Abstract
Interactions between Escherichia coli K1, which causes meningitis in neonates, and macrophages have not been explored well. In this study we found that E. coli K1 was able to enter, survive, and replicate intracellularly in both murine and human macrophage cell lines, as well as in monocytes and macrophages of newborn rats. In addition, we demonstrated that OmpA + E. coli also enters and replicates in human peripheral blood monocytes in vitro. Outer membrane protein A (OmpA) expression on E. coli contributes to binding to macrophages, phagocytosis, and survival within macrophages. Opsonization with either complement proteins or antibody is not required for uptake and survival of the bacteria within the macrophages. Transmission electron microscopy and immunocytochemistry studies with the infected macrophages indicated that OmpA+ E. coli multiplies enormously in a single phagosome and bursts the cell. Internalization of OmpA+ E. coli by RAW 264.7 cells occurred by both actin- and microtubule-dependent processes, which are independent of RGD-mediated integrin receptors. Internalization and intracellular survival within phagocytic cells thus may play an important role in the development of bacteremia, which is crucial for E. coli crossing of the blood-brain barrier.
Escherichia coli is one of the leading gram-negative bacteria that cause neonatal meningitis. The rates of mortality and neurologic sequelae remain high despite advances in antimicrobial therapy (17, 19). The two important steps in the pathogenesis of neonatal meningitis are the development of bacteremia with intravascular growth and passage of bacteria across the blood-brain barrier. Studies have shown that a certain threshold of bacteremia is necessary for the development of meningitis in an experimental rat model of hematogenous meningitis (14, 29). Thus, E. coli must avoid host defense mechanisms and proliferate either in the blood or in tissues to maintain a high level of bacteremia, which leads to the onset of meningitis. Invasion and intracellular survival of E. coli, therefore, represent an important pathogenicity mechanism in this infection. Although several E. coli structures and/or genes have been shown to be essential for interaction with brain microvascular endothelial cells (BMEC) (14, 30, 36, 42), a single layer of cells lining the blood-brain barrier, very little is known about the interaction with phagocytic cells.
Studies with BMEC have suggested that outer membrane protein A (OmpA) of E. coli plays an important role in the invasion by interacting with a 96-kDa glycoprotein on human BMEC (HBMEC) (25, 27). Interaction of OmpA with its receptor induces actin condensation at the E. coli binding site (28). The actin reorganization induced by E. coli depends on activation of several host proteins involved in signaling, including focal adhesion kinase, PI3-kinase, PKC-α, and caveolin-1 (32, 33, 37, 38). The activated molecules accumulate along with actin during the invasion of HBMEC. In addition, it has recently been shown that OmpA also binds to a classical complement fluid phase regulator, C4b-binding protein, to avoid complement-mediated attack (26). The bound C4-binding protein still has the capacity to cleave C4b into C4c and C4d, thus avoiding activation of the downstream complement cascade (unpublished results). In agreement with this, deposition of significantly lower levels of C3, C5, and the membrane attack complex (MAC) on OmpA+ E. coli than on OmpA− E. coli was observed. The lack of significant quantities of C3 (which subsequently is converted to iC3b) on the E. coli surface contributes to inefficient recognition by phagocytic cells. In addition, it has also been observed that normal immunoglobulin G (IgG) does not bind to E. coli K1 very efficiently and thus avoids recognition via Fc-gamma receptors by phagocytic cells. However, neutrophils attack E. coli K1 by targeting OmpA for cleavage via the granule component elastase (3). Under such conditions, entry and survival of E. coli within monocytes and macrophages could play an important role in the course of E. coli infection.
Two other meningitis-causing pathogens in neonates, Listeria and group B Streptococcus (GBS), have been shown to invade and multiply in macrophages (2, 39, 41). However, no previous study has been focused on the E. coli K1 interaction with macrophages. Therefore, we investigated the uptake of E. coli K1 and intracellular replication in both human and murine macrophage cell lines. The results of this study suggest that E. coli phagocytosis by macrophages requires OmpA expression on the surface. In addition, uptake of E. coli occurs in the absence of opsonins via a mechanism that requires both microfilaments and microtubules. OmpA+ E. coli also enters both monocytes and macrophages in the newborn rat model of hematogenous meningitis.
MATERIALS AND METHODS
Bacteria and macrophage cell lines.
E44 is a rifampin-resistant mutant of E. coli K1 strain RS 218 (serotype O18:K1:H7), which was isolated from the cerebrospinal fluid of a neonate with meningitis and invades HBMEC in a cell culture model (30). E91 is a noninvasive derivative of E44 that expresses no OmpA, since the ompA gene is disrupted. E91 was transformed with pUC19 containing the entire ompA gene and the pUC19 plasmid alone to obtain E105 (pOmpA+ E. coli) and E111 (pUC19− E. coli), respectively (30). E44 S− is an invasive strain that lacks the S fimbriae, and noninvasive strains 7A33 and 10A23 are mutants that lack intact ibeA and ibeB genes, respectively (13, 14, 42). HB101 (K-12 capsular polysaccharide), a laboratory strain, is noninvasive in HBMEC. All bacteria were grown in brain heart infusion broth with appropriate antibiotics as necessary. All bacterial media were purchased from Difco Laboratories (Detroit, Mich.). The murine RAW 264.7 and human THP cell lines were obtained from the American Type Culture Collection and were maintained in RPMI supplemented with 10% fetal bovine serum and RPMI 1640 medium supplemented with streptomycin, penicillin, and 5 × 10−5 M mercaptoethanol, respectively. Both cell lines were maintained at 37°C in the presence of 5% CO2. THP-1 cells were differentiated into macrophages by incubating them with phorbol myristate acetate (100 nM) for 48 h for all experimental purposes, and the resulting cells were designated THP-M cells.
Reagents.
Fluorescein isothiocyanate (FITC)-conjugated secondary antibodies, antitubulin antibodies, and rhodamine phalloidin were obtained from Molecular Probes (San Diego, Calif.). Cy3-conjugated secondary antibody was obtained from Zymed Laboratories (San Francisco, Calif.). Normal goat serum and the Vectashield mounting medium with 4′,6′-diamidino-2-phenyindole (DAPI) were obtained from Vector Laboratories, Inc. (Burlingame, Calif.). SuperSignal chemiluminescence reagent and an Immunopure Fab preparation kit were obtained from Pierce Chemical Co. (Rockford, Ill.). All other chemicals were obtained from Sigma Chemical Co (St. Louis, Mo.). Anti-OmpA antibodies were generated as described previously (26). Anti-S-fimbria (anti-Sla) antibodies were kindly provided by Salam Khan of the University of Wurtzburg, Wurtzburg, Germany.
