Abstract
Phase variation in the colonial opacity phenotype of Streptococcus pneumoniae has been implicated as a factor in bacterial adherence, colonization, and invasion in the pathogenesis of pneumococcal otitis media (OM). The purpose of this study was to determine whether S. pneumoniae opacity variants influence the induction of gene expression for proinflammatory mediators in vivo using the rat model of OM. Both the opaque and transparent phenotype variants induced a significant up-regulation in gene expression for interleukin-1α (IL-1α), IL-1β, IL-6, IL-10, tumor necrosis factor alpha, and inducible nitric oxide synthase (iNOS) compared to saline sham-inoculated controls at both 4 and 24 h postinoculation (P < 0.05 in all cases). Furthermore, whereas a significant difference in gene expression was evident for only IL-6 (greater following challenge with the opaque variant) and IL-1β (greater following challenge with the transparent variant) at 4 h, by 24 h the opaque variant cohort demonstrated a significant increase in gene expression for IL-1α, IL-1β, IL-6, IL-10, and iNOS relative to animals inoculated with the transparent phenotype variant (P < 0.05 in all cases). Enzyme-linked immunosorbent assay results confirmed the gene expression data as determined by real-time PCR. Moreover, the concentrations of the opaque variant in the middle ear lavage fluid were a full log higher than those of the transparent variant. The aforementioned results indicate that the opaque phenotype variant is more efficient at survival and multiplication within the middle ear space, resulting in the accumulation of more inflammatory cells and the enhanced expression and production of inflammatory mediators. However, when the data were normalized to account for differences in middle ear bacterial titers, it became apparent that the transparent variant of S. pneumoniae is a more potent inducer of inflammation, triggering the accumulation of more inflammatory cells and substantially greater fold increases in the expression and production of inflammatory mediators. Data from this study indicate that S. pneumoniae opacity variants influence the temporal mRNA expression of inflammatory mediators within the middle ear.
Streptococcus pneumoniae undergoes spontaneous phase variation in colony morphology between a transparent and an opaque colony phenotype. It was Weiser and colleagues who first described the relationship between colonial opacity and nasopharyngeal colonization and adherence in an infant rat model of carriage (24). Transparent variants, which have more cell wall teichoic acid than opaque S. pneumoniae (14), demonstrate an increased ability to adhere to human lung epithelial cells and are selected for during nasopharyngeal colonization in rodent models but are unable to induce sepsis (6). In contrast, opaque variants have more capsular polysaccharide than transparent S. pneumoniae and are characteristically more virulent and associated with invasive disease (14). Given that populations of S. pneumoniae are a heterogenous mixture of opaque and transparent organisms, phase variation provides S. pneumoniae with a unique advantage in vivo (23). Each phase has characteristics which provide a selective advantage for either carriage or systemic infection.
How S. pneumoniae, the primary bacterial pathogen of otitis media (OM), becomes established in the middle ear and induces OM is not completely understood. It has been suggested that the successful attachment to host epithelium during colonization and invasion by S. pneumoniae requires either the recognition of novel receptors or an increase of existing receptors on activated host cells (20). The up-regulation of receptors on respiratory epithelial cells has been reported to occur subsequent to viral infection (10) or following epithelial activation with the chemokine thrombin or with the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1α (IL-1 α) (4, 5, 6, 8). Data from our laboratory's recent study indicate that the middle ear epithelium appears to play a key role in host immune defense by recognizing and subsequently responding to invading pathogens by secretion of proinflammatory cytokines (19). In the case of OM, it is the bacterium-epithelium interaction and the subsequent release of proinflammatory cytokines and other mediators which possibly facilitates the invasion process.
Cytokines have been identified in the middle ear effusions from patients with OM (17, 25). Furthermore, studies utilizing animal models of OM have characterized the expression of inflammatory cytokines and inducible nitric oxide synthase (iNOS) subsequent to challenge with S. pneumoniae (11, 16, 18). Hebda et al. found that the expression levels of TNF-α, IL-6, IL-1, and IL-10 as well as iNOS were significantly induced in the rat model of OM following transbullar inoculation with S. pneumoniae (11). However, to our knowledge, no study has been reported which examines how S. pneumoniae opacity variants impact the initial stages of the host-parasite interaction within the middle ear. Specifically, do transparent and opaque phase variants of S. pneumoniae differentially influence the temporal expression of inflammatory mediators? The purpose of this study was to analyze how S. pneumoniae opacity variants influence cytokine and iNOS expression and production as well as inflammatory cell recruitment and bacterial survival in vivo following transbullar injection by using the rat model of OM.
MATERIALS AND METHODS
Bacteria.
S. pneumoniae 6A (EF3114; kindly provided by B. Andersson, Department of Clinical Immunology, University of Gotebörg, Gotebörg, Sweden) was used for these experiments and has been described in detail previously (1). The isogenic opaque or transparent variants of S. pneumoniae 6A were isolated by Jeffrey Weiser, Children's Hospital of Philadelphia, and confirmed prior to inoculation according to the method established by Weiser et al. (24). Log-phase cultures were prepared from chocolate agar plate subcultures by inoculating Todd-Hewitt broth supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, Mich.) with S. pneumoniae 6A opacity variants grown overnight on chocolate agar and obtained by washing the plates with 5 ml of phosphate-buffered saline (PBS; pH 7.2). After a 3-h incubation, the cultures were centrifuged at 3,500 × g for 20 min, washed twice, and resuspended in PBS. The concentration of S. pneumoniae (in CFU per milliliter) was determined by standard dilution and plate count.
Study design.
