Abstract
An array of mammalian phospho-specific antibodies was used to screen for a host response upon mycobacterial infection, reflected as changes in host protein phosphorylation. Changes in the phosphorylation state of 31 known signaling molecules were tracked after infection with live or heat killed Mycobacterium bovis BCG or after incubation with the mycobacterial cell wall component lipoarabinomannan (LAM). Mycobacterial infection triggers a signaling cascade leading to activation of stress-activated protein kinase and its subsequent downstream target, c-Jun. Mycobacteria were also shown to inhibit the activation of protein kinase C ɛ and to induce phosphorylation of proteins not yet known to be involved in mycobacterial infection, such as the cytoskeletal protein α-adducin, glycogen synthase kinase 3β, and a receptor subunit involved in regulation of intracellular Ca2+ levels. The mycobacterial cell wall component LAM has been identified as a trigger for some of these modulation events.
Mycobacterium tuberculosis is responsible for 2 million to 3 million deaths every year and persists as the leading cause of deaths worldwide due to a single bacterial agent (76). Residing within the host macrophage, M. tuberculosis is able to circumvent the host killing machinery and maintain a state of dormancy that can last for decades (62). Research over the past decades has described M. tuberculosis as a pathogen that is able to adapt to a dynamic and changing host environment (for a review, see reference 45) as well as to actively interfere with and modulate the host killing machinery (53, 68). However, M. tuberculosis is one of the most successful human pathogens of our time, and the mechanism permitting its survival within the host macrophage still remains largely elusive.
Since its discovery more than 30 years ago, a hallmark of mycobacterial infection has been the prevention of phagolysosome fusion (3), enabling the tubercle bacilli to survive and replicate secluded from the endocytic pathway. Additionally, mycobacterial infection is associated with inhibited antigen processing and attenuated gamma interferon activation of the macrophage (15, 43). These cellular events are tightly controlled at the level of signal transduction and are critical for successful clearance of pathogens from the macrophage.
Mycobacterial interference with host signaling pathways has been proposed to be the principal mechanism developed by the bacteria to establish a successful infection. A wide range of host proteins and pathways have been suggested to act as mycobacterial targets, including mitogen-activated protein kinase (MAPK) signaling pathways, JAK-STAT signaling pathways, Ca2+ signaling, NF-κB signaling, and protein kinase C (PKC) signaling (15, 47, 57, 61, 65, 73). These pathways control cellular functions such as cell proliferation, apoptosis, cytokine release, and gene regulation and are an indication of the complexity and scale with which mycobacterial infection might interfere with and possibly counteract host defense mechanisms. The identification of at least 510 protein kinases in the human genome (54) is evidence of the vastness of the eukaryotic signaling network. In such a setting it becomes important to investigate signal transduction as a network of phosphorylation events involving several pathways, rather than focusing on single events.
Large-scale analysis of signaling networks can be performed by using a novel technique in proteomics termed kinome analysis (63). This technique involves the use of an array of phospho-specific antibodies covering kinases and other signaling proteins from major eukaryotic signaling networks known to date. We have used this technique as a tool to identify changes in protein phosphorylation in a human macrophage cell line upon infection with Mycobacterium bovis BCG. We have discovered changes in host signaling pathways that have not previously been described for mycobacterial infection. This includes host proteins involved in regulation of apoptotic pathways, cytoskeletal arrangement, Ca2+ signaling, and macrophage activation.
MATERIALS AND METHODS
Materials.
RPMI 1640, Hanks balanced salt solution (HBSS), protease inhibitors, and phosphatase inhibitors were obtained from Sigma Chemical Co. (St. Louis, Mo.). Anti-phospho-adducin was from Upstate Biotechnology (Lake Placid, N.Y.), and horseradish peroxidase-conjugated anti-rabbit secondary antibody was from Bio-Rad Laboratories (Mississauga, Ontario, Canada). Endotoxin-free lipoarabinomannan (LAM) was generously provided by J. Belisle (Colorado State University, Fort Collins). The LAM used was mannose capped and derived from the virulent H37Rv strain of M. tuberculosis.
Culturing of M. bovis BCG.
M. bovis BCG (ATCC 35734) carrying a plasmid constitutively expressing the green fluorescent protein (19), was used in all experiments. Bacteria were grown in Middlebrook 7H9 broth (Difco, Detroit, Mich.) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose solution (Difco), 0.05% (vol/vol) Tween 80 (Sigma), and 50 μg of hygromycin per ml at 37°C to an A600 of 0.5 on a rotating platform (50 rpm). Bacteria were harvested by centrifugation (5 min, 6,000 × g) at 4°C, and the pellets were resuspended in fresh medium plus 10% glycerol, aliquoted, and kept at −70°C for later use. Frozen stocks were thawed, replenished in fresh medium, and grown for 2 to 4 days before they were used for cell infection.
Infection of THP-1 cells and treatment with LAM.
The monocytic cell line THP-1 (American Type Culture Collection, Manassas, Va.) was cultured in RPMI 1640 supplemented with 10% fetal calf serum (HyClone, Logan, Utah), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were seeded at a density of 105 per cm2 in 10-cm-diameter culture dishes (Corning Inc., Corning, N.Y.) and allowed to adhere and differentiate in the presence of phorbol myristate acetate (PMA) (20 ng/ml) at 37°C in a humidified atmosphere of 5% CO2 for 24 h. The cells were then washed three times with HBSS, and adherent monolayers were exposed to live or killed (80°C, 30 min) BCG at a multiplicity of infection of 50 to 1 in RPMI containing 1% l-glutamine, 10% human serum (purified protein derivative negative), and no antibiotics. After 3 h of incubation at 37°C and 5% CO2, the cells were washed twice with prewarmed HBSS to remove noningested bacteria and reincubated in complete medium at 37°C and 5% CO2 for the indicated time periods. Alternatively, differentiated THP-1 cells were treated with mannose-capped lipoarabinomannan (ManLAM) at a concentration of 1 μg/ml. The infection rate was verified on cells adherent to tissue culture-treated coverslips (Fisher Scientific, Nepean, Ontario, Canada) in 24-well plates. After phagocytosis, cells were fixed for 15 min at 37°C with 2.5% paraformaldehyde-HBSS and then washed three times with HBSS and once with distilled water. Coverslips were then examined with an epifluorescence microscope (Zeiss Axioplan II), and images were taken with a charge-coupled device camera and Empix software.
Kinome analysis by KPSS assay.
