Abstract
We identified a homologue of the alternative oxidase gene in a screen to identify genes that are preferentially transcribed in response to a shift to 37°C in the human-pathogenic yeast Cryptococcus neoformans. Alternative oxidases are nucleus-encoded mitochondrial proteins that have two putative roles: they can function in parallel with the classic cytochrome oxidative pathway to produce ATP, and they may counter oxidative stress within the mitochondria. The C. neoformans alternative oxidase gene (AOX1) was found to exist as a single copy in the genome, and it encodes a putative protein of 401 amino acids. An aox1 mutant strain was created using targeted gene disruption, and the mutant strain was reconstituted to wild type using a full-length AOX1. Compared to both the wild-type and reconstituted strains, the aox1 mutant strain was not temperature sensitive but did have significant impairment of both respiration and growth when treated with inhibitors of the classic cytochrome oxidative pathway. The aox1 mutant strain was also found to be more sensitive to the oxidative stressor tert-butyl hydroperoxide. The aox1 mutant strain was significantly less virulent than both the wild type and the reconstituted strain in the murine inhalational model, and it also had significantly impaired growth within a macrophage-like cell line. These data demonstrate that the alternative oxidase of C. neoformans can make a significant contribution to metabolism, has a role in the yeast's defense against exogenous oxidative stress, and contributes to the virulence composite of this organism, possibly by improving survival within phagocytic cells.
Cryptococcus neoformans is a basidiomycetous fungus and a major human pathogen. This pathogenic fungus has a wide human host range by producing infections in both immunocompromised and immunocompetent hosts (6). Understanding the mechanisms of how this encapsulated yeast can so effectively attack a susceptible host has received renewed attention as the infrastructure for molecular biology of this yeast has matured. Several phenotypic factors in the virulence composite, such as formation of a polysaccharide capsule (7), melanin production (45), urease synthesis (11), phospholipase secretion (10), mannose production (46), high-temperature growth (28), and several signaling molecules and pathways for these factors (44, 28), have been identified at the level of their encoding genes and/or controlling networks.
In the genome era, it has been fashionable to use gene expression profiling of a microorganism under certain environmental conditions to understand its biology. With the use of a global differential gene expression technique, we identified a series of C. neoformans genes which were induced when exposed to mammalian body temperatures (37°C) compared to the gene expression profile at an environmental temperature (25°C). In the sequencing of these transcripts, we identified a gene which appeared to have high homology with genes identified as those of alternative oxidases in other organisms. The functional importance of this gene (AOX1) to the general metabolism, respiration, stress response, and impact on the virulence composite became of particular interest, since there is no homologous pathway in humans and thus it might represent a unique molecular drug target.
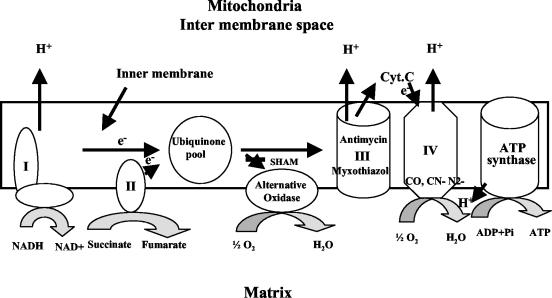
In eukaryotic organisms, energy for growth, development, reproduction, and response to external stresses is derived mainly through ATP production during mitochondrial respiration, and this need for energy is hypothesized to be extremely important to a pathogen's ability to produce disease under stress of the host environment. In animals, during respiration, electron transfer from NADH to molecular oxygen proceeds through sequential protein complexes within the inner mitochondrial membranes. In addition to this sequential cytochrome pathway, higher plants (27), algae (12), yeasts (34), filamentous fungi (20), dimorphic fungi (18), amoebae (17), and trypanosomes (8) possess a unique cyanide-resistant electron transport chain in their mitochondria (Fig. 1). This pathway is composed of a homodimeric protein identified as an alternative oxidase. This cyanide-resistant oxidase catalyzes electron transfer from reduced ubiquinone to oxygen, bypassing the main cytochrome respiratory pathway (42). The pathway is resistant not only to cyanide, but also to myxothiazol and antimycin A. However, the pathway is inhibited by salicylhydroxamic acid (SHAM), disulfiram, and N-propyl galate.
FIG. 1.
Schematic diagram of fungal mitochondrial electron transfer chain during respiration in the inner mitochondrial membrane. Electron transfer complexes (I to IV), alternative oxidase and sites of action of inhibitors, SHAM, myxothiazol, antimycin, and cyanide are shown.
In this study, we describe the identification of the C. neoformans alternative oxidase gene (AOX1) in a screen for genes preferentially transcribed in response to high temperature. Through the use of mutants generated using targeted disruption, we demonstrate that the AOX1 gene has a significant contribution to both cellular metabolism and virulence of this yeast. The role of this gene product in the virulence composite may be due to a contribution of both energy production and resistance to oxidative stress.