E. coli invasion assays.
Both RAW 264.7 cells (∼105 cells) and THP-M cells (∼5 × 104 to 6 × 104 cells) were plated on 24-well plates and were incubated with E. coli in RPMI 1640 for 90 min at 37°C. For the dose-response studies different amounts of bacteria, 104, 105, and 106 cells, were used as final inocula to obtain multiplicities of infection (MOI) of 0.1, 1.0, and 10, respectively. Monolayers were washed three times with RPMI 1640 and incubated in experimental medium containing gentamicin (100 μg/ml) for 1 h to kill the extracellular bacteria. The monolayers were washed again and lysed with 0.5% Triton X-100. The intracellular bacteria were enumerated by plating preparations on sheep blood agar plates. To determine the number of total cell-associated bacteria, the gentamicin step in the experiments described above was omitted. In some experiments, either the bacteria or the macrophages were pretreated with various protein fractions for 30 min before the invasion assays were performed (31). The effects of these reagents on macrophages were assessed by the trypan blue dye exclusion method, and the effects on bacterial viability were tested by colony counting. To monitor intracellular survival and replication inside the macrophages, the bacteria were allowed to infect the macrophages for 1.5 h; after this the cells were incubated for various times in medium containing gentamicin (20 μg/ml), and then the intracellular bacteria were enumerated. The minimum concentration of gentamicin required to inhibit the growth of E. coli is 8 μg/ml. Several investigators have used gentamicin with these cells without any significant effect on the intracellular bacteria for more than 48 h (9, 40). For inhibition studies, either cytochalasin D (CD), nocodazole, or dimethyl sulfoxide (DMSO) was incubated with macrophages for 30 min before the monolayers were infected with bacteria. The chemicals were present throughout the experiment. In the case of the RGD and RGPS peptides, the bacteria were incubated with the peptides for 1 h on ice and then added to the monolayers.
Isolation of rat and human monocytes and spleen macrophages.
Five-day-old rat pups were infected with E. coli strain E44 (OmpA+) or E91 (OmpA−) via the intracardiac route, and after 3 h the blood and tissues were collected. The peripheral blood monocyte populations from both rats and humans were isolated by using the Percoll gradient method, and spleen macrophages were isolated as described previously (12). The spleen tissue was pressed through a nylon mesh with a sterile plunger. The suspension was collected in a petri dish containing RPMI 1640 and washed with RPMI twice before the cells were resuspended in Hanks balanced salt solution for separation by density gradient centrifugation by using 1.070 g of Percoll per ml (the Percoll was centrifuged at 30,000 × g for 15 min at 4°C to form the gradient). The cells were layered on top of the gradient and centrifuged at 400 × g for 20 min at 4°C. The macrophage band was aspirated, washed with Hanks balanced salt solution, and resuspended in RPMI containing 20 μM glutamine, 10 μg of gentamicin per ml, and 10% heat-inactivated newborn calf serum. Both the monocytes and macrophages were seeded onto eight-well chamber slides for immunostaining.
Immunofluorescence staining.
THP-M or RAW 264.7 cells were grown in eight-well chamber slides and infected with 105 CFU of E. coli for various times, and this was followed by washing with RPMI. For differential staining of the bacteria, the monolayers were kept at 4°C in the presence of excess amounts of primary antibody for 30 min. Then the monolayers were washed with cold RPMI and incubated with either FITC- or horseradish peroxidase-conjugated secondary antibody for 30 min. After the monolayers were washed four times with RPMI, they were fixed with 2% paraformaldehyde for 15 min. Then the monolayers were incubated with 0.1% Triton X-100 in 3% normal goat serum in phosphate-buffered saline (NGS-PBST) for 1 h; this was followed by incubation with primary antibody in NGS-PBST or rhodamine phalloidin for 30 min at room temperature. The primary antibody was probed with Cy3-conjugated secondary antibody. The cells were washed again, the chambers were removed, and the slides were mounted in an antifade solution containing DAPI. The cells were viewed with a Leica (Wetzlar, Germany) DMRA microscope with Plan-apochromat 40×/1.25 NA and 63×/1.40 NA oil immersion objective lenses. Images were acquired with a SkyVision-2/VDS digital charge-coupled device camera (12-bit; 1,280 by 1,024 pixels) in unbinned or 2 × 2 binned models into the EasyFISH software, saved as 16-bit monochrome images, and merged as 24-bit RGB TIFF images (Applied Spectral Imaging, Inc., Carlsbad, Calif.)
Transmission electron microscopy (TEM).
OmpA+ E. coli strain E44 and OmpA− E. coli strain E91 were allowed to invade macrophages as described above. At specific times (1, 4, 8, and 24 h), the cells were washed four times with prewarmed RPMI and fixed with 2% glutaraldehyde in 0.1 M phosphate-buffered saline (pH 7.4) for 1 h. After the cells were rinsed four times with phosphate-buffered saline, they were stained with methylene blue for 15 min. The cells were then rinsed, postfixed with 2% OsO4 for 1 h, rinsed again, dehydrated with graded ethanol solutions, and embedded in polypropylene oxide. Ultrathin sections were cut at right angles to the culture cell layer, mounted on collodion one-hole grids, stained with uranyl acetate and lead citrate, and examined with a Phillips CM12 transmission electron microscope.
RESULTS
Uptake of E. coli K1 by RAW 264.7 and THP-M macrophages in the absence of serum.
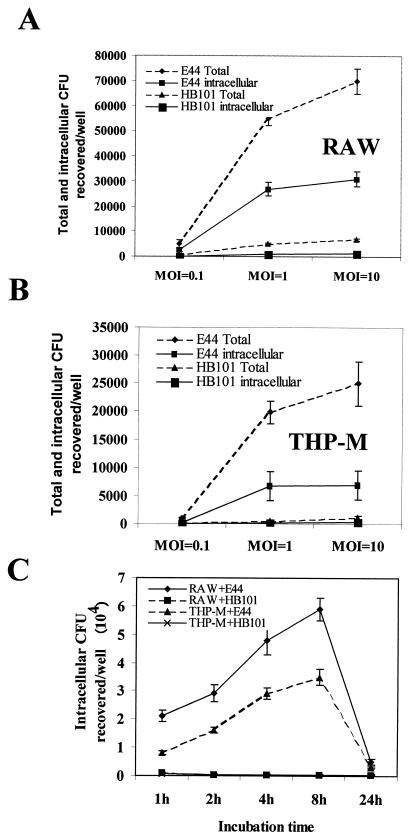
The ability of E. coli K1 to enter macrophages, both RAW 264.7 and differentiated THP-1 (THP-M) cells, was examined. After several pilot experiments, we found that an MOI of 1 (bacteria/cell ratio, 1:1) and 1.5 h of incubation at 37°C were optimal conditions for phagocytosis. In addition, the invasion assays were also carried out with 10-fold less (MOI,0.1) and 10-fold more (MOI, 10) bacteria to determine the effect of inoculum on invasion. E. coli strain E44 entered RAW 264.7 cells with an invasion frequency of 20 to 25%, which was 20-fold greater than the invasion frequency of E. coli strain HB101 (2.5 × 104 ± 0.2 × 104 CFU/well for E44 versus 0.11 × 104 ± 0.01 × 104 CFU/well for HB101; P < 0.001) (Fig. 1A) with an MOI of 1. The phagocytosis of E44 was slightly greater when the assays were carried out with the higher MOI (MOI, 10). These experiments also revealed that the entry process was rapid as 10% of the total inoculum could be recovered from antibiotic-treated macrophages at 15 min postinfection. Similarly, the uptake of E44 by THP-M cells resulted in an invasion frequency of 8 to 10%, whereas strain HB101 exhibited an invasion frequency of less than 1% (0.9 × 104 ± 0.2 × 104 CFU/well for E44 versus 0.8 × 103 ± 0.05 × 103 CFU/well for HB101; P < 0.001) (Fig. 1B). The differences in internalization between these two bacteria appeared to be due to the greater ability of E. coli K1 to associate with RAW 264.7 cells (∼50 to 60% of the inoculum) compared to the ability of HB101 to associate with RAW 264.7 cells (∼5% of the inoculum) at an MOI of 1 (Fig. 1A). Interestingly, the total number of cell-associated bacteria was further increased with a greater MOI, in contrast to the phagocytosis results, suggesting that the additional binding might have been due to nonspecific interactions. Similar results were obtained with THP-M cells, although the total number of cell-associated bacteria was 2.5 times less than the number observed with RAW 264.7 cells (Fig. 1B). These results suggest that E. coli K1 specifically binds to and enters macrophages.