Two separate Kinetworks Phospho Site Screen (KPSS) (Kinexus, Vancouver, Canada) analyses were performed. In the initial screen, THP-1 cells were infected with live or heat-killed M. bovis BCG, and in the second screen, THP-1 cells were infected with live M. bovis BCG or treated with purified ManLAM from M. tuberculosis H37Rv at a concentration of 1 μg/ml. Untreated, PMA-differentiated THP-1 cells were used as a control in both experiments. Kinome analyses (KPSS) were performed as previously described (78) and according to the instructions of the manufacturer (Kinexus). In brief, cells were homogenized at 4°C in a buffer containing 20 mM Tris-HCl (pH 7.0), 2 mM EGTA, 5 mM EDTA, 30 mM sodium fluoride, 40 mM β-glycerophosphate (pH 7.2), 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and 1 mM phenylmethylsulfonyl fluoride and spun down at 100,000 × g in a Beckman GS 6R tabletop ultracentrifuge. Protein content was estimated by using the Bradford assay (Bio-Rad). Three hundred micrograms of total protein for each sample was divided equally on a 20-lane Immunetics mutiblotter. Each channel was probed with up to three primary antibodies from an array of 31 phospho-specific antibodies, which were selected so as to avoid overlapping cross-reactivity with target protein. The blots were developed with ECL Plus reagent (Amersham Biosciences), and signals were captured with a Fluor-S-MultiImager and quantified with Quantity One software (Bio-Rad).
Western analysis of adducin.
PMA-differentiated THP-1 cells were infected with live BCG or challenged with purified LAM (concentration range, 0.01 to 1.0 μg/ml). The cells were then incubated for 2, 12, or 24 h. The blots were probed with anti-phospho-adducin and horseradish peroxidase-conjugated anti-rabbit antibodies and developed by enhanced chemiluminescence.
RESULTS AND DISCUSSION
In this study we have examined the effect of mycobacterial infection on human macrophage signal transduction, reflected as changes in the phosphorylation profiles of 31 known signaling molecules. M bovis BCG organisms were used in this study to infect the human monocytic cell line THP-1, which is one of the common human cell lines used in mycobacterial infection studies. PMA treatment induces differentiation of THP-1 cells into a macrophage-like cell line that displays most of the human monocyte-derived macrophage phenotypes with regard to morphology, expression of membrane receptors, cytokine secretion, and induction of several proto-oncogenes (for a review, see reference 4).
Although genotypically M. bovis BCG is a deletion version of M. tuberculosis (8), it retains the ability to survive intracellularly (59) and it successfully prevents macrophage phagosome maturation (41, 75). Therefore, M. bovis BCG is a suitable model for the study of interactions between mycobacteria and the host cell. We have chosen to investigate events occurring after 24 h, thereby excluding early events associated with initial phagocytotic uptake of the bacteria. We rationalize that at this time point, the bacteria have established themselves in their host environment to a greater extent than earlier, offering an opportunity to assess the host response to bacteria residing within phagosomes.
A unique multiphosphoprotein analysis, termed kinome analysis (63), was used as a tool to identify changes in phosphorylation of key host proteins upon infection. The advantage of this technique is the ability to quantitatively track single amino acid phosphorylations. Cells were subjected to a simultaneous screen for the phosphorylation status of 31 host phosphoproteins. Two separate screens were performed; first, THP-1 cells were infected with either live or heat-killed M. bovis BCG (Fig. 1), and second, THP-1 cells were infected with live bacteria or treated with purified ManLAM (Fig. 2). Untreated, differentiated THP-1 cell were used as controls in both screens. ManLAM is expressed by both M. tuberculosis and M. bovis BCG and differs in terms of structure and immunogenicity from LAMs expressed by avirulent strains of mycobacteria (23). ManLAM from BCG and M. tuberculosis has been found to inhibit the production of proinflammatory cytokines such as interleukin-12 and tumor necrosis factor alpha (53). Experiments using latex beads coated with ManLAM have further supported a role for this molecule in the persistence of virulent mycobacteria within macrophages. In this manner, ManLAM was recently shown to interfere with a phosphatidylinositol 3-kinase-dependent pathway between the trans-Golgi network and the phagosomal compartment, inhibiting the acquisition of lysosomal markers and resulting in decreased phagosomal maturation (31).
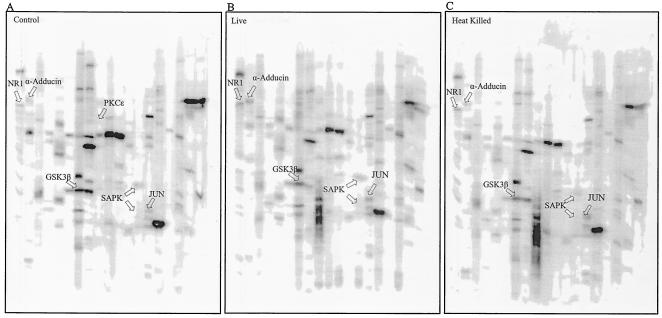
FIG. 1.
Multiphosphoprotein analysis of THP-1 cells infected with live and heat-killed M. bovis BCG. Each sample is represented as a phosphoprotein fingerprint. Accurate intensity values for each band are the accumulated signal obtained over a given scan time for each blot. These are shown as numerical values in Table 1. The data shown here represent protein bands visualized as a snapshot of the scan time and may therefore not represent exact band intensities. The three gels represent untreated THP-1 cells (A), THP-1 cells infected with live M. bovis BCG (B), and THP-1 cells infected with heat-killed M. bovis BCG (C). Each lane was probed with one or more antibodies. The proteins indicated by arrows are discussed in the text. Lanes 1 and 21 in each panel contained molecular size standards. Antibodies against the phosphorylated proteins were as follows: lanes 2, NR1 (S896); lanes 3, adducin (S724) and CDK1 (Y15); lanes 4, CREB (S133); lanes 5, ERK1/2 (T202/Y204) (T183/Y185) and p70 S6K (T389); lanes 6, RSK (T360/S364), RAF1 (S259), and MEK1/2 (S217/S221); lanes 7, GSK3α (S21), GSK3β (S9), and PKB (T308); lanes 8, GSK3α (Y279/Y216); lanes 9, PKR1 (T451); lanes 10, PKCɛ (S719); lanes 11, PKCα (S567); lanes 12, PKCα/β (T638) and SRC (Y529); lanes 13, PKCδ (T505); lanes 14, SAPK (T183/Y185); lanes 15, MSK 1/2 (S376) and JUN (S73); lanes 16, JAK2 (Y1007/Y1008) and p38MAPK (T180/Y182); lanes 17, STAT1 (S701); lanes 18, STAT3 (S727); lanes 19, RB (S780); lanes 20, RB (S807/S811).
FIG. 2.