Fifty-five male Sprague-Dawley rats (225 to 250 g) were randomly assigned to three cohorts and anesthetized by intramuscular injection with ketamine hydrochloride (80 mg/kg of body weight) and xylazine (8 mg/kg). OM was then induced by the direct bilateral inoculation of the middle ears, with 30 μl of a suspension containing 2 × 105 CFU of either the transparent or opaque phenotype variants of S. pneumoniae 6A in sterile pyrogen-free saline as previously described (16). Inoculations were made through the bony wall of the cephalid bullae, which was accessed through a neutral midline incision and blunt dissection. A control cohort was sham inoculated with 30 μl of diluent alone (saline), and an additional six rats were used as normal controls without injections.
At 4 and 24 h postinoculation, rats were anesthetized and then sacrificed by the intracardiac injection of an overdose of xylazine, and the middle ear mucosa were harvested by in situ lysis as previously described by our laboratory (3). For half of the animals from each cohort, the bullae were exposed and the middle ear epithelium was harvested by in situ lysis with 50 μl of lysis buffer from an RNeasy Mini kit (Qiagen, Valencia, Calif.). This process was repeated three times, and the lysates were aspirated, pooled, and stored at −70°C. Total RNA was isolated by using an RNeasy Mini kit according to the manufacturer's instructions (Qiagen). The purity of the isolated RNA was estimated by spectrophotometric determination of the 260- to 280-nm absorption ratio, and the RNAs were stored at −80°C until analyzed by real-time PCR.
For the remaining animals from each cohort, the middle ear space was lavaged prior to in situ lysis of the epithelium to allow assessment of S. pneumoniae and inflammatory cell titers. The bullae were exposed, the middle ear space was rinsed three times with 50 μl of sterile pyrogen-free saline, the washings were aspirated and pooled, and the inflammatory cell concentration (in cells per cubic millimeter) for each sample was determined by use of a hemocytometer. Also, middle ear lavage samples were cultured overnight at 37°C on chocolate agar plates in an incubator supplemented with humidity and 5% CO2, and the concentration of S. pneumoniae was determined by standard dilution and plate count. Following lavage, in situ lysis of the middle ear epithelium and total RNA extraction were carried out as described above. The study was repeated once.
Quantitation of cytokine transcripts from middle ear epithelium by real-time PCR.
Real-time PCR allows for the rapid, accurate, and precise quantitation of gene transcripts (9, 12). Real-time PCR assays were performed to specifically quantitate IL-1α, IL-1β, IL-6, IL-10, iNOS, and TNF-α transcripts as our investigators have described previously (19). Briefly, total cellular RNA was extracted using an RNeasy Mini kit (Qiagen), and cDNAs were synthesized using the Superscript preamplification system (Gibco BRL). Each cDNA sample was used as a template for a real-time PCR amplification mixture containing forward and reverse primers and probes for the target cytokine and chemokine genes and for 18S rRNA (internal control) and 2× TaqMan Universal PCR Master Mix obtained from Applied Biosystems (Foster City, Calif.). Real-time PCR amplifications were performed on an Applied Biosystems Prism 7700 sequence detector according to the manufacturer's instructions. Predicted cycle threshold (CT) values were exported directly into Excel worksheets for analysis. Relative changes in gene expression were determined using the 2−ΔΔCΤ method as described elsewhere (15) and reported as the fold difference relative to a calibrator cDNA (normal control rats, uninoculated) prepared in parallel with the experimental cDNAs. Primers and probes for IL-1α, IL-1β, IL-6, IL-10, iNOS, and TNF-α were designed using the computer program Primer Express (Applied Biosystems) and synthesized by Perkin-Elmer/Applied Biosystems. The primers and probes for each gene are listed in Table 1.
TABLE 1.
Primer and probe sequences used for real-time PCR
| Gene | GenBank accession no. | Primer and probe sequence |
|---|---|---|
| iNOS | NM012611 | Forward: 5′ TGG TCC AAC CTG CAG GTC TT 3′ |
| Reverse: 5′ CAG TAA TGG CCG ACC TGA TGT 3′ | ||
| Probe: 5′ TGC CCG GAG CTG TAG CAC TGC AT 3′ | ||
| IL-6 | NM012589 | Forward: 5′ TCC AAA CTG GAT ATA ACC AGG AAA T 3′ |
| Reverse: 5′ TTG TCT TTC TTG TTA TCT TGT AAG TTG TTC TT 3′ | ||
| Probe: 5′ AAT CTG CTC TGG TCT TCT GGA GTT CCG TTT CTA 3′ | ||
| IL-10 | NM012854 | Forward: 5′ GAA GCT GAA GAC CCT CTG GAT ACA 3′ |
| Reverse: 5′ CCT TTG TCT TGG AGC TTA TTA AAA TCA 3′ | ||
| Probe: 5′ CGC TGT CAT CGA TTT CTC CCC TGT GA 3′ | ||
| IL-1α | NM017019 | Forward: 5′ AGC CCA TGA TTT AGA AGA GAC CAT 3′ |
| Reverse: 5′ TGA TGA ACT CCT GCT TGA CGA T 3′ | ||
| Probe: 5′ CAG ATC AGC ACC TCA CAG CTT CCA GAA TAA TT 3′ | ||
| IL-1β | NM031512 | Forward: 5′ CCA AGC ACC TTC TTT TCC TTC A 3′ |
| Reverse: 5′ AGC CTG CAG TGC AGC TGT CTA A 3′ | ||
| Probe: 5′ CAG GTC GTC ATC ATC CCA CGA GTC A 3′ | ||
| TNF-α | NM012675 | Forward: 5′ GAC CCT CAC ACT CAG ATC ATC TTC T 3′ |
| Reverse: 5′ TTG TCT TTG AGA TCC ATG CCA TT 3′ | ||
| Probe: 5′ ACG TCG TAG CAA ACC ACC AAGCGG A 3′ |
Quantitation of cytokine proteins in the middle ear lavage samples by ELISA.