FIG. 1.
Entry and intracellular survival of E. coli K1 in RAW 264.7 and THP-M cells. (A and B) Monolayers of both RAW 264.7 (RAW) (A) and THP-M (B) cells were incubated with either OmpA+ E. coli strain E44 or E. coli HB101 by using various MOI (0.1, 1.0, and 10) for 1.5 h in the absence of serum. For enumeration of total cell-associated bacteria, the monolayers were washed, dissolved in 0.5% Triton X-100 in saline, and counted by plating dilutions on blood agar. For intracellular bacteria, the monolayers were washed and then incubated in medium containing gentamicin (100 μg/ml) to kill the extracellular bacteria. Then the bacteria were released by Triton X-100 and enumerated. (C) In some experiments, the intracellular survival of the bacteria was monitored as described in Materials and Methods. Each experiment was performed at least four times in triplicate, and the error bars indicate standard deviations.
To monitor intracellular bacterial survival for longer times, invasion experiments were performed for 1.5 h with an MOI of 1; the monolayers were then washed to remove unbound bacteria and were incubated with medium containing gentamicin for the remaining time as described in Materials and Methods. The results showed that the number of CFU increased for up to 8 h postinfection, suggesting that E44 multiplied inside the macrophages, and this was followed by an 85% decrease in survival at 24 h (Fig. 1C). In contrast, HB101 strains showed no multiplication in these cells at any time. The extracellular medium was also examined for the growth of bacteria in the presence of gentamicin, and this examination revealed no significant difference between 1 and 24 h, suggesting that the increased level of intracellular bacteria was not due to increased internalization of bacteria that multiplied in the medium. The integrity of the E. coli-infected cells was examined by light microscopy throughout the 24-h period by using the trypan blue exclusion method (data not shown). Significant numbers of macrophages were stained with trypan blue at 24 h, suggesting that the bacteria which multiplied were released from the cells into the medium and subsequently exposed to the antibiotic. This could be the reason that significantly fewer viable bacteria were recovered after 24 h. In contrast, HB101-infected monolayers did not exhibit any damage.
Phagocytosis of nonopsonized and opsonized E. coli.
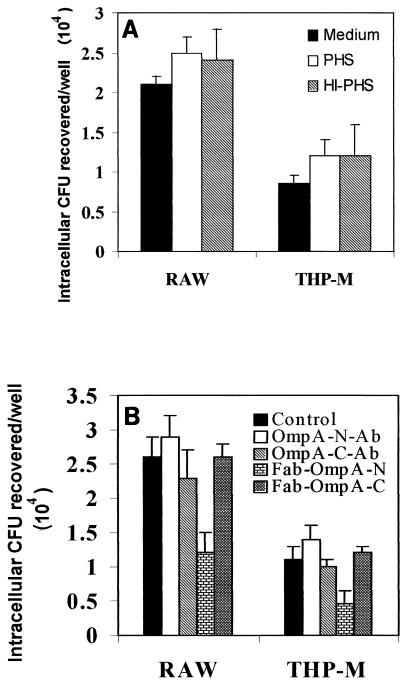
To examine the effect of opsonins on the uptake or survival of E. coli, the bacteria were opsonized with fresh normal or heat-inactivated human serum or were treated with medium alone prior to invasion assays. As shown in Fig. 2A, the number of viable opsonized E. coli cells and the number of nonopsonized E. coli cells did not differ. The heat-inactivated serum also resulted in no difference in invasion, suggesting that complement had no major effect on bacterial uptake. Similar results were obtained with E. coli which had been opsonized with anti-OmpA antibodies recognizing either N-terminal or C-terminal portions of OmpA (Fig. 2B). Interestingly, Fab fragments of OmpA-N antibody resulted in a 50% decrease in the uptake of E. coli by macrophages. In contrast, the Fab fragments of OmpA-C antibody resulted in no such blocking activity, indicating that extracellular domains of OmpA of E. coli interact with macrophages. To further determine whether the inhibition of E. coli internalization by anti-OmpA-N antibody Fab fragments was due to reduced binding of the bacteria, we examined the total cell-associated bacteria. The Fab fragments resulted in a >50% reduction in the total number of cell-associated bacteria compared to the data for Fab fragments from control antibody (1.2 × 104 ± 0.5 × 104 CFU/well for Fab-treated E. coli versus 2.6 × 104 ± 0.65 × 104 CFU/well for control Fab fragments in RAW 264.7 cells; P < 0.05), suggesting that OmpA also contributes to the binding of E. coli to macrophages. Intracellular survival and multiplication of opsonized bacteria were also assessed for 24 h, and the results showed that there was no difference in survival and multiplication when the data were compared to the data for cells treated with medium alone (data not shown).
FIG. 2.
Effect of opsonization on phagocytosis of E. coli K1 by macrophages. RAW 264.7 (RAW) and THP-M cells were infected with E. coli K1 strain E44, which was allowed to invade for 1.5 h. Then the macrophages were washed and incubated in medium containing gentamicin (20 μg/ml) for 3 h. (A) E. coli K1 was pretreated with 10 to 20% normal pooled human serum (PHS) or heat-inactivated serum (HI-PHS). (B) Bacteria were incubated with either rabbit preimmune serum (Control) or rabbit serum containing antibodies to the N-terminal portion of OmpA (OmpA-N-Ab) or to the C-terminal portion OmpA (OmpA-C-Ab) or with the Fab fragments of the antibodies (Fab-OmpA-N and Fab-OmpA-C) for 1 h at 4°C prior to invasion assays. The error bars indicate standard deviations for values from three separate experiments carried out in triplicate.