(Portions of this work were presented at the Fifth International Conference on Cryptococcus and Cryptococcosis, March 2002.)
MATERIALS AND METHODS
Strains and media.
C. neoformans strain H99 (serotype A) and strain H99R (spontaneous ura5 auxotroph derived from H99 by plating on 5-fluoroorotic acid agar) were grown on yeast-peptone-dextrose (YPD) medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] dextrose) with continuous shaking at 30°C. Null mutants were cultured on defined minimal medium containing 0.68% yeast nitrogen base without amino acids (Sigma, St. Louis, Mo.), 1% (wt/vol) dextrose, 0.2% yeast synthetic dropout medium (lacking uracil), and 1.6% agar. The reconstituted strains were maintained on YPD media supplemented with 100 μg of nourseothricin (clonNAT; Werner Bioagents, Jena, Germany)/ml. For animal and macrophage studies, strains were grown in YPD broth with shaking for 18 to 24 h at 30°C. Cells were harvested with centrifugation at 1,500 × g for 10 min and washed three times with phosphate-buffered saline (PBS), and the resuspended cells were counted by utilizing a hemacytometer.
Cloning of the AOX1 gene.
A subtractive cDNA library using differential PCR amplification (PCR Select; Clontech, Palo Alto, Calif.) was used to select for cDNA transcripts of C. neoformans preferentially expressed at an exposure temperature of 25°C versus 37°C. Briefly, strain H99 was grown in YPD broth for 1, 4, 8, and 24 h at either 25 or 37°C in a shaking incubator. Total RNA from H99 cells grown under each temperature condition was isolated using Trizol reagent (Invitrogen, Carlsbad, Calif.), and the RNA from each temperature was pooled and used in the differential PCR amplification according to the manufacturer's protocol. Clones from the pool of cDNAs preferentially expressed at 37 versus 25°C were screened for intensity of hybridization using labeled total RNA from H99 cells grown at the two temperatures and pulsed with [32P]dATP. Several of the clones having an approximately threefold or greater hybridization profile with labeled total RNA from the H99 cells grown at 37°C compared to that with labeled total RNA from cells grown at 25°C were cloned into a pBlueScript SK vector and sequenced. The deduced sequence identified from one clone suggested that it was a partial transcript of a gene with similarities to an alternative oxidase gene from other species. Further analyses from cryptococcal databases (http://www-sequence.stanford.edu; http://www.genome.ou.edu; http://cneo.genetics.duke.edu) identified several fragments, and these were utilized to construct overlapping contigs. From these sequences, a genomic fragment of 3,497 bp was identified which contained a full-length AOX1. Putative introns in the coding region were identified both by comparing the putative amino acid sequence with those of other plant and fungal alternative oxidase genes and by identifying the 5′ and 3′ splice sites of GTNNGY and YAG, respectively. MacVector 6.0.1 (Oxford Molecular Group) software was used for amino acid comparisons.
Disruption of AOX1 gene and creation of an aox1 mutant.
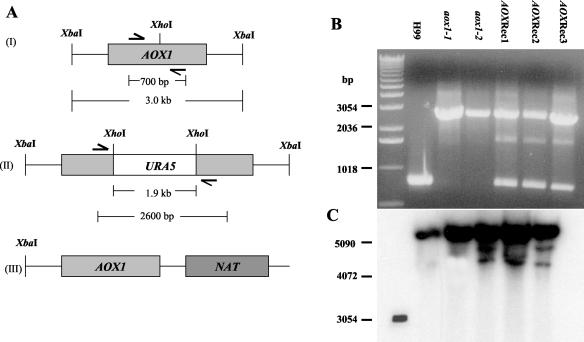
In order to generate a knockout construct for AOX1, a 2.18-kb fragment containing the entire coding sequences of AOX1 was amplified from H99 genomic DNA using primers P1 (CGATGAACGAACTGCGATATC) and P2 (GAGACAACGGATGCATCGACG) and ligated into a pBluescript SK vector lacking an intact XhoI site [Fig. 2A (I)]. A disruption construct was made by digesting the vector containing the 2.18-kb AOX1 fragment with XhoI, and a 1,900-bp XhoI-cut C. neoformans URA5 gene was ligated into this site [Fig. 2A (II)]. The plasmid containing the disruption construct was then used to transform a ura5 auxotrophic H99 strain of C. neoformans using biolistic DNA delivery (38). Transformants were grown on uracil-dropout media. Individual transformants were screened with colony PCR using 5′ primer ATTAGCTAGCGTTTACACTTCTGC and 3′ primer CAAGGTTCCGCCGACCATG. For Southern analysis, 20 μg of genomic DNA from H99 and aox1 strains was digested for 2 h at 37°C with XbaI. Digested DNA was transferred from gel to nylon membrane as described previously (11). The blot was probed with a [32P]dCTP-labeled 2.18-kb genomic AOX1 fragment using a random primer labeling kit (Gibco BRL, Gaithersburg, Md.).