Middle ear lavage samples were centrifuged at 500 × g and frozen at −70°C. Concentrations of IL-1β, IL-6, and ΤNF-α in middle ear lavage samples were measured by use of commercial enzyme-linked immunosorbent assay (ELISA) kits (Quantikine; R & D Systems, Minneapolis, Minn.), according to the manufacturer's instructions. Middle ear lavage samples from saline sham-inoculated animals served as the control.
Statistical analysis.
Data are expressed as the arithmetic mean ± the standard error of the mean (SEM). Statistical analysis was performed by use of either the Mann-Whitney rank sum test (for S. pneumoniae concentration in middle ear lavage samples) or the Student t test for all other comparisons. In all cases, a P level of <0.05 was set as the measure of significance.
RESULTS
Effect of opacity phenotype on S. pneumoniae survival in the middle ear.
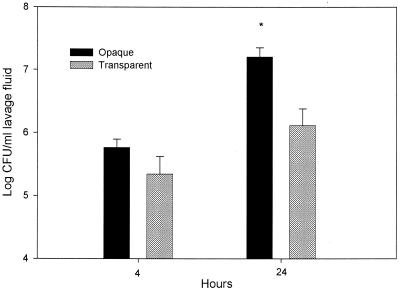
To ascertain whether opacity variants influence S. pneumoniae survival, the number of organisms present in the middle ear following direct transbullar inoculation with S. pneumoniae was determined. At both 4 and 24 h postinoculation, middle ear lavage samples contained higher concentrations of opaque organisms (Fig. 1). There was a statistically significant difference between the number of opaque and transparent organisms at 24 h postinoculation (P < 0.05). The opaque phenotype variant had more than 12 times the number of viable transparent S. pneumoniae by 24 h postinoculation. The experiment was repeated once, and similar results were obtained.
FIG. 1.
Survival of the transparent and opaque phenotype variants of S. pneumoniae in the middle ears of rats following transbullar inoculation. Each data point represents the geometric mean of CFU of S. pneumoniae (± SEM) per milliliter of middle ear lavage fluid. These results are from a single experiment, with a total of four to eight middle ear samples obtained from two to four animals for each cohort (opaque and transparent), at each time point. *, P < 0.05 for the comparison.
Effect of opacity phenotype on middle ear inflammatory cell response.
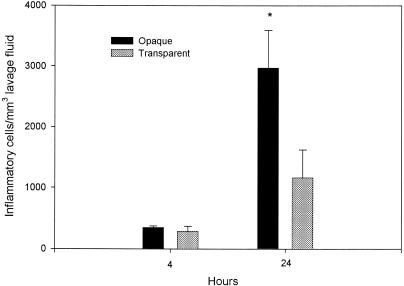
To determine whether phase variants of S. pneumoniae differentially induce inflammatory cell recruitment, cell counts were performed on middle ear lavage samples. Whereas there was little difference at 4 h, by 24 h postinoculation there were significantly more inflammatory cells in the middle ear lavage samples from rats inoculated with the opaque phenotype of S. pneumoniae compared to that in animals inoculated with the transparent phenotype (P < 0.05) (Fig. 2). Also, at 4 and 24 h following inoculation, lavage samples collected from rats inoculated with transparent or opaque phenotype variants had significantly more inflammatory cells compared to those from saline sham-inoculated controls (P < 0.001 in all cases). All middle ear lavage samples from sham-inoculated controls had fewer than 10 cells per mm3 for both the 4- and 24-h samples. The experiment was repeated, and similar results were obtained.
FIG. 2.
Accumulation of inflammatory cells in the middle ears of rats following transbullar inoculation of transparent or opaque variants of S. pneumoniae. Each data point represents the mean concentration of inflammatory cells (± SEM) per milliliter of middle ear lavage fluid. These results are from a single experiment, with a total of four to eight middle ear lavage samples obtained from two to four animals for each cohort (opaque and transparent), at each time point. *, P < 0.05 for the comparison (opaque versus transparent).
Effect of opacity phenotype on the kinetics of cytokine and iNOS gene expression in the rat middle ear following direct transbullar inoculation with opaque or transparent variants of S. pneumoniae.