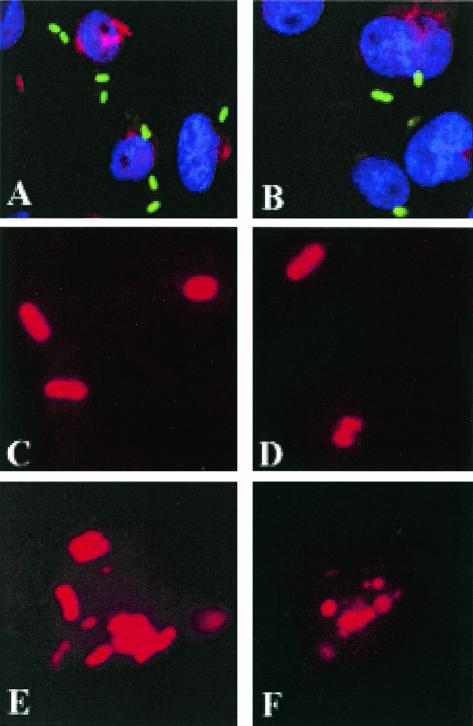
Since OmpA appears to contribute both to binding to and to entry of macrophages, OmpA+ and OmpA− E. coli strains were used to examine intracellular bacteria by labeling with an anti-S-fimbria antibody after preparations were opsonized with normal human serum. In these experiments, the extracellular bacteria were stained with either FITC-conjugated or horseradish peroxidase-conjugated secondary antibody prior to permeabilization of the cells. The intracellular bacteria were stained with Cy3-conjugated secondary antibody after incubation with anti-Sfa antibody. As shown in Fig. 3A, OmpA+ E. coli bound to significant numbers of RAW 264.7 cells (green), and some of the bacteria were internalized by 15 min (red). In contrast, very few OmpA− E. coli cells either bound to or entered RAW 264.7 cells (Fig. 3B). The internalized bacteria were monitored further for growth over a 4.5-h period. One hour after invasion (total time, 2.5 h), OmpA+ E. coli had a morphology similar to that of bacteria at 1.5 h (Fig. 3C), whereas OmpA− E. coli exhibited signs of fragmentation (diffuse and irregular border) (Fig. 3D). After 3 h (total time, 4.5 h), OmpA+ E. coli had multiplied significantly, although some bacteria showed signs of degradation (Fig. 3E). In contrast, we observed only fragmented bacterial staining with OmpA− E. coli (Fig. 3F). The OmpA+ E. coli pretreated with Fab fragments of OmpA-N antibody also showed no apparent degradation inside the macrophages. Interestingly, the bacteria also appeared to multiply inside the cells, suggesting that Fab fragments of anti-OmpA-N antibody block only the binding of E. coli to macrophages and do not promote killing inside the cells.
FIG. 3.
Immunocytochemistry of intracellular E. coli K1 in RAW 264.7 cells. Monolayers of RAW 264.7 cells were fixed after various periods of infection with either OmpA+ E. coli or OmpA− E. coli pretreated with normal human serum. Differential staining of extracellular (green) and intracellular (red) bacteria was performed as described in Materials and Methods. (A and B) Magnification, ×40. (C to F) Magnification, ×100.
OmpA expression is necessary for entry and survival of E. coli in macrophages.
Previous studies on the interaction of E. coli with BMEC suggested that several E. coli factors are responsible for invasion (14, 30, 36, 42). Thus, we set out to examine the role of these molecules in E. coli uptake and survival of the bacteria in macrophages. As shown in Table 1, E. coli invasion of both RAW 264.7 and THP-M cells was significantly affected by the absence of OmpA. The OmpA− E. coli strain was internalized approximately fivefold less than the OmpA+ E. coli strain (P < 0.001). The uptake of E. coli strain 10A23, in which the ibeA gene is disrupted, was only 50 to 60% of the uptake of E44 in both these cell lines. Interestingly, the K1 capsule-deficient and IbeB mutants phagocytosed with greater frequency, and there were corresponding increases in the survival rates in these cells. In addition, we compared the total numbers of cell-associated bacteria using these strains, and the results indicated that OmpA− E. coli bound to much less than 10% of the inoculum, whereas IbeA− E. coli showed 60% binding compared to E44 (data not shown). To confirm the role of OmpA in E. coli entry into macrophages, the OmpA− E. coli strain was complemented with the ompA gene (pOmpA+ E. coli) and then used in invasion assays. The numbers of pOmpA+ E. coli cells that entered both types of macrophages were similar to the numbers of wild-type E. coli cells when the data were compared to the data for the OmpA− E. coli strain containing the plasmid alone (pUC19− E. coli). These results strongly suggest that OmpA expression contributes to both binding of E. coli K1 to macrophages and uptake of E. coli K1 by macrophages.
TABLE 1.
Intracellular survival of various strains of E. coli in macrophages
| E. coli strain | Phenotype | Survival (104 CFU/well) ina:
|
|||||
|---|---|---|---|---|---|---|---|
| RAW 264.7 cells
|
THP-M cells
|
||||||
| 1 h | 4 h | 24 h | 1 h | 4 h | 24 h | ||
| E44 | OmpA+ K1+ Sfa+ IbeA+ IbeB+ | 3.1 ± 0.4 | 8.8 ± 1.5 | 1.1 ± 0.5 | 1.2 ± 1.0 | 2.7 ± 0.5 | 0.5 ± 0.5 |
| E91 | OmpA− K1+ Sfa+ IbeA+ IbeB+ | 0.5 ± 0.5 | 0.7 ± 0.2 | 0.1 ± 0.2 | 0.2 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| E66 | OmpA+ K1− Sfa+ IbeA+ IbeB+ | 6.0 ± 0.9 | 12.4 ± 1.6 | 1.6 ± 0.5 | 3.0 ± 0.7 | 5.1 ± 0.9 | 0.7 ± 0.2 |
| E44 S− | OmpA+ K1+ Sfa− IbeA+ IbeB+ | 3.9 ± 0.6 | 7.2 ± 0.7 | 0.7 ± 0.6 | 1.4 ± 0.4 | 2.9 ± 0.7 | 0.9 ± 0.4 |
| 10A23 | OmpA+ K1+ Sfa+ IbeA−IbeB+ | 1.9 ± 0.5 | 3.2 ± 0.6 | 0.2 ± 0.3 | 0.7 ± 0.4 | 1.1 ± 0.4 | 0.2 ± 0.2 |
| 7A33 | OmpA+ K1+ Sfa+ IbeA+ IbeB− | 6.1 ± 1.2 | 12.1 ± 0.9 | 1.1 ± 0.8 | 1.8 ± 0.6 | 3.4 ± 0.7 | 0.9 ± 0.4 |
| E105 | pOmpA+ K1+ Sfa+ IbeA+ IbeB+ | 3.8 ± 0.9 | 9.1 ± 0.7 | 1.7 ± 0.7 | 1.4 ± 0.7 | 3.7 ± 0.4 | 0.7 ± 0.8 |
| E111 | OmpA− K1+ Sfa+ IbeA+ IbeB+ | 0.4 ± 0.5 | 0.6 ± 0.4 | 0.1 ± 0.2 | 0.3 ± 0.3 | 0.9 ± 0.2 | 0.1 ± 0.3 |
The values are means ± standard deviations.