Multiphosphoprotein analysis of THP-1 cells infected with M. bovis BCG or exposed to ManLAM. As for Fig. 1, accurate intensity values for each band are the accumulated signal obtained over a given scan time for each blot. These are shown as numerical values in Table 1. The data shown here represent protein bands visualized as a snapshot of the scan time and may therefore not represent exact band intensities. The second screen, presented here, shows three separate blots: untreated THP-1 cells (A), THP-1 cells infected with M. bovis BCG (B), and THP-1 cells treated with purified ManLAM (C). The proteins indicated by arrows are discussed in the text. Lanes 1 and 21 in each panel contained molecular size standards. Antibodies against the phosphorylated proteins were as follows: lanes 2, NR1 (S896); lanes 3, adducin (S724), gamma (S662), CDK1 (Y15), and SRC (Y529); lanes 4, p38MAPK (T180/Y182) and STAT5 (Y694); lanes 5, SRC (Y418) and PKCα (T567); lanes 6, RSK (T360/S364) and SAPK (T183/Y185); lanes 7, PKCβ (T368), MEK3 (S189/T193), and MEK6 (S207/T211); lanes 8, ERK1 (T202/Y204), ERK2 (T185/Y204), and p70 S6K (T389); lanes 9, PKCɛ (S719) and SMAD1 (S463/S465); lanes 10, STAT3 (S727); lanes 11, JUN (S73); lanes 12, RAF1 (S259) and STAT1 (S701); lanes 13, CREB (S133), PKBα (T308), and PKCδ (T505); lanes 14, PKBα (S473); lanes 15, GSK3α (S21), GSK3β (S9), and MSK1/2 (S376); lanes 16, PKR1 (T451); lanes 17, GSK3α (Y279) and GSKβ (Y216); lanes 18, RB (S780); lanes 19, MEK1/2 (S221/S225); lanes 20, RB (S807/S811).
Changes in phosphorylation were measured based on the band intensity for individual phosphoproteins. Any change in phosphorylation of greater than 25% between control and treated cells was considered significant. This is justified by the high sensitivity of the screens in determining the phosphorylation state, as well as the level of each phosphoprotein present within the cells. According to the manufacturer (Kinexus), a change in phosphorylation of less than 25% could be due to experimental variation. As seen in Fig. 1, several proteins displayed significant changes in phosphorylation. These include α-adducin, the N-methyl-d-aspartate (NMDA) glutamate receptor subunit NR1, the c-Jun oncoprotein, stress-activated-protein kinase (SAPK) (also known as c-Jun N-terminal kinase), retinoblastoma protein (RB), and glycogen synthase kinase 3β (GSK3β). To examine whether the observed phosphorylation changes could be due to an effect of mycobacterial cell wall components, a second screen was applied to ManLAM-treated cells along with cells infected with live bacteria (Fig. 2). As observed in the first screen, α-adducin, c-Jun, SAPK, GSK3β, and NR1 showed increased phosphorylation in THP-1 cells exposed to live mycobacteria. Interestingly, most of these results were also seen for cells exposed to purified ManLAM.
The two screens were compared in terms of the relative fold change in phosphorylation between live infection and untreated control cells (Table 1). This discussion is limited to proteins that for each screen show more than a 25% change in phosphorylation between infected and control cells. Values for cells infected with heat-killed bacteria and cells treated with ManLAM are shown as fold changes relative to the respective control cells (Table 1). As seen in Table 1, none of the changes in phosphorylation induced by ManLAM were reproduced in cells infected with heat-killed bacteria. Since common purification methods for LAM involve heat killing of M. tuberculosis (39), one would assume that ManLAM is unaffected by heat treatment and that heat-killed bacteria retain LAM in its original structure. However, several factors could explain the differences in host cell response to ManLAM in its purified form and ManLAM in heat-killed bacteria. First, mycobacteria may shed LAM as a result of heat treatment, reducing the amount of LAM encountered by the host cell. Second, heat may induce conformational changes of bacterial surface molecules that can hinder interaction of LAM with host cell receptors. Third, heat treatment followed by killing of BCG most probably alters the active release of ManLAM once the bacteria reside within the host cell. Active release of ManLAM by live bacteria causes exposure of the whole ManLAM molecule to the host cell. It is possible that the lipid moiety of ManLAM, which is normally embedded in the mycobacterial cell wall, is required for the biological functions of LAM in its interaction with host cell signaling elements. Lipopolysaccharide, which is present on the surface of gram-negative bacteria, is an example of a molecule that shows structural similarity to LAM. The endotoxic activity of lipopolysaccharide is predominantly associated with its free form and particularly the lipid component, lipid A, which needs to be exposed for full activity (2).
TABLE 1.
Comparison of phosphorylated host proteins
| Protein
|
Epitope(s) | Signala
|
||||
|---|---|---|---|---|---|---|
| Full name | Abbreviation | Control | Fold change
|
|||
| Live avg | LAM | HK | ||||
| Alpha adducin | α-Adducin | S724 | 1 | 1.75b | 2.62 | 0.38 |
| Gamma adducin | γ-Adducin | S662 | 1 | 0.00 | 2.15 | NDc |
| Cyclic AMP response element binding protein | CREB | S133 | 1 | 0.00 | 1.00 | 0.86 |
| Cyclin-dependent kinase 1 (cdc2) | CDK1 | Y15 | 1 | 0.37 | 2.07 | 0.00 |
| Double-standard RNA-dependent protein kinase | PKR | T451 | 1 | 1.16 | 1.88 | 0.57 |
| Extracellular regulated kinase 1 | ERK1 | T202/Y204 | 0 | 0.51 | 0.00 | 1.01 |
| Extracellular regulated kinase 2 | ERK2 | T185/Y204 | 0 | 0.65 | 0.00 | 0.79 |
| Glycogen synthase kinase 3 alpha | GSK3α | Y279 | 1 | 1.29 | 1.44 | 0.82 |
| Glycogen synthase kinase 3 alpha | GSK3α | S21 | 1 | 0.92 | 1.79 | 0.36 |
| Glycogen synthase kinase 3 beta | GSK3β | Y216 | 1 | 1.57b | 1.52 | 0.65 |
| Glycogen synthase kinase 3 beta | GSK3β | S9 | 0 | 0.00 | 0.00 | 0.36 |
| Mitogen-activated protein kinase kinase 1/2 | MEK1/2 | S221/S225 | 1 | 1.85 | 1.69 | 0.39 |
| Mitogen-activated protein kinase kinase 3 | MEK3 | S189/T193 | 0 | 0.00 | 0.00 | ND |
| Mitogen-activated protein kinase kinase 6 | MEK6 | S207/T211 | 0 | 0.00 | 0.00 | ND |
| Mitogen- and stress-activated protein kinase 1/2 | MSK1/2 | S376 | 1 | 1.46 | 2.69 | 0.48 |
| Mitogen- and stress-activated protein kinase 1/2 | MSK1/2 | S376 | 1 | 2.71 | 4.01 | 0.33 |