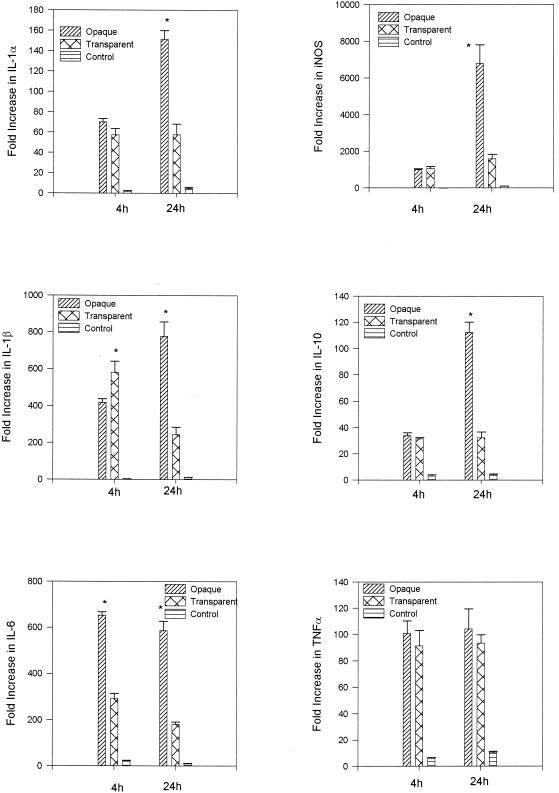
Real-time PCR was used to examine the role of S. pneumoniae opacity variants in the induction of a panel of proinflammatory cytokines (IL-1α, IL-1β, IL-6, IL-10, and TNF-α) and iNOS genes in the rat middle ear in vivo during the course of experimental OM. By 4 h after transbullar inoculation with S. pneumoniae 6A opaque or transparent variants, both S. pneumoniae opacity variants induced a significant up-regulation of these genes relative to the saline sham control (P < 0.001 in all cases) (Fig. 3). When comparing gene expression levels between the opaque and transparent variants, at 4 h a significant difference in gene expression levels was observed only for IL-1β and IL-6. Whereas IL-1β expression was significantly increased following transbullar challenge with the transparent phenotype (P < 0.05), IL-6 expression was significantly increased subsequent to challenge with the opaque phenotype of S. pneumoniae (P < 0.001) (Fig. 3).
FIG. 3.
Induction of gene expression as measured by real-time PCR on total RNA samples prepared by the direct in situ lysis of the middle ear space at 4 and 24 h following inoculation of transparent and opaque variants of S. pneumoniae. Results are the mean fold increase in IL-1α, IL-1β, IL-6, IL-10, iNOS, and TNF-α transcript levels (± SEM) from duplicate samples from two separate experiments. *, P < 0.05 for the comparison (opaque versus transparent).
Just as at 4 h, 24 h following the transbullar challenge with S. pneumoniae 6A opaque and transparent variants both S. pneumoniae opacity variants induced a significant up-regulation of gene expression relative to the saline sham control (P < 0.001 in all cases) (Fig. 3). Also, at this time the induction of gene expression for IL-1α, IL-1β, IL-6, IL-10, and iNOS was significantly different between the cohorts challenged with the S. pneumoniae opaque and transparent variants. The amount of mRNA transcripts for IL-1β, IL-6, and IL-10 were all greater than threefold higher, and those of IL-1α and iNOS were greater than two- and fourfold higher, respectively, in the S. pneumoniae opaque variant cohort compared to levels in the S. pneumoniae transparent cohort (P < 0.05 in all cases). In contrast, at this time there was no statistical difference in TNF-α gene expression induced by the opaque and transparent phenotype variants of S. pneumoniae.
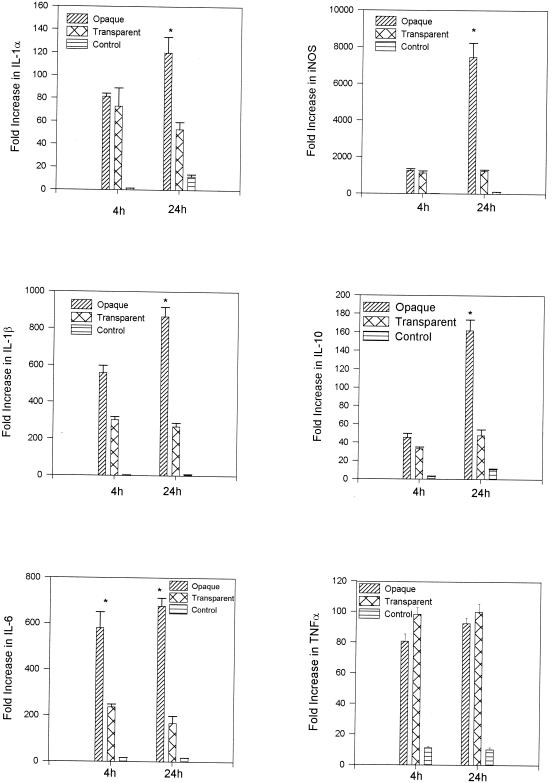
The above gene expression results were generated using total RNA samples prepared by direct in situ lysis of the middle ear space. RNA samples prepared by direct in situ lysis of the epithelium (nonlavaged) included gene transcripts produced from both white blood cells as well as from cells making up the mucosal surface. To permit examination of that proportion of gene expression which could be attributed to the mucosal surface, total RNA samples for gene expression analysis were also prepared by in situ lysis of the middle ear epithelium subsequent to lavage of the middle ear cavity. For these samples, at 4 and 24 h following transbullar inoculation with S. pneumoniae 6A opaque or transparent variants, both S. pneumoniae opacity variants induced a significant up-regulation of these cytokine genes relative to the levels in saline sham controls (P < 0.001 in all cases) (Fig. 4). Also, with the exception of TNF-α, expression levels were higher for all genes following challenge with the opaque phenotype. Furthermore, by 4 h postinoculation, IL-6 expression levels were significantly different, with the S. pneumoniae opaque variant inducing a 2.5-fold increase in IL-6 gene expression relative to the transparent variant (P < 0.001).
FIG. 4.
Induction of gene expression as measured by real-time PCR on total RNA samples prepared by in situ lysis of the middle ear space subsequent to lavage at 4 and 24 h following inoculation of transparent and opaque variants of S. pneumoniae. Results are the mean fold increase in IL-1α, IL-1β, IL-6, IL-10, iNOS, and TNF-α transcript levels (± SEM) from duplicate samples from two separate experiments. *, P < 0.05 for the comparison (opaque versus transparent).