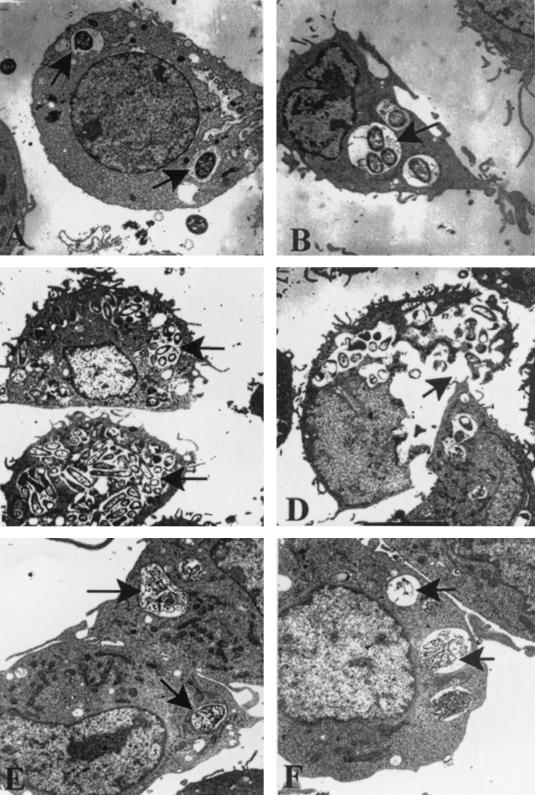
TEM of macrophages infected with E. coli for various times revealed that several OmpA+ E. coli cells entered these cells; however, each phagosome contained only a single bacterium at 1 h postinfection (Fig. 4A). By 4 h it was clear that more than one E. coli cell was present inside some of the phagosomes (Fig. 4B). The OmpA+ E. coli-infected cells exhibited large phagosomes, some of which appeared to originate from fusion, leading to the formation of spacious phagosomes (Fig. 4C). At 24 h postinfection, the macrophages burst due to the enormous growth of the bacteria (Fig. 4D). In contrast, very few OmpA− E. coli cells entered the macrophages, and most of them were degraded after 1 h inside the phagosomes (Fig. 4E). Complete degradation of OmpA− E. coli cells was observed by 4 h postinfection (Fig. 4F). The damage to the macrophages at 24 h resulted in release of all of the intact bacteria into the medium, which then may have been killed by gentamicin present in the medium. This observation is consistent with the survival data, which showed that there was a significant decrease in the survival of the bacteria at 24 h. These results also corroborate the immunostaining data, which showed that that there was significant multiplication of OmpA+ E. coli at 4 to 8 h postinfection.
FIG. 4.
TEM of macrophages infected with OmpA+ E. coli. RAW 264.7 cells were infected with either OmpA+ E. coli (A to D) or OmpA− E. coli (E and F) and processed as described for intracellular survival experiments in Materials and Methods. Samples were taken at 1 h (A and E), 4 h (B and F), 8 h (C), and 24 h (D) postinfection. Arrows indicate either intact or degraded bacteria. Magnification: ×18,900 (A, B, and E), ×11,400 (C), ×15,000 (D), and ×24,000 (F).
OmpA+ E. coli enters monocytes and macrophages in the newborn rat model of hematogenous meningitis.
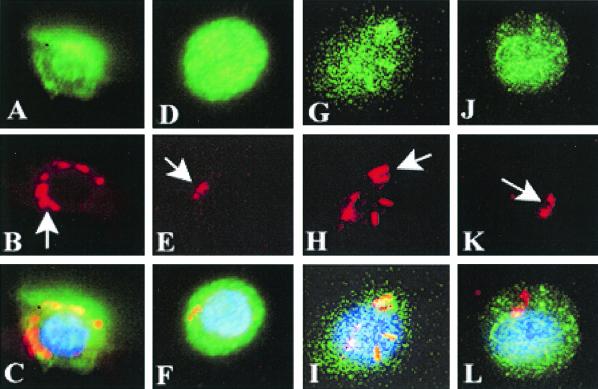
The newborn rat model of hematogenous meningitis has several similarities with human disease, such as age dependence, a required level of bacteremia, and hematogenous infection of meninges (16). Previous studies also showed that a certain threshold level of bacteremia is required for E. coli to cross the blood-brain barrier, suggesting that E. coli must survive and multiply within the host (14, 29). Thus, we examined whether circulating E. coli invades either monocytes or macrophages in this animal model. Rat pups that were 5 days old were infected with either OmpA+ or OmpA− E. coli by intracardiac injection. After 3 h, blood was collected by cardiac puncture, and monocytes were isolated. Spleens were recovered from the same animals, and macrophages were isolated by the Percoll gradient method. After both monocytes and macrophages were seeded into eight-well chamber slides, the cells were stained with anti-CD64 antibody and the bacteria were stained with anti-Sfa antibody. We observed that approximately 10% of the monocytes contained OmpA+ E. coli, whereas less than 1% of the cells contained OmpA− E. coli. Similarly, OmpA− E. coli was present in less than 1% of the spleen macrophages, whereas ∼50% of the spleen macrophages were infected with OmpA+ E. coli. Representative images, with a single cell in each situation to show the details of the bacteria associated with the cells, are shown in Fig. 5. Multiple OmpA+ E. coli cells entered both monocytes and spleen macrophages, while for the most part one bacterium per cell was observed with OmpA− E. coli (Fig. 5B and H). OmpA− E. coli appeared to be fragmented inside the macrophages and monocytes (Fig. 5E and K). All these cells showed positive staining with anti-CD64 antibody. These results, which are in good agreement with the in vitro data, suggest that OmpA+ E. coli has the capacity to enter both monocytes and macrophages, to survive, and to multiply in vivo.
FIG. 5.
OmpA+ E. coli entry into monocytes and spleen macrophages in neonatal rats. Five-day-old rat pups were infected with either OmpA+ E. coli strain E44 or OmpA− E. coli strain E91 by intracardiac injection. After 3 h, both monocytes and spleen macrophages were isolated as described in Materials and Methods. The cells were seeded into eight-well chamber slides, fixed, and stained with anti-CD64 antibody (A, D, G, and J) and anti-Sfa antibody for bacteria (B, E, H, and K). Overlays of these pairs of images are shown in panels C, F, I, and L. Note the fragmentation of OmpA− E. coli taken up by the cells (E and K).
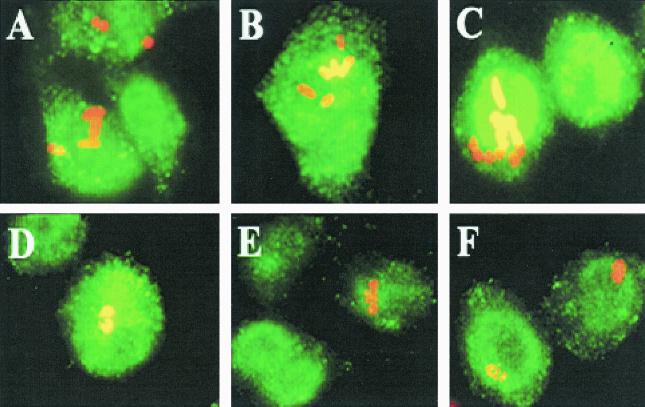
Uptake and replication of OmpA+ E. coli in human monocytes in culture.
