macrophages. Glycosomes are known to provide an enclosed environment separate from the cytoplasm for a variety of essential activities, such as glycolysis and β-oxidation of fatty acids, which generate O2˙− as a by-product. When O2˙− reaches toxic levels, it may inactivate important proteins essential for parasite survival or it may exacerbate oxidative damage through its conversion into and/or reaction with other ROS and RNS. A defense against glycosomal O2˙− has not been clearly defined. There was a brief report of Cu/ZnSOD activity using SOD inhibitors within the glycosomes of L. donovani (7), but a Cu/ZnSOD gene or protein has never been isolated.
To further elucidate the functional role of LcFeSODB1 in the survival of Leishmania, we generated a single-allele knockout of the Lcfesodb1 gene. Analysis of the Lcfesodb1 single-allele knockout (Δsodb1hyg) parasites revealed a significant decrease in the survival of these parasites within macrophages and when exposed to paraquat in culture. The infection data (Fig. 7) suggest that the defect in Δsodb1hyg parasite growth could be due to the inability of the parasites to deal with their own endogenous superoxide. If the defect in Δsodb1hyg parasite growth was due to intracellular killing by macrophage-derived superoxide produced by the respiratory burst which occurs in the early stage of infection, we would have observed the same initial infection level as for the wild-type control parasites, followed by a transient decrease in the infection level. Figure 7A and B show that the initial infection level of the Δsodb1hyg parasites is low and remains low over a period of 48 h. Our findings that the Δsodb1hyg parasites exhibit a decreased level of survival compared to wild-type control parasites upon exposure to paraquat, which increases superoxide levels in the cytosol, suggests that the knockout parasites have a defect in protection from endogenous superoxide toxicity, which further supports our infection studies suggesting that the Δsodb1hyg parasites have an impaired ability to deal with their own endogenous superoxide levels.
The results show that LcFeSODB1 and LcFeSODB2 both act within the glycosomes, and we propose that their role is to protect glycosomal proteins involved in key metabolic processes from O2˙− toxicity. These findings also highlight the importance of the glycosomes in parasite survival. It is noteworthy that macrophage-derived O2˙− still has an important role in contributing to the intracellular killing of Leishmania (8, 13, 21, 26, 33), perhaps by its conversion into and/or reaction with other ROS and RNS. While this paper was in preparation, an antisense knockout against FeSOD from Leishmania tropica (38% identical in amino acid sequence to LcFeSODB1) was published demonstrating that FeSOD is important for survival in mouse macrophages, apparently providing protection against macrophage-derived superoxide (9). Interestingly, our results suggest that LcFeSODB1 does not provide protection against macrophage-derived O2˙−. Although we were only able to create a single-allele knockout of the Lcfesodb1 gene, we were able to demonstrate its importance for survival, and because of the absence of FeSODs in mammals, LcFeSODB is undoubtedly an attractive target for future drug design.
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (to L.G.) and by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship (to K.A.P.).
We thank Wendy Paramchuk for generating the Lcfesodb1 single-allele knockout parasites and Vasanthakrishna Mundodi and Ashwini Kucknoor for assistance in RNA isolation and blotting.
Editor: F. C. Fang
REFERENCES
- 1.Bannister, J. V., W. H. Bannister, and G. Rotilio. 1987. Aspects of the structure, function, and applications of superoxide dismutase. Crit. Rev. Biochem. 22:111-180. [DOI] [PubMed] [Google Scholar]
- 2.Beaman, B. L., C. M. Black, F. Doughty, and L. Beaman. 1985. Role of superoxide dismutase and catalase as determinants of pathogenicity of Nocardia asteroides: importance in resistance to microbicidal activities of human polymorphonuclear neutrophils. Infect. Immun. 47:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, D. H., B. K. Na, M. S. Seo, H. R. Song, and C. Y. Song. 2000. Purification and characterization of iron superoxide dismutase and copper-zinc superoxide dismutase from Acanthamoeba castellanii. J. Parasitol. 86:899-907. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Cruz, A., C. M. Coburn, and S. M. Beverley. 1991. Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. USA 88:7170-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey, R., and S. C. Datta. 1994. Leishmanial glycosomes contain superoxide dismutase. Biochem. J. 301:317-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gantt, K. R., T. L. Goldman, M. L. McCormick, M. A. Miller, S. M. Jeronimo, E. T. Nascimento, B. E. Britigan, and M. E. Wilson. 2001. Oxidative responses of human and murine macrophages during phagocytosis of Leishmania chagasi. J. Immunol. 167:893-901. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, S., S. Goswami, and S. Adhya. 2003. Role of superoxide dismutase in survival of Leishmania within the macrophage. Biochem. J. 369:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannaert, V., M. Callens, F. R. Opperdoes, and P. A. Michels. 1994. Purification and characterization of the native and the recombinant Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 225:143-149. [DOI] [PubMed] [Google Scholar]