FIG. 2.
(A) Disruption of C. neoformans AOX1 by homologous recombination. (I) Restriction map of the genomic fragment containing the alternative oxidase gene (AOX1). (II) A 1.9-kb URA5 gene is ligated into the XhoI site within AOX1. The solid arrows flanking the XhoI sites designate the sites of the PCR primers that were used to screen for disruption of the native gene. (III) Construct used to complement aox1 strain with AOX1. A nourseothricin resistance cassette (NAT) was ligated into the plasmid containing AOX1 and was used as a selectable marker. (B) PCR analysis using primers shown in panel A (I) gives a 700-bp fragment in H99. In aox1 strains (aox1-1 and aox1-2), this fragment is displaced by 2,600 bp due to insertion of URA5. The fragments from reconstituted strains (AOXRec1 to AOXRec3) demonstrate the disrupted native locus as well as that corresponding to the wild-type copy of AOX1. (C) Southern analysis of genomic DNA cut with XbaI confirms that AOX1 has been disrupted in the knockout mutant and that the reconstituted AOX1 strains show random ectopic insertion of AOX1.
Reconstitution of aox1 mutant.
A 3.2-kb genomic fragment containing the full-length AOX1 was amplified and subcloned into a plasmid. A 1.7-kb cassette conferring resistance to the antibiotic nourseothricin (25) was ligated into a filled-in NcoI site of this plasmid [Fig. 2A (III)]. The plasmid was used to transform the aox1 mutant using the biolistic DNA delivery method. Transformants were selected on YPD plates supplemented with 100 μg of nourseothricin/ml. Both PCR and Southern analyses identified the presence of the intact wild-type AOX1. This strain (AOXRec1) showed ectopic insertion of the entire AOX1 and was selected for further studies (Fig. 2B and C).
Northern analysis.
Strains H99, aox1, and AOXRec1 were grown in YPD broth at 30°C for 18 h. Yeast cells were pelleted, suspended, and divided equally for exposures to specific environmental temperatures (30 versus 37°C) for 4 versus 24 h. Cells were pelleted and crushed with glass beads, and total RNA was isolated with the use of the Trizol reagent. Twenty micrograms of total RNA was used for each lane. Gel electrophoresis, RNA transfer, and blotting were performed by standard Northern techniques, and the blot was probed with the 2.18-kb 32P-labeled AOX1 fragment.
Measurement of respiration rate.
Wild-type (H99), aox1, and AOXRec1 strains were grown at 30°C for 24 h in YPD broth. Cells were harvested, and oxygen consumption was measured with a Clark-type oxygen electrode in a 2.0-ml water-jacketed chamber. All measurements were conducted at 30°C. The assay buffer consisted of 10 mM potassium phosphate [pH 7.0], 10 mM KCl, 5 mM MgCl2, 0.3 M mannitol, and 0.1% (wt/vol) bovine serum albumin. Respiratory inhibitors (myxothiazol at 6 μM and SHAM at 2 mM) were added just before measurement of respiration rate. The respiratory rate of the yeasts was calculated in nanomoles of O2 per milliliter per minute.
Growth inhibition assay.
For the quantitative in vitro growth response to respiratory inhibitors, all three strains were grown in YPD broth for 24 h. Cells were pelleted, suspended, and counted in a hemacytometer. The yeast cells were adjusted with YPD to 104 cells/ml. The adjusted yeast cells were then treated with the respiratory inhibitors, 1 mM potassium cyanide, and 2 mM SHAM in YPD broth for 24 h at 30°C, and then yeasts were quantitatively subcultured onto YPD agar plates for measurements of viable colonies per milliliter of broth.
Impact of oxidative or nitrosative challenge on yeast growth.
The three yeast strains (H99, aox1, and AOXRec1) were grown in YPD broth overnight at 30°C with shaking, washed three times in PBS, and counted in a hemacytometer. Yeasts were then added at a concentration of 104 CFU to a 10-ml Falcon tube containing 2 ml of RPMI 1640 and either 0.1 mM tert-butyl hydroperoxide, 1 mM 2,2′-(hydroxynitrosohydrozono) bis-ethanamine (Cayman Chemicals), or media alone. Triplicate cultures were grown for 48 h at 30°C with shaking, and then aliquots were taken for quantitative cultures on YPD agar.
Macrophage-cryptococcus growth assay.