Having demonstrated that OmpA+ E. coli enters cells and replicates in the rat model of hematogenous meningitis, we set out to examine whether this bacterium can replicate in isolated human monocytes in vitro. Human monocytes were isolated from heparinized blood and seeded into eight-well chamber slides. After 8 h the wells were washed to remove unbound cells, infected with either OmpA+ or OmpA− E. coli, and monitored for intracellular growth as described above. Staining of only intracellular bacteria showed that significant numbers of OmpA+ E. coli cells entered the monocytes by 1 h, in contrast to the results obtained with OmpA− E. coli cells (Fig. 6A and D). Approximately 20% of the seeded monocytes were infected with OmpA+ E. coli, whereas only 2 to 3% of the cells were infected with OmpA− E. coli. At 4 and 8 h postinfection, the OmpA+ E. coli strain had multiplied significantly (Fig. 6B and C). In contrast, monocytes infected with OmpA− E. coli showed fragmentation of bacteria without a clear rod shape (Fig. 6E and F). Very few cells contained intact OmpA− E. coli even at 8 h postinfection. These cells may have represented the OmpA− E. coli cells that entered or phagocytosed at a later time and may have been in the process of degradation. All these cells showed positive reactivity to anti-CD64 antibody. These results are in agreement with the data obtained in immunocytochemical studies of infected RAW 264.7 cells, suggesting that entry and replication of OmpA+ E. coli in human monocytes in vitro is similar to entry and replication in macrophage cell lines.
FIG. 6.
Phagocytosis and intracellular replication of OmpA+ E. coli in human monocytes in vitro. Monocytes from human peripheral blood were isolated and seeded onto eight-well chamber slides. After 8 h, the attached monocytes were infected with either OmpA+ E. coli (A, B, and C) or OmpA− E. coli (D, E, and F) and monitored for intracellular growth at 1 h (A and D), 4 h (B and E), and 8 h (C and F) as described in Materials and Methods. Only internal bacteria were stained as described in the legend to Fig. 3 with anti-S-fimbria antibody, and the monocytes were stained with anti-CD64 antibody. The primary antibodies were recognized by Cy3- and FITC-conjugated secondary antibodies, respectively.
OmpA+ E. coli internalization by macrophages requires both actin and microtubules.
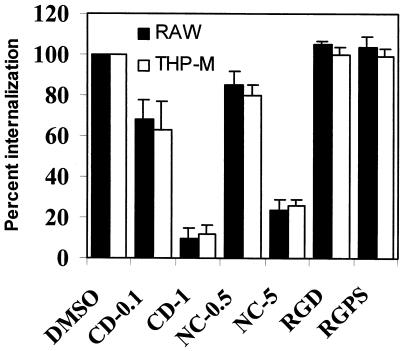
To understand the mechanisms of E. coli K1 entry into macrophages, inhibitors of cytoskeletal architecture were initially used in the invasion assays. As shown in Fig. 7, CD (an inhibitor of actin polymerization) blocked the phagocytosis of OmpA+ E. coli by both macrophage cell lines in a dose-dependent manner; there was >90% inhibition at a concentration of 1 μg/ml compared to DMSO-treated macrophages (1.1 × 104 ± 0.5 × 104 CFU/well for untreated cells versus 0.13 × 104 ± 0.01 × 104 CFU/well for CD-treated THP-M cells; P < 0.001). Similarly, nocodazole, an inhibitor of microtubule polymerization, significantly blocked the entry by 80% at a concentration of 5 μM. We observed that there was no significant difference in the total numbers of cell-associated E. coli cells for CD-, nocodazole-, and DMSO-treated macrophages (data not shown), ruling out the possibility that the absence of internalization of the bacteria was due to inefficient binding of bacteria. The cells were slightly rounded due to exposure to these inhibitors; however, they showed no signs of detachment from the substratum. In addition, the bacteria were also incubated with either RGD or RGPS peptide prior to invasion assays. These peptides did not have an inhibitory effect on the phagocytosis of OmpA+ E. coli by THP-M and RAW 264.7 cells. These findings suggest that the uptake of OmpA+ E. coli by macrophages is not dependent on RGD-mediated integrin receptor structures but is dependent on intact actin filaments and microtubules.
FIG. 7.
Effects of CD (0.1 and 1.0 μg), nocodazole (0.5 and 5.0 μg), and RGD peptides (100 μM) on E. coli entry into macrophages. RAW 264.7 (RAW) and THP-M cells were treated with inhibitors as described in Materials and Methods prior to E. coli invasion assays. The levels of intracellular bacteria were calculated and were expressed as relative internalization values; the level of entry into DMSO-treated cells was defined as 100%. The experiments were carried out at least two times, and the error bars indicate standard deviations.
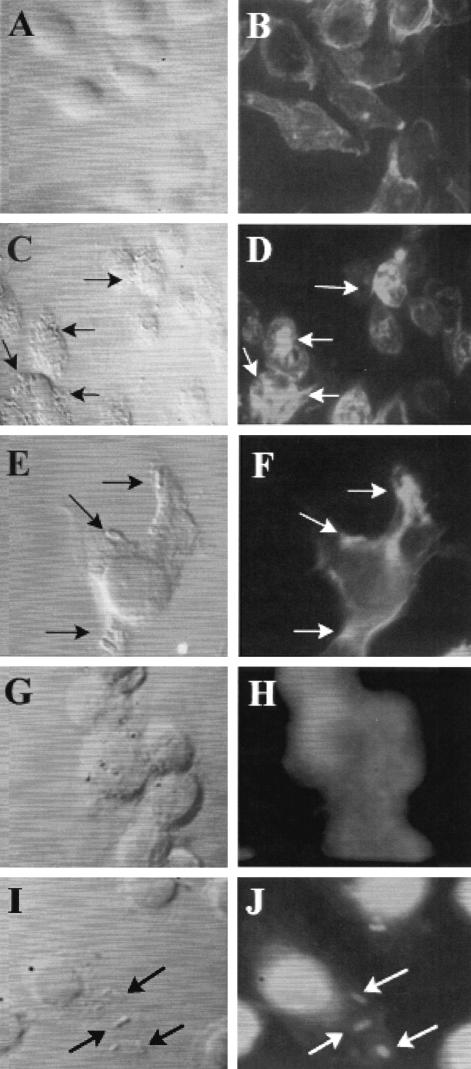
Since an intact cytoskeleton is important for E. coli entry, the actin distribution pattern of the infected macrophages was assessed after fixation and staining with rhodamine phalloidin. Uninfected RAW 264.7 cells exhibited diffuse staining of actin throughout the cells (Fig. 8A and B). In contrast, OmpA+ E. coli-infected cells exhibited a strong accumulation of actin beneath the E. coli adherence site (Fig. 8C and D) in certain areas, whereas other areas did not have such an accumulation despite the presence of bacteria. This phenomenon could have reflected the fact that only a fraction of the bacteria were in an adherence mode, whereas other bacteria were in the process of invasion. A similar phenomenon in E. coli invasion of BMEC has been observed (24). In some instances, a single macrophage extending pseudopods to engulf the bacteria was observed, and at this site there was also a strong accumulation of actin (Fig. 8E and F). In contrast, OmpA− E. coli-infected RAW 264.7 cells showed no detectable condensation of actin beneath the bacteria (data not shown), suggesting that actin reorganization is important for the phagocytosis of OmpA+ E. coli. To further confirm that the inhibition of E. coli uptake by CD-treated RAW 264.7 cells was due to the absence of actin accumulation at the E. coli entry site, CD-treated and OmpA+ E. coli-infected RAW 264.7 cells were also stained for actin. These cells showed no accumulation of actin beneath the bacteria (data not shown), suggesting that CD inhibits E. coli invasion by blocking the actin reorganization necessary for invasion.