At 24 h, the levels of induction of IL-1α, IL-1β, IL-6, IL-10, and iNOS were significantly different between the cohorts challenged with S. pneumoniae opaque and transparent variants. The amount of mRNA transcripts of IL-1α and IL-10 genes were two- and threefold higher, respectively, and that of IL-1β, IL-6, and iNOS genes were three-, four-, and sixfold higher, respectively, in the S. pneumoniae opaque variant cohort compared to expression levels in the S. pneumoniae transparent cohort (P < 0.05 in all cases) (Fig. 4). Also, just as at 4 h, there was no statistical difference in TNF-α gene expression induced by either phenotype. Though not statistically significant, TNF-α was the only gene expressed at a higher level following transbullar challenge with transparent S. pneumoniae compared to challenge with opaque S. pneumoniae.
It should be noted that comparisons of gene expression levels for saline sham-inoculated controls versus noninoculated normal controls were included for these studies. Fold increases in gene expression levels following inoculation with saline alone served as the inoculation control, providing information on how the physical trauma of the inoculation procedure impacted upon gene expression. Increases in gene expression for the saline sham controls (greater than a 10-fold increase in gene expression relative to the noninoculated control) were observed for IL-6 at 4 h and IL-1β, iNOS, and TNF-α at 24 h (Fig. 3). For total RNA samples prepared by in situ lysis of the epithelium following lavage, low levels of gene expression were observed in the saline sham-inoculated controls for IL-1α and IL-10 at 24 h and for TNF-α, IL-6, and iNOS at both 4 and 24 h (Fig. 4). However, as noted above, the induction of gene expression was significantly greater following the inoculation of either opacity variant for all genes tested compared to the saline controls, at all times assayed.
Kinetics of cytokine production in the rat middle ear following transbullar inoculation with opaque or transparent phenotypic variants of S. pneumoniae.
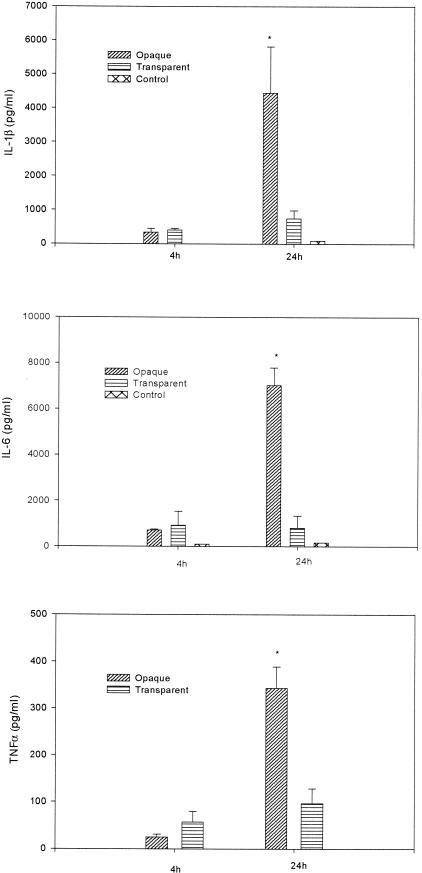
The significant induction of cytokine gene expression in the rat middle ear in vivo during the course of experimental OM was confirmed by quantitating the secretion of TNF-α, IL-1β, and IL-6 in middle ear lavage samples by use of ELISAs. At 4 and 24 h postinoculation, the concentration of cytokines in the lavage samples for controls was 96 and 164 pg/ml for IL-6 and 8 and 84 pg/ml for IL-1β, respectively (Fig. 5). No secreted TNF-α was detected in middle ear lavage samples at either time point. The minimum detectable dose of TNF-α using the Quantikine M kit was 5 pg/ml.
FIG. 5.
Concentrations of cytokines in middle ear lavage samples. Results are the mean concentrations of IL-1β, IL-6, and TNF-α (± SEM) in middle ear lavage samples from two duplicate wells from a single experiment. *, P < 0.05 for the comparison (opaque versus transparent).
By comparison, at both 4 and 24 h after transbullar inoculation with S. pneumoniae 6A opaque or transparent variants, both S. pneumoniae opacity variants induced a significant increase in the production of these cytokines relative to saline sham-inoculated controls (P < 0.001 in all cases) (Fig. 5). Furthermore, a comparison of protein expression levels at 24 h following transbullar challenge with S. pneumoniae indicated that animals inoculated with the opaque variant had significantly higher levels of TNF-α, IL-1β, and IL-6 in the lavage samples (four-, six-, and ninefold more, respectively) than animals inoculated with the transparent phenotype (P < 0.05 for all comparisons).
Analysis of inflammatory cell response and gene expression and production following normalization of data to account for differences in middle ear S. pneumoniae populations.
As previously noted, there were significant differences in the populations of opaque and transparent organisms within the middle ear cavity following transbullar inoculation. In fact, there were 3- and 12-fold more opaque S. pneumoniae organisms in the middle ear by 4 and 24 h postinoculation, respectively. As we were interested in examining whether opaque and transparent variants differentially induce the inflammatory cell response as well as the expression and production of inflammatory mediators, the data were normalized to account for differences in bacterial populations and the results were expressed as the unit change per 100,000 CFU of S. pneumoniae.
For inflammatory cell response data, following normalization of the data mean inflammatory cell counts (in cells per cubic millimeter of middle ear lavage fluid [mean ± SEM]) were 129.1 ± 38.6 and 89.7 ± 35.4 following inoculation with the transparent variant at 4 and 24 h and 59.8 ± 4.1 and 18.6 ± 3.9 following inoculation with the opaque variant at 4 and 24 h. Thus, there were approximately two- and fivefold more white blood cells at 4 and 24 h, respectively, following inoculation with the transparent variant.