- 11.Hart, D. T., and F. R. Opperdoes. 1984. The occurrence of glycosomes (microbodies) in the promastigote stage of four major Leishmania species. Mol. Biochem. Parasitol. 13:159-172. [DOI] [PubMed] [Google Scholar]
- 12.Hassan, H. M., and I. Fridovich. 1979. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254:10846-10852. [PubMed] [Google Scholar]
- 13.Jackson, S. H., J. I. Gallin, and S. M. Holland. 1995. The p47phox mouse knock-out model of chronic granulomatous disease. J. Exp Med. 182:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapler, G. M., C. M. Coburn, and S. M. Beverley. 1990. Stable transfection of the human parasite Leishmania major delineates a 30-kilobase region sufficient for extrachromosomal replication and expression. Mol. Cell. Biol. 10:1084-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keyer, K., A. S. Gort, and J. A. Imlay. 1995. Superoxide and the production of oxidative DNA damage. J. Bacteriol. 177:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korshunov, S. S., and J. A. Imlay. 2002. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram-negative bacteria. Mol. Microbiol. 43:95-106. [DOI] [PubMed] [Google Scholar]
- 17.Lynch, R. E., and I. Fridovich. 1978. Permeation of the erythrocyte stroma by superoxide radical. J. Biol. Chem. 253:4697-4699. [PubMed] [Google Scholar]
- 18.Masoro, E. J., I. Shimokawa, and B. P. Yu. 1991. Retardation of the aging processes in rats by food restriction. Ann. N. Y. Acad. Sci. 621:337-352. [DOI] [PubMed] [Google Scholar]
- 19.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 20.McKean, P. G., R. Delahay, P. F. Pimenta, and D. F. Smith. 1997. Characterisation of a second protein encoded by the differentially regulated LmcDNA16 gene family of Leishmania major. Mol. Biochem. Parasitol. 85:221-231. [DOI] [PubMed] [Google Scholar]
- 21.Murray, H. W. 1982. Cell-mediated immune response in experimental visceral leishmaniasis. II. Oxygen-dependent killing of intracellular Leishmania donovani amastigotes. J. Immunol. 129:351-357. [PubMed] [Google Scholar]
- 22.Muyombwe, A., M. Olivier, M. Ouellette, and B. Papadopoulou. 1997. Selective killing of Leishmania amastigotes expressing a thymidine kinase suicide gene. Exp. Parasitol. 85:35-42. [DOI] [PubMed] [Google Scholar]
- 23.Paramchuk, W. J., S. O. Ismail, A. Bhatia, and L. Gedamu. 1997. Cloning, characterization and overexpression of two iron superoxide dismutase cDNAs from Leishmania chagasi: role in pathogenesis. Mol. Biochem. Parasitol. 90:203-221. [DOI] [PubMed] [Google Scholar]
- 24.Parker, M. W., and C. C. Blake. 1988. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 229:377-382. [DOI] [PubMed] [Google Scholar]
- 25.Piddington, D. L., F. C. Fang, T. Laessig, A. M. Cooper, I. M. Orme, and N. A. Buchmeier. 2001. Cu, Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollock, J. D., D. A. Williams, M. A. Gifford, L. L. Li, X. Du, J. Fisherman, S. H. Orkin, C. M. Doerschuk, and M. C. Dinauer. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202-209. [DOI] [PubMed] [Google Scholar]
- 27.Sacks, D., and S. Kamhawi. 2001. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 55:453-483. [DOI] [PubMed] [Google Scholar]
- 28.Schnell, S., and H. M. Steinman. 1995. Function and stationary-phase induction of periplasmic copper-zinc superoxide dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 177:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih, S., H. Y. Hwang, D. Carter, P. Stenberg, and B. Ullman. 1998. Localization and targeting of the Leishmania donovani hypoxanthine-guanine phosphoribosyltransferase to the glycosome. J. Biol. Chem. 273:1534-1541. [DOI] [PubMed] [Google Scholar]
- 30.Sommer, J. M., Q. L. Cheng, G. A. Keller, and C. C. Wang. 1992. In vivo import of firefly luciferase into the glycosomes of Trypanosoma brucei and mutational analysis of the C-terminal targeting signal. Mol. Biol. Cell 3:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streit, J. A., J. E. Donelson, M. W. Agey, and M. E. Wilson. 1996. Developmental changes in the expression of Leishmania chagasi gp63 and heat shock protein in a human macrophage cell line. Infect. Immun. 64:1810-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilks, K. E., K. L. Dunn, J. L. Farrant, K. M. Reddin, A. R. Gorringe, P. R. Langford, and J. S. Kroll. 1998. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect. Immun. 66:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, M. E., K. A. Andersen, and B. E. Britigan. 1994. Response of Leishmania chagasi promastigotes to oxidant stress. Infect. Immun. 62:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, C. H., J. J. Tsai-Wu, Y. T. Huang, C. Y. Lin, G. G. Lioua, and F. J. Lee. 1998. Identification and subcellular localization of a novel Cu, Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 439:192-196. [DOI] [PubMed] [Google Scholar]
- 35.Youn, H. D., E. J. Kim, J. H. Roe, Y. C. Hah, and S. O. Kang. 1996. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 318:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarley, J. H., B. E. Britigan, and M. E. Wilson. 1991. Hydrogen peroxide-mediated toxicity for Leishmania donovani chagasi promastigotes. Role of hydroxyl radical and protection by heat shock. J. Clin. Investig. 88:1511-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]