FIG. 8.
Actin and microtubule nucleation at the sites of E. coli entry into RAW 264.7 cells. Monolayers of RAW 264.7 cells, either uninfected (A and B) or infected with OmpA+ E. coli (C to F), were fixed and stained for actin with rhodamine phalloidin. In some experiments, uninfected RAW 264.7 cells (G and H) or cells infected with OmpA+ E. coli (I and J) were stained with antitubulin antibody, followed by Cy3-conjugated secondary antibody. The morphology of the cells was revealed by using transmitted light with a blue filter. The arrows indicate either the location of the bacteria or the corresponding actin or tubulin condensation.
Since nocodazole also blocked the entry of E. coli into macrophages, we next examined the distribution of microtubules by using antitubulin antibody and infected macrophages. The tubulin staining was more intense in the nucleus and diffuse in the cytoplasm in uninfected RAW 264.7 cells (Fig. 8G and H). OmpA+ E. coli-infected RAW 264.7 cells exhibited accumulation of tubulin beneath the bacteria, whereas OmpA− E. coli-infected cells showed no such staining (Fig. 8I and J). Nocodazole treatment completely blocked the accumulation of tubulin associated with the bacteria (data not shown). Taken together, these results suggest that both microtubules and microfilaments are important for E. coli K1 invasion of macrophages.
DISCUSSION
In this study we established a macrophage model of internalization and intracellular survival for E. coli K1 that causes meningitis in neonates. Our results indicate that OmpA expression is necessary for effective binding to and subsequent uptake of E. coli by both human and murine macrophages. In addition, OmpA+ E. coli survives and replicates within the macrophages, whereas OmpA− E. coli, which may enter the macrophages by an OmpA-independent process to a lesser extent, is unable to survive or multiply intracellularly. The requirement for OmpA on the surface of E. coli is similar to the requirement for E. coli interaction with HBMEC (29). Moderate inhibition of OmpA+ E. coli invasion of macrophages due to the absence of the IbeA protein indicates that IbeA might also contribute to some extent to the phagocytosis. Our studies with HBMEC showed that OmpA binds to a gp96-like receptor on endothelial cells via both the carbohydrate and the protein backbone (27). Thus, we are examining whether the bacteria use similar receptor structures on macrophages to enter. In agreement with our finding that OmpA is important for binding to macrophages, Soulas et al. have shown that purified OmpA from Klebsiellapneumoniae (which exhibits 88% sequence similarity to OmpA of E. coli K1) also binds to and activates macrophages (35). Interestingly, purified OmpA binds very effectively to both THP-1 cells (monocytes) and RAW 264.7 cells (macrophages) but not to U937 cells (promonocytes) and HL60 cells (promyelocytes), suggesting that the receptors for OmpA are differentially expressed on these cells. The phagocytosis of OmpA+ E. coli by isolated human peripheral blood monocytes and the replication of this organism within these cells strongly suggest that native phagocytic cells are also susceptible to E. coli infection. This concept is supported by our in vivo demonstration that mouse monocytes and macrophages phagocytose E. coli K1. However, the number of spleen macrophages infected with OmpA+ E. coli is significantly greater than the number of monocytes infected, suggesting that macrophages might be the primary target for E. coli K1. The possibility that macrophages from different body locations have different susceptibilities cannot be ruled out. It is interesting to speculate that that E. coli might infect the central nervous system through phagocyte-facilitated invasion, also known as the “Trojan horse” mechanism. Several facultative intracellular bacteria that naturally infect the central nervous system, such as Brucella, Listeria, and Mycobacterium, use this mode of entry (1, 4, 6, 7, 11, 22).
Macrophages discriminate between infectious agents and themselves by a restricted number of receptors that recognize structures shared by large group of bacteria (40). These receptors include endocytic receptors (mannose and scavenger receptors) and signaling receptors (Toll-like receptors) (6, 23, 24). In addition, bacteria opsonized with complement proteins and IgG molecules are recognized by complement receptors and Fc-gamma receptors, respectively (1, 2, 20, 21). The present study revealed that in the absence of opsonins E. coli enters and survives in macrophages very effectively, indicating that E. coli uptake does not require either antibody or complement. This phenomenon is in sharp contrast with what happens with GBS; opsonization renders the latter bacteria more susceptible to intracellular degradation (2, 40). An absence of either complement or antibody on the surface of the bacteria leads to ineffective phagocytosis by macrophages. In addition, a lack of specific antibodies to either the K1 capsule or other membrane proteins creates IgG-free bacteria that interact directly with macrophages. It is known that GBS and Bordetella pertussis directly bind to β2 integrins, whereas E. coli internalization is not mediated by RGD binding integrins (15, 34). Interestingly, nonopsonized E. coli uptake by macrophages requires actin polymerization, suggesting that receptor structures other than complement and Fc receptors might interact with OmpA for internalization. However, the dramatic actin-dependent extensions of the plasma membrane around OmpA+ E. coli resemble Fc receptor-mediated phagocytosis rather than the sinking phenomenon observed in CR3-mediated phagocytosis (39). CR3 can mediate nonopsonic phagocytosis of particles by binding to β-glucan-containing particles and can produce different downstream effects for phagocytosis (40). Further studies are in progress to identify how E. coli enters macrophages.
An important finding of this study is that the infected macrophages contained several megaphagosomes that probably originated from the fusion of small phagosomes. Under such conditions, the movement of phagosomes and fusion with one another and with other organelles are tightly regulated. Previous studies have shown that phagosomes move bidirectionally along with microtubular tracts in macrophages (5). Nocodazole treatment of macrophages prior to infection with Helicobacter pylori significantly inhibited megasome formation (18). Our data also showed that microtubule depolymerization by nocodazole significantly blocked the phagocytosis of OmpA+ E. coli by macrophages, suggesting that megasome formation might be important for survival of OmpA+ E. coli. Several mechanisms, such as inhibition of respiratory burst, resistance to lysosomal enzymes, and attenuation of phagosomal acidification, have been reported to be used by other intracellular bacteria to evade intraphagosomal killing (8, 10). Although the nature of megasomes containing OmpA+ E. coli is not known yet, our data show that OmpA+ E. coli down regulates superoxide production in macrophages, whereas OmpA− E. coli cannot do this (unpublished results). In addition, it is possible that E. coli K1 expresses a unique complement of genes that collectively allow the bacteria to prosper in the hostile environment of macrophages.
In summary, our findings showed that E. coli K1 enters macrophages in an OmpA-dependent manner, probably to maintain a high degree of bacteremia for the onset of meningitis. The phagocytosis of E. coli by macrophages requires intact microfilaments and microtubules. Further studies are in progress to identify the receptor for OmpA on macrophages. A better understanding of the mechanisms of interaction between OmpA and its receptor could help in developing new therapeutic strategies.
Acknowledgments
We thank Luiz Bermudez and Martine Torres for critical reading of the manuscript. We thank Sheng-He Huang for providing E. coli strains E44S−, 7A33, and 10A23.
This work was supported by NIH grant AI 40567 (to N.V.P).