| N-Methyl-d-aspartate glutamate receptor subunit 1 | NR1 | S896 | 1 | 1.93b | 1.24 | 0.91 |
| Oncogene JUN | JUN | S73 | 1 | 2.08b | 1.30 | 1.21 |
| Oncogene Raf 1 | RAF1 | S259 | 1 | 1.28 | 3.01 | 0.20 |
| Oncogene Raf 1 | RAF1 | S259 | 1 | 1.78 | 4.71 | 0.28 |
| Oncogene SRC | SRC | Y529 | 1 | 3.18 | 1.35 | 0.83 |
| Oncogene SRC | SRC | Y418 | 0 | 0.00 | 0.00 | ND |
| p38 alpha mitogen-activated protein kinase | p38MAPK | T180/Y182 | 1 | 4.91 | 1.10 | 0.74 |
| Protein kinase B alpha (Akt1) | PKBα | S473 | 0 | 0.00 | 0.00 | ND |
| Protein kinase B alpha (Akt1) | PKBα | T308 | 1 | 1.08 | 1.60 | 0.63 |
| Protein kinase C alpha | PKCα | S657 | 1 | 1.61 | 1.93 | 0.55 |
| Protein kinase C alpha/beta | PKCα/β | T638/641 | 1 | 1.69 | 2.82 | 0.51 |
| Protein kinase C delta | PKCδ | T505 | 1 | 1.28 | 2.59 | 0.43 |
| Protein kinase C epsilon | PKCε | S719 | 1 | 0.00b | 1.21 | 0.00 |
| Retinoblastoma 1 | RB | S780 | 1 | 0.82 | 1.85 | 0.64 |
| Retinoblastoma 1 | RB | S807/S811 | 1 | 0.35 | 2.05 | 0.54 |
| Ribosomal S6 kinase 1 | RSK1 | T360/S364 | 0 | 0.53 | 0.00 | 0.31 |
| S6 kinase p70 | p70 S6K | T389 | 1 | 2.40 | 3.93 | 0.57 |
| S6 kinase p70 | p70 S6K | T389 | 1 | 2.60 | 4.27 | 0.71 |
| Signal transducer and activator of transcription 1 | STAT1 | S701 | 1 | 2.05 | 3.02 | 0.80 |
| Signal transducer and activator of transcription 3 | STAT3 | S727 | 1 | 1.93 | 2.95 | 0.00 |
| Signal transducer and activator of transcription 5 | STAT5 | Y694 | 0 | 0.00 | 0.00 | ND |
| SMA- and MAD-related protein 1 | SMAD1 | S463/465 | 0 | 0.00 | 0.00 | ND |
| Stress-activated protein kinase (JNK) | SAPK | T183/Y185 | 1 | 2.55b | 3.07 | 0.68 |
| Stress-activated protein kinase (JNK) | SAPK | T183/Y185 | 1 | 3.36b | 1.85 | 0.80 |
The trace quantity of each protein band is measured by the area under its intensity profile curve and corrected for the individual scan times (recorded time before saturation occurs). Values for the control samples have been set to 1 or 0. A value 0 indicates that no immunoreactive signal was detected for this protein in either of the two screens. Live average is the average value, expressed as fold change, for the difference in phosphorylation between live infection and respective control samples for both screens. Values for LAM and heat-killed bacteria (HK) show the fold change relative to their respective control samples.
Proteins that showed a similar and greater-than-25% increase in phosphorylation for both screens.
ND, not determined.
Five host signaling proteins, i.e., NR1, SAPK, c-Jun, GSK3β, and α-adducin (Fig. 1 and 2 and Table 1) showed more than a 25% increase in phosphorylation over that for the control untreated cells in both screens. Conversely, the basal level of PKCɛ phosphorylation was completely attenuated in cells infected with either live or killed bacteria, indicating a possible deactivation of this protein upon infection.
NR1 is a principle subunit of the NMDA receptors, which represent a major class of glutamate-gated ion channels in the central nervous system (60). These receptors regulate intracellular levels of Ca2+ in neuronal cells and also in nonneuronal cell lines when these cells are transfected with recombinant receptors (17, 34, 35, 74). A complex regulatory machinery involving several protein kinases and phosphatases controls the function of NMDA receptors in neurons (for a review, see reference 55). S896 of the NR1 subunit is phosphorylated upon infection with BCG and has been identified as a specific phosphorylation site for PKC (71). Controversy exists with regard to the consequences of PKC phosphorylation of the NR1 subunit. PKC phosphorylation has been shown to both enhance and inhibit NMDA receptor currents, depending on cell type and compositional variation of the receptor itself (for a review, see reference 33). In our system, cells infected with live bacteria show a twofold average increase in the phosphorylation of NR1 S896 above levels for cells infected with heat-killed bacteria and cells exposed to ManLAM (Table 1). The possible role of NR1 expression in THP-1 cells is not clear; to our knowledge, NMDA receptors have not yet been described for THP-1 cells or any other cells from the hemopoietic cell linage. Our results indicate that an NR1 homologue could be present in THP-1 cells, possibly constituting an NMDA-related receptor. If so, this receptor might have a function similar to that described for neurons, i.e., regulation of intracellular Ca2+ levels. Given that M. tuberculosis is able to inhibit Ca2+ signaling in human macrophages, correlating with its intracellular survival (56), it is tempting to speculate that mycobacterial interference with a potential NMDA-related receptor could be a mediating factor for the inhibition of host Ca2+ signaling upon infection.
SAPK showed a two to threefold increase in phosphorylation in cells infected with live bacteria or exposed to ManLAM compared to heat-killed and untreated cells (Table 1). SAPK is a member of the MAPK family and is encoded by three genes, SAPK1, SAPK2, and SAPK3 (37). All SAPK genes are expressed as 46- and 54-kDa protein kinases (37), and both SAPK isoforms showed a similar increase in phosphorylation in our system. Several transcription factors, including ATF-2, Ets, and c-Jun have been identified as downstream targets for SAPK (for a review, see reference 48). Specifically, SAPK has been found to bind the c-Jun transactivation domain and phosphorylate it on S63 and S73, thereby enhancing transcriptional activity (25, 42). c-Jun is a central component of activator protein 1 complexes (16), which upon transcriptional activation have been associated with a variety of cellular functions, including cell proliferation, tumorigenesis, and apoptosis (for a review, see reference 28). In this study we show increased phosphorylation of c-Jun S73 in cells infected with live bacteria, as well as in cells exposed to ManLAM. Activation of SAPK upon mycobacterial infection has been shown to be an early response in several model systems, often in concert with p38 MAPK and ERK 1/2 activation (11, 73). Even though a regulatory role of the SAPK signaling pathway has been suggested for the production of nitric oxide in mouse macrophages (14), the role of SAPK in mycobacterial infection remains unclear. Our results are the first to indicate that LAM triggers a signaling cascade leading to activation of SAPK and its downstream effector c-Jun.