Also, for real-time PCR studies, analysis of the normalized data indicates that the transparent variant generated a greater fold increase in gene expression than the opaque variant for every inflammatory mediator examined at both 4 and 24 h (Table 2). This was also true for ELISA results. Following normalization of the data, mean cytokine production (in picograms per milliliter) following inoculation with the transparent variant was 25.9 ± 10.1 and 7.5 ± 2.4 for TNF-α, 181.2 ± 23.1 and 56.7 ± 18.2 for IL-1β, and 416.8 ± 283.2 and 62.3 ± 40.5 for IL-6 at 4 and 24 h, respectively. Following inoculation with the opaque variant, normalized mean cytokine production (in picograms per milliliter) was 4.3 ± 1.1 and 2.1 ± 0.3 for TNF-α, 57.7 ± 19.0 and 27.8 ± 8.6 for IL-1β, and 121.7 ± 7.8 and 43.9 ± 4.8 for IL-6 at 4 and 24 h, respectively. Thus, by 4 h postinoculation, production of IL-1β, IL-6, and TNF-α were found to be three-, three-, and sixfold higher, respectively, following challenge with the transparent variant (compared to the opaque variant) of S. pneumoniae. For the 24-h samples, the fold differences in production for these three cytokines were 2, 1, and 4, respectively.
TABLE 2.
Fold increase in gene expression per 100,000 CFU of S. pneumoniae relative to normal (noninoculated) controls
| Inflammatory mediator | S. pneumoniae phenotype variant | Fold increase in gene expression
|
|||
|---|---|---|---|---|---|
| Nonlavageda
|
Lavageda
|
||||
| 4 h | 24 h | 4 h | 24 h | ||
| IL-1α | Opaque | 12.1 | 0.9 | 14.1 | 0.8 |
| Transparent | 26.1 | 4.4 | 33.2 | 4.1 | |
| IL-1β | Opaque | 72.0 | 4.9 | 96.6 | 5.4 |
| Transparent | 264.2 | 18.8 | 138.0 | 20.6 | |
| IL-6 | Opaque | 112.6 | 3.7 | 100.1 | 4.2 |
| Transparent | 132.6 | 14.0 | 107.6 | 12.9 | |
| IL-10 | Opaque | 5.9 | 0.7 | 7.9 | 1.0 |
| Transparent | 14.3 | 2.5 | 15.1 | 3.7 | |
| iNOS | Opaque | 173.3 | 42.5 | 217.3 | 46.6 |
| Transparent | 489.0 | 123.0 | 513.4 | 94.8 | |
| TNF-α | Opaque | 17.4 | 0.7 | 13.9 | 0.6 |
| Transparent | 41.6 | 7.2 | 44.7 | 7.7 | |
Total RNA samples for the analysis of gene expression were prepared by in situ lysis of the middle ear epithelium either directly (nonlavaged) or following lavage to remove inflammatory cells at 4 and 24 h postinoculation with 2 × 105 CFU of opaque or transparent S. pneumoniae.
DISCUSSION
Previous studies have provided evidence of a link between pneumococcal opacity phase variation and nasopharyngeal colonization and virulence (21). In this study, our data indicate that there was a significant difference in survival of transparent and opaque phenotype variants of S. pneumoniae within the middle ear. Twenty-four hours following transbullar challenge, there were significantly more opaque S. pneumoniae organisms in middle ear lavage samples than in animals challenged with the transparent variant of S. pneumoniae. Interestingly, lavage samples collected at 4 h postinoculation were not significantly different, with only 3-fold more opaque than transparent S. pneumoniae. However, by 24 h postinoculation there were 12-fold more opaque than transparent pneumococci.
The question arises as to what factor(s) can account for the observed differences in survival of the two opacity phenotype variants in the middle ear. Most notably, structural differences between the two phase variants may be responsible for the more rapid clearance of the transparent variant. As noted in the introduction, transparent S. pneumoniae variants have been shown to have more cell wall teichoic acid than opaque variants of S. pneumoniae (14). As reviewed by Weiser, choline in the form of phosphorylcholine binds to teichoic acid and is the target for the acute-phase reactant C-reactive protein (CRP) (23). CRP facilitates opsonophagocytosis in the absence of specific antibody, and only transparent pneumococci appear to bind significant amounts of human CRP (23). In children with chronic recurrent OM, CRP can be detected in 57% of middle effusion samples with an average level of 39 μg/liter (H. H. Tong and T. F. DeMaria, unpublished data). However, the exact role and significance of CRP within the rat model of OM have not been determined.
A second structural difference between the opacity phase variants of S. pneumoniae which could influence in vivo survival involves the antiphagocytic capsular polysaccharide. Opaque S. pneumoniae variants have more capsular polysaccharide than related transparent S. pneumoniae variants (14) and are more resistant to phagocytosis (13). Thus, given that the primary mechanism of clearance for the pneumococcus is opsonophagocytosis, differences in middle ear survival for the two phenotype variants can best be described as the result of a combination of two factors: (i) the increased susceptibility of the transparent variant to opsonophagocytosis (possibly due to CRP), in conjunction with (ii) the increased resistance of the opaque variant to opsonophagocytosis by virtue of its thicker polysaccharide capsule.