Editor: J. N. Weiser
REFERENCES
- 1.Aderem, A., and D. M. Underhill. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17:593-623. [DOI] [PubMed] [Google Scholar]
- 2.Antal, J. M., J. V. Cunningham, and K. J. Goodrum. 1992. Opsonin-independent phagocytosis of group B streptococci: role of complement receptor type 3. Infect. Immun. 60:1114-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belaaouaj, A., K. S. Kim, and S. D. Shapiro. 2000. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 289:1185-1188. [DOI] [PubMed] [Google Scholar]
- 4.Bermudez, L. E., L. S. Young, and H. Enkel. 1991. Interaction of Mycobacterium avium complex with human macrophages: roles of membrane receptors and serum proteins. Infect. Immun. 59:1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blocker, A., G. Griffiths, J. C. Olivo, A. A. Hymann, and F. F. Severin. 1998. A role of microtubule dynamics in phagosome movement. J. Cell Sci. 111:3030-3312. [DOI] [PubMed] [Google Scholar]
- 6.Celli, J., and B. B. Finlay. 2002. Bacterial avoidance of phagocytosis. Trends Microbiol. 10:232-237. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, J. D. 2000. Bacterial inhibition of phagocytosis. Cell Microbiol. 2:379-386. [DOI] [PubMed] [Google Scholar]
- 8.Falkow, S., R. R. Isberg, and D. A. Portnoy. 1992. The interaction of bacteria with mammalian cells. Annu. Rev. Cell Biol. 8:333-363. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Prada, C. M., E. B. Zelazowska, M. Nikolich, T. L. Hadfield, R. M. Roop II, G. L. Robertson, and D. L. Hoover. 2003. Interactions between Brucella melitensis and human phagocytes: bacterial surface O-polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect. Immun. 71:2110-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finlay, B. B., and S. Falkow. 1989. Common themes in microbial pathogenicity. Microbiol. Rev. 53:210-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfine, H., and S. J. Wadsworth. 2002. Macrophage intracellular signaling induced by Listeria monocytogenes. Microbes Infect. 13:1335-1343. [DOI] [PubMed] [Google Scholar]
- 12.Handel-Fernandez, M. E., and D. M. Lopez. 2000. Isolation of macrophages from tissues, p. 1-16. In D. M. Paulnock (ed.), Macrophages: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 13.Huang, S. H., Z. S. Wan, Y. H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 7:1071-1078. [DOI] [PubMed] [Google Scholar]
- 14.Huang, S. H., C. A. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes, R. O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed]
- 16.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of E. coli meningitis in the rat. J. Clin. Investig. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koedel, U., and H. W. Pfister. 1999. Models of experimental bacterial meningitis: role and limitations. Infect. Dis. Clin. N. Am. 13:549-577. [DOI] [PubMed] [Google Scholar]
- 18.Lee-Ann, H., L. S. Schlesinger, and B. Kang. 2000. Virulent strains of Helicobacter pylori demonstrated delayed phagocytosis and stimulated phagosome fusion in macrophages. J. Exp. Med. 191:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leib, S. L., and M. G. Tauber. 1999. Pathogenesis of bacterial meningitis. Infect. Dis. Clin. N. Am. 13:527-547. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl, G., U. Sjobring, and E. Johnsson. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr. Opin. Immunol. 12:44-51. [DOI] [PubMed] [Google Scholar]
- 21.Linehan, S. A., and D. W. Holden. 2003. The interplay between Salmonella typhimurium and its macrophage host—what can it teach us about innate immunity? Immunol. Lett. 85:183-192. [DOI] [PubMed] [Google Scholar]
- 22.McDonough, K. A., Y. Kress, and B. R. Bloom. 1993. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect. Immun. 61:2763-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Means, T. K., S. Wang, E. Lien, A. Yoshimura, D. T. Golenbock, and M. J. Fenton. 1999. Human Toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. 163:3920. [PubMed] [Google Scholar]
- 24.Pieters, J. 2001. Evasion of host cell defense mechanisms by pathogenic bacteria. Curr. Opin. Immunol. 13:37-44. [DOI] [PubMed] [Google Scholar]
- 25.Prasadarao, N. V., P. K. Srivastava, R. S. Rudrabhatla, K. S. Kim, S. H. Huang, and S. K. Sukumaran. 2003. Cloning and expression of the Escherichia coli K1 outer membrane protein A receptor, a gp96 homologue. Infect. Immun. 71:1680-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasadarao, N. V., A. M. Blom, B. O. Villoutreix, and L. C. Linsangan. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 11:6352-6360. [DOI] [PubMed] [Google Scholar]
- 27.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasadarao, N. V., C. A. Wass, M. F. Stins, H. Shimada, and K. S. Kim. 1999. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion. Infect. Immun. 67:5775-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAc beta 1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasadarao, N. V., C. A. Wass, S. H. Huang, and K. S. Kim. 1999. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 67:1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy, M. A., C. A. Wass, K. S. Kim, D. D. Schlaepfer, and N. V. Prasadarao. 2000. Involvement of focal adhesion kinase in Escherichia coli invasion of human brain microvascular endothelial cells. Infect. Immun. 68:6423-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy, M. A., N. V. Prasadarao, C. A. Wass, and K. S. Kim. 2000. Phosphatidylinositol 3-kinase activation and interaction with focal adhesion kinase in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J. Biol. Chem. 275:36769-36774. [DOI] [PubMed] [Google Scholar]
- 34.Saukkonen, K., C. Cabellos, M. Burroghs, S. Prasad, and E. Tuomanen. 1991. Integrin-mediated localization of Bordetella pertussis within macrophages: role in pulmonary colonization. J. Exp. Med. 173:1143-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulas, C., T. Baussant, J. P. Aubry, Y. Delneste, N. Barillat, G. Caron, T. Renno, J. Y. Bonnefoy, and P. Jeannin. 2000. Outer membrane protein A (OmpA) binds to and activates human macrophages. J. Immunol. 165:2335-2340. [DOI] [PubMed] [Google Scholar]
- 36.Stins M. F., N. V. Prasadarao, L. Ibric, C. A. Wass, P. Luckett, and K. S. Kim. 1994. Binding characteristics of S fimbriated Escherichia coli to isolated brain microvascular endothelial cells. Am. J. Pathol. 145:1228-1236. [PMC free article] [PubMed] [Google Scholar]
- 37.Sukumaran, S. K., M. J. Quon, and N. V. Prasadarao. 2002. Escherichia coli K1 internalization via caveolae requires caveolin-1 and protein kinase C alpha interaction in human brain microvascular endothelial cells. J. Biol. Chem. 278:50716-50724. [DOI] [PubMed] [Google Scholar]
- 38.Sukumaran, S. K., and N. V. Prasadarao. 2002. Regulation of protein kinase C in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J. Biol. Chem. 277:12253-12262. [DOI] [PubMed] [Google Scholar]
- 39.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825-852. [DOI] [PubMed] [Google Scholar]
- 41.Valentin-Weigand, P., P. Benkel, M. Rohde, and G. S. Chhatwal. 1996. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64:2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, Y., and K. S. Kim. 2002. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr. Res. 51:559-563. [DOI] [PubMed] [Google Scholar]