The phosphoprotein GSK3β showed an average increase of 57% for Y216 phosphorylation in THP-1 cells infected with live bacteria and in cells exposed to LAM. GSK3 has been shown to play an essential role in the regulation of cell fate in both pro- and antiapoptotic manners. For example, GSK3β gene disruption in mice caused hepatocyte apoptosis and correlated with impaired antiapoptotic NF-κB signaling (44). In contrast, phosphatidylinositol 3-kinase-mediated activation of protein kinase B (Akt) was found to induce cell survival by inhibiting GSK3 (22). In agreement with the latter observation, Akt has been shown to inhibit GSK3β activity through phosphorylation of its Ser9 residue (27, 29). Conversely, phosphorylation of GSK3β Y216 is critical for full activation of the enzyme (46) and has been shown to induce apoptosis (10). Our results demonstrate activation of GSK3β through increased phosphorylation of Y216 upon infection with live bacteria and exposure to ManLAM. Furthermore, our results showed no significant activation of Akt or Ser9 phosphorylation of GSK3β in infected cells (Table 1) suggesting that mycobacterial infection primes THP-1 cells for apoptosis via activation of GSK3β, while the antiapoptotic pathway remains silent.
The two phosphoscreens showed no phosphorylation of PKCɛ in cells infected with live bacteria. In contrast, control cells and cells exposed to LAM showed substantial PKCɛ phosphorylation. The function of PKC and the specificities of the different isoforms in macrophage signaling and mycobacterial infection are still unclear. However, PKCɛ has been shown to be important in macrophage activation and defense against bacterial infection (13). Macrophages from PKCɛ−/− mice showed severely attenuated responses to lipopolysaccharide and gamma interferon, characterized by a dramatic decrease in the generation of nitric oxide, tumor necrosis factor alpha, and interleukin-1β (13). Our results indicate attenuation of PKCɛ activity, which could be regarded as a strategy employed by mycobacteria to avoid activation of the macrophage.
A major finding in this study is the phosphorylation of the cytoskeletal protein α-adducin (Table 1). Phosphorylation of α-adducin showed an average increase of 75% in cells infected with live bacteria compared to those infected with heat-killed bacteria. Owing to its status as a unique protein that has not previously been investigated with respect to mycobacterial infection, α-adducin was selected for further analysis. Western blot analysis of α-adducin phosphorylation confirmed our screening results and showed that α-adducin is phosphorylated in THP-1 cells infected with live M. bovis BCG but not in cells infected with heat-killed M. bovis BCG (Fig. 3A). Furthermore, adducin phosphorylation increased with time, to reach a maximum at 24 h (Fig. 3B). Treatment of cells with purified ManLAM caused an increase in α-adducin phosphorylation, and further investigation showed that ManLAM induces phosphorylation in a dose-dependent manner (Fig. 3C). We found that a concentration of as low as 0.01 μg/ml was sufficient to induce phosphorylation of α-adducin.
FIG. 3.
Western analysis of adducin phosphorylation in THP-1 cells. (A) Adducin is phosphorylated in cells infected with BCG but not in cells infected with heat-killed bacteria. Panel 1, Western blot of THP-1 cells infected with live or heat-killed (HK) BCG for 24 h; panel 2, densitometry analysis of panel 1. (B) Time-dependent phosphorylation of adducin by M. bovis BCG infection. Panel 1, Western blot of THP-1 cells infected with BCG for 2, 12, or 24 h; panel 2, densitometry analysis of panel 1. (C) Adducin phosphorylation by purified ManLAM. Panel 1, Western blot of THP-1 cells treated with increasing concentrations of purified ManLAM; panel 2, densitometry analysis of panel 1. Error bars represent standard errors from at least three independent experiments. Equal loading of protein was verified by India ink staining.
Adducin is expressed as a tetramer of either α/β or β/γ subunits (26, 50). A myristolated alanine-rich C kinase-related domain (1, 12), present in the C termini of the adducin subunits, constitutes the major phosphorylation site for PKC (58). The S724 of α-adducin detected in our screens lies within the myristolated alanine-rich C kinase domain, suggesting PKC as an upstream kinase acting on α-adducin. In support of this, we observed increased phosphorylation of α-adducin upon treatment of cells with the PKC activator PMA (results not shown).
Phosphorylation of adducin by PKC has been shown to interfere with organization of the actin filament network through inhibition of actin capping and prevention of spectrin recruitment to the ends of actin filaments (58). In a pathogenic setting, the cytoskeletal network, including microtubules, microfilaments, and actin filaments, plays a crucial role in facilitating fusion between early endosomes, phagosomes, and other organelles of the endocytic pathway (for a review, see reference 9). Although the participation of actin filaments in phagocytosis of microorganisms has been well characterized and studied in several model systems, the involvement of actin and actin-binding proteins in the maturation and fusion of phagosomes is still unclear. Nevertheless, several lines of evidence assign an important role for actin filaments in the postphagocytotic fate of nascent phagosomes. For example, treatment with the actin-depolymerizing agent cytochalasin D has been shown to inhibit phagosomal transport and fusion events along the endosomal pathway (49, 72). Furthermore, actin, as well as several actin-binding proteins, has been found in association with mature phagosomes (24, 52, 67). Unambiguous evidence for the role of actin in phagosomal motility was provided by video microscopy of murine macrophages, which showed the formation of actin-rich rocket tails behind latex bead-containing phagosomes (77). These reports suggest a role for the actin filament network beyond the initial phagocytic uptake, during phagosomal motility and processing.
The actin network of phagocytic cells has been identified as a target for a wide range of pathogenic bacteria, including Salmonella, Shigella, Listeria, and Legionella (18, 21, 40, 70). These pathogens have been shown to manipulate the actin cytoskeleton in several ways to either enhance their uptake by mammalian cells or facilitate their own movement through host cell cytoplasm and eventually into neighboring cells. Therefore, it can be hypothesized that pathogenic species of mycobacteria would take advantage of such a mechanism. Indeed, virulent strains of Mycobacterium avium have been shown to disrupt the host actin network, correlating with a delay in the acquisition of endocytic markers by mycobacterium-containing phagosomes (36). It was shown that disruption of the actin network occurred at 24 h postinfection and was maintained up to 15 days following infection. In agreement with this observation, our results also demonstrate a time-dependent mechanism for the phosphorylation of adducin, with the highest phosphorylation occurring at 24 h postinfection (Fig. 3B)
As proposed by Guerin and de Chastellier (36), actin filaments, along with their associated proteins, may function as a network surrounding nascent and maturing phagosomes. Here, organelles of the endocytic machinery are brought into proximity to facilitate fusion events and intermingling of contents between compartments. By disrupting such a network, invading mycobacteria can possibly prevent exchange of content between organelles and inhibit eventual fusion with lysosomes. In this context, adducin phosphorylation, mediated by mycobacterial factors such as LAM, could possibly lead to the disruption or reorganization of the actin cytoskeleton, enabling exclusion of the phagosome from the endocytic pathway.