A previous study utilizing the rat model of OM documented the ability of pneumococci to induce an inflammatory cell response within the middle ear (18). We found that inoculation with the opaque variant rather than the transparent one resulted in the accumulation of significantly more inflammatory cells in the middle ear 24 h after inoculation. Most likely, this difference can be explained in terms of the differences in bacterial survival within the middle ear. As discussed in a review by Tuomanen, a major result of bacterial replication in the middle ear is the influx of neutrophils (22). More specifically, a threshold number of bacteria are required to induce the production of IL-6, IL-1, and TNF-α (by the mucosa), and it is the production of these cytokines which drives the influx of neutrophils into the middle ear (22). It is conceivable that increased titers of bacteria resulted in enhanced cytokine production, with the result being the accumulation of more inflammatory cells. When our data were normalized to account for differences in bacterial numbers, it became apparent that the transparent variant actually resulted in the accumulation of more inflammatory cells per viable bacterium.
Our results indicate that transparent and opaque variants of S. pneumoniae induce a significant up-regulation of gene expression within the middle ear. These results are consistent with those obtained by Hebda et al. (11). They found that expression of TNF-α, IL-6, IL-10, and iNOS genes was significantly induced following the transbullar inoculation of S. pneumoniae. Our data also suggest that the opaque variant is more capable of inducing gene expression and production of inflammatory mediators within the middle ear following transbullar inoculation. However, there were 3- and 12-fold more opaque than transparent S. pneumoniae organisms in middle ear lavage samples at 4 and 24 h, respectively; therefore, we chose to normalize the gene expression and production data per the number of S. pneumoniae organisms.
When the data were normalized to account for differences in middle ear bacterial titers and the results were expressed as the fold change per 100,000 S. pneumoniae organisms, it became apparent that the transparent variant of S. pneumoniae is a more potent inducer of gene expression and production, triggering substantially greater fold increases in the expression and production of inflammatory mediators. In fact, for the normalized data, inoculation of the transparent variant resulted in a greater fold increase for every gene, at both time points assayed.
It is still an open issue to interpret the normalization of data using bacterial titers in middle ear lavage samples. In order to normalize the data, the assumption must be made that the response to each bacterial load is the same. This may not always be the case. For instance, it is possible that the transparent phenotype variant, as a result of enhanced susceptibility to killing by the inflammatory cell response, as well as autolysis, results in a lower number of organisms at 24 h postinoculation. Alternately, it is also possible that more inflammation per viable bacterium for the transparent phenotype variant results in fewer viable bacteria remaining at 24 h. Either scenario, alone or in tandem, would result in the normalized inflammatory cell response per viable bacterium to be higher for the transparent group. Furthermore, this same argument can also be applied to the normalization of the gene expression and production results. In any case, until a better model system is devised to control for differences in survival between the two phenotype variants in vivo, normalization of the data provides a means by which differences in inflammatory cell response and gene expression and production data can be compared between transparent and opaque cohorts.
The link between increased cytokine production and adhesion requires further comment. Our results indicate that of the six genes assayed, the largest fold increase (relative to the opaque variant) in expression occurred for IL-1α and TNF-α. Our investigators reported previously that TNF-α and IL-1α increase S. pneumoniae 6A (predominantly transparent) adherence to chinchilla tracheal epithelium (20). It is possible that increased adhesion by the transparent phenotype results in increased inflammation. It is also possible that increased inflammation results in receptor up-regulation and therefore increased attachment. As previously noted, the adherence of the transparent phenotype to human type II lung cells and vascular endothelial cells was shown to be increased following activation with IL-1α or TNF-α, whereas adherence of the opaque phenotype was not (7). Our results suggest that transparent variants are capable of inducing a greater cytokine response. In particular, the increased production of IL-1 and TNF-α in response to the transparent variant could result in the activation and expression of receptors, which would further enhance and select for the binding of the transparent variant to host epithelial receptors. In this way, populations of transparent variants could self modulate adherence to the mucosal or epithelial surface.
It is interesting to speculate about the relative contribution inflammatory cells and epithelial cells make on the overall gene expression levels observed within the middle ear following exposure to phase variants of S. pneumoniae. A comparison of gene expression results between lavaged and nonlavaged data sets would allow information to be gleaned regarding this topic. Unfortunately, a statistical analysis of differences cannot be made, as the data sets were generated in separate independent experiments. However, a comparison of trends in the data reveals that overall gene expression levels for lavaged and nonlavaged samples were very similar in terms of fold differences relative to those in the noninoculated control. This is true for both normalized and nonnormalized data sets. These results indicate that over the first 24 h, the mucosal surface appears to be the major contributor to gene transcript levels.
Another way to glean information regarding the relative contribution of white blood cells and the mucosal surface to the expression and production of cytokines within the middle ear involves analysis of ELISA data collected early in the infectious process. In studies by Sato et al., using the chinchilla model of OM, they found that 93 and 50% of middle ear fluid samples were positive for IL-6 and IL-1β production, respectively, by 2 h postinoculation with S. pneumoniae (18). As the middle ear is relatively devoid of resident lymphocytes and has no lymphoid tissue (2, 16), it is likely that the early production of IL-6, IL-1β, and possibly TNF-α was the product of cells lining the mucosal surface. In our study, it was especially interesting that by 4 h postinoculation, for the normalized ELISA results, production of IL-1β, IL-6, and TNF-α was found to be 3.1-, 3.4-, and 6-fold higher, respectively, following challenge with the transparent variant of S. pneumoniae than with the opaque form. These data suggest that the mucosal surface is active in the production of cytokines and that the transparent variant is a more potent inducer of this production.