Changes in phosphorylation seen for SAPK, c-Jun, α-adducin, and GSK3β upon infection with M. bovis BCG have been reproduced in cells exposed to ManLAM, suggesting ManLAM to be the mediating factor. In contrast, changes in phosphorylation of PKCɛ and NR1 were not reproduced in cells exposed to ManLAM, indicating that another bacterial component is responsible. In murine macrophages, phagosomes containing M. bovis BCG were shown to be permeable to dextrans as large as 70 kDa (69), and a number of mycobacterial surface proteins are released from phagosomes into subcellular compartments (7). Thus, the notion that mycobacteria are capable of releasing proteins into the host cell cytoplasm led us to hypothesize that mycobacteria actively interfere with host signaling elements to promote their own survival. With the presence of genes for 11 eukaryotic-like protein serine kinases and four protein phosphatases in the genome of M. tuberculosis (5), it is tempting to speculate that these proteins might be exported intracellulary and interfere with signal transduction cascades within the host cell. Eukaryotic-like protein kinases and phosphatases have previously been implicated in the virulence of other pathogens such as Yersinia pseudotuberculosis and Salmonella enterica serovar Typhimurium, both of which translocate bacterial proteins into the host cell cytoplasm, resulting in disruption of the host cell cytoskeleton (32, 38). Interestingly, a mycobacterial phosphatase, PtpA, is present in the mycobacterial genome without a corresponding substrate within the bacterium (20), further supporting the hypothesis of cross-interaction between bacterial and host cell signaling elements.
In conclusion, we have presented a unique set of results based on a simultaneous screen of key host proteins upon mycobacterial infection of a human macrophage-like cell line. We have shown changes in phosphorylation of host proteins that are novel to the field of cellular mycobacteriology. As described above, some of the signaling proteins shown to be activated, such as SAPK, c-Jun, and GSK3β, have previously been implicated in the regulation of apoptotic pathways. It has been well established that macrophages infected with mycobacteria have increased rates of apoptosis in vitro (30, 64, 66). The observation that virulent strains induce less apoptosis than avirulent and attenuated strains (6, 51) has reinforced the conception that apoptosis functions as a host cell defense mechanism in mycobacterial infection and that virulent strains have developed strategies to promote host cell survival. M. bovis BCG was recently shown to induce apoptosis in THP-1 cells (64). In agreement with this, activation of the JNK-c-Jun signaling pathway and GSK3β could be interpreted as proapoptotic signaling as part of the host cell defense. However, since we have not analyzed downstream effects of the reported phosphorylation events, we cannot draw any final conclusions regarding the ultimate outcome of these events. On the other hand, we have presented evidence to suggest an active attenuation of macrophage defense through mycobacterial inhibition of PKCɛ and possible interference with host Ca2+ signaling. Furthermore, we propose a potential mechanism by which mycobacteria interfere with the actin cytoskeleton as a means to exclude the phagosome from the endocytic pathway. Taken together, our results present evidence in accordance with previous reports indicating that a number of host signaling pathways are modulated upon mycobacterial infection. Whether this is due to a single modulation event affecting several downstream signaling pathways or to direct modulation of different signaling cascades remains to be elucidated. Further research into these pathways will lead to increased knowledge about the complex interplay between mycobacteria and the host cell.
Acknowledgments
We thank Julian Davies and Yaffa Mizrahi for their critical reading of the manuscript.
This work was supported by Canadian Institute of Health Research (CIHR) grants MOP-43941 (to Y.A.-G.) and MOP-43891 (to Z.H.) and by funding from the TB Veterans Charitable Foundation. Y.A.-G. is a CIHR and British Columbia Lung Association Scholar. Z.H. was supported by salary awards from CIHR and the Michael Smith Foundation for Health Research. Colorado State University and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, supplied material under contract NO1 AI-75320 (Tuberculosis Research Materials and Vaccine Testing).
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Aderem, A. 1992. The MARCKS brothers: a family of protein kinase C substrates. Cell 71:713-716. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, C., and E. T. Rietschel. 2001. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7:167-202. [PubMed] [Google Scholar]
- 3.Armstrong, J. A., and P. D. Hart. 1971. Response of cultured macrophages to M. tuberculosis with observations of fusion of lysosomes with phagosomes. J. Exp. Med. 134:713-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auwerx, J. 1991. The human leukemia cell line, THP-1: a multifaceted model for the study of monocyte-macrophage differentiation. Experientia 47:22-31. [DOI] [PubMed] [Google Scholar]
- 5.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 6.Balcewicz-Sablinska, M. K., J. Keane, H. Kornfeld, and H. G. Remold. 1998. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-alpha. J. Immunol. 161:2636-2641. [PubMed] [Google Scholar]
- 7.Beatty, W. L., and D. G. Russell. 2000. Identification of mycobacterial surface proteins released into subcellular compartments of infected macrophages. Infect. Immun. 68:6997-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 9.Beron, W. A.-D. C. M. L. S. PD. 1995. Membrane trafficking along the phagocytic pathway. Trends Cell Biol. 5:100-104. [DOI] [PubMed]
- 10.Bhat, R. V., J. Shanley, M. P. Correll, W. E. Fieles, R. A. Keith, C. W. Scott, and C. M. Lee. 2000. Regulation and localization of tyrosine216 phosphorylation of glycogen synthase kinase-3beta in cellular and animal models of neuronal degeneration. Proc. Natl. Acad. Sci. USA 97:11074-11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya, A., S. Pathak, M. Kundu, and J. Basu. 2002. Mitogen-activated protein kinases regulate Mycobacterium avium-induced tumor necrosis factor-alpha release from macrophages. FEMS Immunol. Med. Microbiol. 34:73. [DOI] [PubMed] [Google Scholar]
- 12.Blackshear, P. J. 1993. The MARCKS family of cellular protein kinase C substrates. J. Biol. Chem. 268:1501-1504. [PubMed] [Google Scholar]
- 13.Castrillo, A., D. J. Pennington, F. Otto, P. J. Parker, M. J. Owen, and L. Bosca. 2001. Protein kinase Cepsilon is required for macrophage activation and defense against bacterial infection. J. Exp. Med. 194:1231-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, E. D., K. R. Morris, J. T. Belisle, P. Hill, L. K. Remigio, P. J. Brennan, and D. W. Riches. 2001. Induction of inducible nitric oxide synthase-NO* by lipoarabinomannan of Mycobacterium tuberculosis is mediated by MEK1-ERK, MKK7-JNK, and NF-κB signaling pathways. Infect. Immun. 69:2001-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, J., X. D. Fan, S. W. Hunter, P. J. Brennan, and B. R. Bloom. 1991. Lipoarabinomannan, a possible virulence factor involved in persistence of Mycobacterium tuberculosis within macrophages. Infect. Immun. 59:1755-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 17.Choi, D. W. 1987. Ionic dependence of glutamate neurotoxicity. J. Neurosci. 7:369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cossart, P. 1995. Actin-based bacterial motility. Curr. Opin. Cell Biol. 7:94-101. [DOI] [PubMed] [Google Scholar]
- 19.Cowley, S. C., and Y. Av-Gay. 2001. Monitoring promoter activity and protein localization in Mycobacterium spp. using green fluorescent protein. Gene 264:225-231. [DOI] [PubMed] [Google Scholar]
- 20.Cowley, S. C., R. Babakaiff, and Y. Av-Gay. 2002. Expression and localization of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Res. Microbiol. 153:233-241. [DOI] [PubMed] [Google Scholar]
- 21.Coxon, P. Y., J. T. Summersgill, J. A. Ramirez, and R. D. Miller. 1998. Signal transduction during Legionella pneumophila entry into human monocytes. Infect. Immun. 66:2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross, D. A., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785-789. [DOI] [PubMed] [Google Scholar]
- 23.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 24.Defacque, H., M. Egeberg, A. Habermann, M. Diakonova, C. Roy, P. Mangeat, W. Voelter, G. Marriott, J. Pfannstiel, H. Faulstich, and G. Griffiths. 2000. Involvement of ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J. 199-212. [DOI] [PMC free article] [PubMed]
- 25.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 26.Dong, L., C. Chapline, B. Mousseau, L. Fowler, K. Ramsay, J. L. Stevens, and S. Jaken. 1995. 35H, a sequence isolated as a protein kinase C binding protein, is a novel member of the adducin family. J. Biol. Chem. 270:25534-25540. [DOI] [PubMed] [Google Scholar]
- 27.Dudek, H., S. R. Datta, T. F. Franke, M. J. Birnbaum, R. Yao, G. M. Cooper, R. A. Segal, D. R. Kaplan, and M. E. Greenberg. 1997. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275:661-665. [DOI] [PubMed] [Google Scholar]
- 28.Dunn, C., C. Wiltshire, A. MacLaren, and D. A. Gillespie. 2002. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 14:585-593. [DOI] [PubMed] [Google Scholar]
- 29.Franke, T. F., D. R. Kaplan, and L. C. Cantley. 1997. PI3K: downstream AKTion blocks apoptosis. Cell 88:435-437. [DOI] [PubMed] [Google Scholar]
- 30.Fratazzi, C., R. D. Arbeit, C. Carini, and H. G. Remold. 1997. Programmed cell death of Mycobacterium avium serovar 4-infected human macrophages prevents the mycobacteria from spreading and induces mycobacterial growth inhibition by freshly added, uninfected macrophages. J. Immunol. 158:4320-4327. [PubMed] [Google Scholar]
- 31.Fratti, R. A., J. Chua, I. Vergne, and V. Deretic. 2003. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. USA 100:5437-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 33.Grant, E. R., B. J. Bacskai, N. J. Anegawa, D. E. Pleasure, and D. R. Lynch. 1998. Opposing contributions of NR1 and NR2 to protein kinase C modulation of NMDA receptors. J. Neurochem. 71:1471-1481. [DOI] [PubMed] [Google Scholar]
- 34.Grant, E. R., B. J. Bacskai, D. E. Pleasure, D. B. Pritchett, M. J. Gallagher, S. J. Kendrick, L. J. Kricka, and D. R. Lynch. 1997. N-Methyl-d-aspartate receptors expressed in a nonneuronal cell line mediate subunit-specific increases in free intracellular calcium. J. Biol. Chem. 272:647-656. [DOI] [PubMed] [Google Scholar]
- 35.Grimwood, S., E. Gilbert, C. I. Ragan, and P. H. Hutson. 1996. Modulation of 45Ca2+ influx into cells stably expressing recombinant human NMDA receptors by ligands acting at distinct recognition sites. J. Neurochem. 66:2589-2595. [DOI] [PubMed] [Google Scholar]
- 36.Guerin, I., and C. de Chastellier. 2000. Disruption of the actin filament network affects delivery of endocytic contents marker to phagosomes with early endosome characteristics: the case of phagosomes with pathogenic mycobacteria. Eur. J. Cell Biol. 79:735-749. [DOI] [PubMed] [Google Scholar]
- 37.Gupta, S., T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Derijard, and R. J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760-2770. [PMC free article] [PubMed] [Google Scholar]
- 38.Hakansson, S., E. E. Galyov, R. Rosqvist, and H. Wolf-Watz. 1996. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol. Microbiol. 20:593-603. [DOI] [PubMed] [Google Scholar]
- 39.Hamasur, B., G. Kallenius, and S. B. Svenson. 1999. A new rapid and simple method for large-scale purification of mycobacterial lipoarabinomannan. FEMS Immunol. Med. Microbiol. 24:11-17. [DOI] [PubMed] [Google Scholar]
- 40.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 41.Hasan, Z., C. Schlax, L. Kuhn, I. Lefkovits, D. Young, J. Thole, and J. Pieters. 1997. Isolation and characterization of the mycobacterial phagosome: segregation from the endosomal/lysosomal pathway. Mol. Microbiol. 24:545-553. [DOI] [PubMed] [Google Scholar]
- 42.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7:2135-2148. [DOI] [PubMed] [Google Scholar]
- 43.Hmama, Z., R. Gabathuler, W. A. Jefferies, G. de Jong, and N. E. Reiner. 1998. Attenuation of HLA-DR expression by mononuclear phagocytes infected with Mycobacterium tuberculosis is related to intracellular sequestration of immature class II heterodimers. J. Immunol. 161:4882-4893. [PubMed] [Google Scholar]
- 44.Hoeflich, K. P., J. Luo, E. A. Rubie, M. S. Tsao, O. Jin, and J. R. Woodgett. 2000. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 406:86-90. [DOI] [PubMed] [Google Scholar]
- 45.Honer zu Bentrup, K., and D. G. Russell. 2001. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9:597-605. [DOI] [PubMed] [Google Scholar]
- 46.Hughes, K., E. Nikolakaki, S. E. Plyte, N. F. Totty, and J. R. Woodgett. 1993. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 12:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussain, S., B. S. Zwilling, and W. P. Lafuse. 1999. Mycobacterium avium infection of mouse macrophages inhibits IFN-gamma Janus kinase-STAT signaling and gene induction by down-regulation of the IFN-gamma receptor. J. Immunol. 163:2041-2048. [PubMed] [Google Scholar]