To our knowledge, this is the first study to demonstrate differences in the ability of S. pneumoniae opacity phase variants to differentially induce proinflammatory gene expression in vivo. These studies indicate that the transparent variant appears to be a more potent inducer of the expression and production of inflammatory mediators within the middle ear. However, how S. pneumoniae opacity variants target the middle ear epithelial cells and trigger them to activate cytokine responses has not yet been fully elucidated. Future studies with opacity variants of S. pneumoniae using a combination of carefully designed in vitro and in vivo studies will allow for an analysis of the mechanisms responsible for differences in bacterial adherence and multiplication. This information will permit us to further define how phase variation impacts the host-parasite interaction at a molecular and cellular level during the initial stages of OM.
Acknowledgments
This study was supported, in part, by a grant from the National Institute on Deafness and Other Communication Disorders, National Institutes of Health (RO1 DC3105-06).
Editor: J. N. Weiser
REFERENCES
- 1.Andersson, B., B. M. Gray, H. C. Dillon, A. Bahrmand, and C. Svanborg-Eden. 1988. Role of adherence of Streptococcus pneumoniae in acute otitis media. Pediatr. Infect. Dis. 7:476-480. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, J. M., and P. L. Ogra. 1980. Mucosal immune system: implications in otitis media with effusion. Ann. Otol. Rhinol. Laryngol. 89(Suppl.):326-332. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y. P., H. H. Tong, M. A. James, and T. F. DeMaria. 2001. Detection of mucin gene expression in normal rat middle ear mucosa by reverse transciptase-polymerase chain reaction. Acta Otolaryngol. 121:45-51. [DOI] [PubMed] [Google Scholar]
- 4.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 5.Cundell, D. R., H. R. Masure, and E. I. Tuomanen. 1995. The molecular basis of pneumococcal infection: a hypothesis. Clin. Infect. Dis. 21(Suppl. 3):S204-S211. [DOI] [PubMed] [Google Scholar]
- 6.Cundell, D. R., B. J. Pearce, J. Sandros, A. M. Naughton, and H. R. Masure. 1995. Peptide permeases from Streptococcus pneumoniae affect adherence to eukaryotic cells. Infect. Immun. 63:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundell, D. R., J. N. Weiser, J. Shen, A. Young, and E. I. Tuomanen. 1995. Relationship between colonial morphology and adherence of Streptococcus pneumoniae. Infect. Immun. 63:757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geelen, S., C. Bhattacharyya, and E. Tuomanen. 1993. The cell wall mediates pneumococcal attachment to and cytopathology in human endothelial cells. Infect. Immun. 61:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995. [DOI] [PubMed] [Google Scholar]
- 10.Hakansson, A., A. Kidd, G. Wadell, H. Sabharwal, and C. Svanborg. 1994. Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect. Immun. 62:2707-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebda, P. A., C. M. Alper, W. J. Doyle, G. J. Burckart, W. F. Diven, and A. Zeevi. 1998. Upregulation of messenger RNA for inflammatory cytokines in middle ear mucosa in a rat model of acute otitis media. Ann. Otol. Rhinol. Laryngol. 107:501-507. [DOI] [PubMed] [Google Scholar]
- 12.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. O., S. Romero-Steiner, U. B. Sorensen, J. Bloom, M. Carvalho, S. Barnard, G. Carlone, and J. N. Weiser. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae.. Infect. Immun. 67:2327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 15.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta Delta CT) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 16.Melhus, A., and A. F. Ryan. 2000. Expression of cytokine genes during pneumococcal and nontypeable Haemophilus influenzae acute otitis media in the rat. Infect. Immun. 68:4024-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ophir, C., T. Hahn, and A. Schattner. 1988. Tumor necrosis factor in middle ear effusion. Arch. Otolaryngol. Head Neck Surg. 114:1256-1258. [DOI] [PubMed] [Google Scholar]
- 18.Sato, K., C. L. Liebeler, M. K. Quartey, C. T. Le, and G. S. Giebink. 1999. Middle ear fluid cytokine and inflammatory cell kinetics in the chinchilla otitis media model. Infect. Immun. 67:1943-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong, H. H., Y. Chen, M. James, J. Van Deusen, D. B. Welling, and T. F. DeMaria. 2001. Expression of cytokine and chemokine genes by human middle ear epithelial cells induced by formalin-killed Haemophilus influenzae or its lipooligosacharide htrB and rfaD mutants. Infect. Immun. 69:3678-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong, H. H., L. M. Fisher, G. M. Kosunick, and T. F. DeMaria. 1999. Effect of tumor necrosis factor α and interleukin 1-α on the adherence of Streptococcus pneumoniae to chinchilla tracheal epithelium. Acta Otolaryngol. 119:78-82. [DOI] [PubMed] [Google Scholar]
- 21.Tong, H. H., J. N. Weiser, M. A. James, and T. F. DeMaria. 2001. Effect of influenza A virus infection on nasopharyngeal colonization and otitis media induced by transparent or opaque phenotype variants of Streptococcus pneumoniae in the chinchilla model. Infect. Immun. 69:602-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuomanen, E. I. 2001. Pathogenesis of pneumococcal inflammation: otitis media. Vaccine 19:S38-S40. [DOI] [PubMed] [Google Scholar]
- 23.Weiser, J. N. 2000. Phase variation of Streptococcus pneumoniae, p. 225-231. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 24.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yellon, R. F., G. Leornard, P. T. Marucha, R. Craven, R. J. Carpenter, W. B. Lehmann, J. A. Burleson, and D. L. Kreutzer. 1991. Characterization of cytokines present in middle ear effusions. Laryngoscope 101:165-169. [DOI] [PubMed] [Google Scholar]