- 48.Ip, Y. T., and R. J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr. Opin. Cell Biol. 10:205-219. [DOI] [PubMed] [Google Scholar]
- 49.Jahraus, A., M. Egeberg, B. Hinner, A. Habermann, E. Sackman, A. Pralle, H. Faulstich, V. Rybin, H. Defacque, and G. Griffiths. 2001. ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol. Biol. Cell 12:155-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joshi, R., D. M. Gilligan, E. Otto, T. McLaughlin, and V. Bennett. 1991. Primary structure and domain organization of human alpha and beta adducin. J. Cell Biol. 115:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keane, J., H. G. Remold, and H. Kornfeld. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016-2020. [DOI] [PubMed] [Google Scholar]
- 52.Kersken, H., J. Vilmart-Seuwen, M. Momayezi, and H. Plattner. 1986. Filamentous actin in Paramecium cells: mapping by phalloidin affinity labeling in vivo and in vitro. J. Histochem. Cytochem. 34:443-454. [DOI] [PubMed] [Google Scholar]
- 53.Knutson, K. L., Z. Hmama, P. Herrera-Velit, R. Rochford, and N. E. Reiner. 1998. Lipoarabinomannan of Mycobacterium tuberculosis promotes protein tyrosine dephosphorylation and inhibition of mitogen-activated protein kinase in human mononuclear phagocytes. Role of the Src homology 2 containing tyrosine phosphatase 1. J. Biol. Chem. 273:645-652. [DOI] [PubMed] [Google Scholar]
- 54.Kostich, M., J. English, V. Madison, F. Gheyas, L. Wang, P. Qiu, J. Greene, and T. M. Laz. 2002. Human members of the eukaryotic protein kinase family. Genome Biol. 3:RESEARCH0043. [DOI] [PMC free article] [PubMed]
- 55.Lu, W. Y., M. F. Jackson, D. Bai, B. A. Orser, and J. F. MacDonald. 2002. In CA1 pyramidal neurons of the hippocampus protein kinase C regulate calcium-dependent inactivation of NMDA receptors. J. Neurosci. 20:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malik, Z. A., G. M. Denning, and D. J. Kusner. 2000. Inhibition of Ca(2+) signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. J. Exp. Med. 191:287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik, Z. A., S. S. Iyer, and D. J. Kusner. 2001. Mycobacterium tuberculosis phagosomes exhibit altered calmodulin-dependent signal transduction: contribution to inhibition of phagosome-lysosome fusion and intracellular survival in human macrophages. J. Immunol. 166:3392-3401. [DOI] [PubMed] [Google Scholar]
- 58.Matsuoka, Y., X. Li, and V. Bennett. 1998. Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons. J. Cell Biol. 142:485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monahan, I. M., J. Betts, D. K. Banerjee, and P. D. Butcher. 2001. Differential expression of mycobacterial proteins following phagocytosis by macrophages. Microbiology 147:459-471. [DOI] [PubMed] [Google Scholar]
- 60.Moriyoshi, K., M. Masu, T. Ishii, R. Shigemoto, N. Mizuno, and S. Nakanishi. 1991. Molecular cloning and characterization of the rat NMDA receptor. Nature 354:31-37. [DOI] [PubMed] [Google Scholar]
- 61.Morris, K. R., R. D. Lutz, H. S. Choi, T. Kamitani, K. Chmura, and E. D. Chan. 2003. Role of the NF-κB signaling pathway and κB cis-regulatory elements on the IRF-1 and inducible nitric oxide synthase promoter regions in mycobacterial lipoarabinomannan induction of nitric oxide. Infect. Immun. 71:1442-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parrish, N. M., J. D. Dick, and W. R. Bishai. 1998. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107-112. [DOI] [PubMed] [Google Scholar]
- 63.Pelech, S., and H. Zhang. 2002. Plasticity of the kinomes in monkey and rat tissues. Sci. STKE 162:PE50. [Online.] [DOI] [PubMed] [Google Scholar]
- 64.Riendeau, C. J., and H. Kornfeld. 2003. THP-1 cell apoptosis in response to mycobacterial infection. Infect. Immun. 71:254-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roach, S. K., and J. S. Schorey. 2002. Differential regulation of the mitogen-activated protein kinases by pathogenic and nonpathogenic mycobacteria. Infect. Immun. 70:3040-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rojas, M., M. Olivier, P. Gros, L. F. Barrera, and L. F. Garcia. 1999. TNF-alpha and IL-10 modulate the induction of apoptosis by virulent Mycobacterium tuberculosis in murine macrophages. J. Immunol. 162:6122-6131. [PubMed] [Google Scholar]
- 67.Stockem, W., H. U. Hoffmann, and B. Gruber. 1983. Dynamics of the cytoskeleton in Amoeba proteus. I. Redistribution of microinjected fluorescein-labeled actin during locomotion, immobilization and phagocytosis. Cell Tissue Res. 232:79-96. [DOI] [PubMed] [Google Scholar]
- 68.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 69.Teitelbaum, R., M. Cammer, M. L. Maitland, N. E. Freitag, J. Condeelis, and B. R. Bloom. 1999. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc. Natl. Acad. Sci. USA 96:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Theriot, J. A. 1995. The cell biology of infection by intracellular bacterial pathogens. Annu. Rev. Cell Dev. Biol. 11:213-239. [DOI] [PubMed] [Google Scholar]
- 71.Tingley, W. G., M. D. Ehlers, K. Kameyama, C. Doherty, J. B. Ptak, C. T. Riley, and R. L. Huganir. 1997. Characterization of protein kinase A and protein kinase C phosphorylation of the N-methyl-d-aspartate receptor NR1 subunit using phosphorylation site-specific antibodies. J. Biol. Chem. 272:5157-5166. [DOI] [PubMed] [Google Scholar]
- 72.Toyohara, A., and K. Inaba. 1989. Transport of phagosomes in mouse peritoneal macrophages. J. Cell Sci. 94:143-153. [DOI] [PubMed] [Google Scholar]
- 73.Tse, H. M., S. I. Josephy, E. D. Chan, D. Fouts, and A. M. Cooper. 2002. Activation of the mitogen-activated protein kinase signaling pathway is instrumental in determining the ability of Mycobacterium avium to grow in murine macrophages. J. Immunol. 168:825-833. [DOI] [PubMed] [Google Scholar]
- 74.Tymianski, M., M. P. Charlton, P. L. Carlen, and C. H. Tator. 1993. Source specificity of early calcium neurotoxicity in cultured embryonic spinal neurons. J. Neurosci. 13:2085-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Via, L. E., D. Deretic, R. J. Ulmer, N. S. Hibler, L. A. Huber, and V. Deretic. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272:13326-13331. [DOI] [PubMed] [Google Scholar]
- 76.World Health Organization.2002. World health report. World Health Organization, Geneva, Switzerland.
- 77.Zhang, F., F. S. Southwick, and D. L. Purich. 2002. Actin-based phagosome motility. Cell Motil. Cytoskeleton 53:81-88. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, H., X. Shi, Q. J. Zhang, M. Hampong, H. Paddon, D. Wahyuningsih, and S. Pelech. 2002. Nocodazole-induced p53-dependent c-Jun N-terminal kinase activation reduces apoptosis in human colon carcinoma HCT116 cells. J. Biol. Chem. 277:43648-43658. [DOI] [PubMed] [Google Scholar]