The MH-S murine alveolar macrophage cell line (American Type Culture Collection, Manassas, Va.) was maintained in RPMI 1640 containing 10% fetal calf serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 4.5 g of glucose/liter, 1.5 g of bicarbonate/liter, 0.05 mM 2-mercaptoethanol, and penicillin and streptomycin at 37°C with 5% CO2. Macrophages were harvested from monolayers using 0.25% trypsin-0.03% EDTA, and viable cells were determined by trypan blue exclusion and counted in a hemacytometer. The macrophage concentration was adjusted to 5 × 105cells/ml, and in experiments using activated macrophages, the cells were primed with 100 units of murine gamma interferon/ml and stimulated with 0.15 μg of lipopolysaccharide (LPS)/ml just prior to mixing with yeasts. One hundred microliters of the macrophage suspensions was placed into 96-well plates. Cryptococci that had been washed three times in PBS were counted in a hemacytometer and adjusted to 5 × 105 yeast cells/ml using cell culture media, and 100 μl was added to the MH-S cells at a multiplicity of infection (effector-to-target ratio) of 1:1. Control wells containing only macrophages or yeasts were included in all experiments. All experiments used 10 μg of 18B7, an immunoglobulin G1 anti-GXM monoclonal antibody, per ml; the antibody was added to the yeast inocula as an opsonin and was generously provided by Arturo Casadevall. The macrophage and yeast mixtures were allowed to incubate for 2 hours before washing with three exchanges of culture media to remove extracellular yeasts. After 24 h, quantitative cultures were performed by aspirating the media from each well and then lysing the remaining macrophages with two exchanges of 100 μl of 1% sodium dodecyl sulfate (SDS) in water. The aspirated media and SDS washes were combined and cultured on YPD agar containing chloramphenicol for yeast colony counts. All experiments were done in triplicate and repeated twice.
Animal models. (i) Rabbits
A total of 108 cryptococci from strain H99, aox1, or AOXRec1 were inoculated into the cisterna magna of each of nine corticosteroid-treated New Zealand White male rabbits (three for each strain), as previously described (30). Rabbits were immunosuppressed daily with cortisone acetate injections (5 mg/kg of body weight), and cerebrospinal fluid (CSF) was sampled from rabbits sedated with xylasine and ketamine on days 4, 7, and 10 after initiation of infection. CSF dilutions in PBS were plated onto YPD agar plates for measurement of colony counts per milliliter of CSF.
(ii) Mice.
The three yeast strains were used to infect 4- to 6-week-old female, A/Jcr mice (NCI/Charles River Laboratories) using nasal inhalation. Ten mice were infected with 5 × 105 CFU of the H99, aox1, and AOXRec1 strains in a volume of 50 μl via nasal inhalation, as described previously (11). The mice were fed ad libitum and monitored with twice-daily inspections. Mice that appeared moribund or in pain were sacrificed using CO2 inhalation.
Statistics.
In macrophage and oxidant challenge experiments, yeast counts were compared by a one-way analysis of variance with Bonferroni's correction posttest. Survival data from the murine experiments were analyzed by the Kruskal-Wallis test, and a Student t test was performed for comparison of CSF yeast counts. A test comparison with a P value of <0.05 was considered to be significant.
Nucleotide sequence accession number.
The genomic sequence of the C. neoformans AOX1 has been submitted to GenBank with the accession number AF502293.
RESULTS
Isolated AOX1 contains all conserved motifs and sequences.
In the identification of differentially expressed genes for H99 when exposed to temperatures of 25 or 37°C, we cloned a 400-bp partial transcript which was truncated and up-regulated with growth at 37°C compared to that at 25°C. The transcript possessed sequence homology with an alternative oxidase gene from several other yeast species. With the use of C. neoformans genomic sequences in various databases (Stanford, Oklahoma, and Duke University) and further Southern analyses of C. neoformans DNA, we identified a full-length AOX1 gene of C. neoformans and confirmed that only one copy of this gene resides in the C. neoformans genome. AOX1 from C. neoformans is encoded by a 1,415-bp gene that contains four introns. Alignment of known alternative oxidase amino acid residues from different fungi with the predicted protein sequence of C. neoformans revealed several highly conserved regions within the C. neoformans AOX1. The most highly conserved areas are clustered in the central regions of the protein. The potential metal binding sites are highly conserved within the C. neoformans AOX1. The protein also contains the proposed active catalytic site defined by two pairs of helices forming a four-helix bundle. Two antiparallel helices, one in each pair, contain a critical EXXH (Glu-X-X-His) motif (1). These motifs supply residues for iron coordination (19). It can be noted that the sequences contain a hydrophobic linker in between the two helical pairs. Since the sequence includes several conserved positively charged residues, the linker is proposed to act as a membrane anchoring region (19). Like the Candida albicans AOX1, the C. neoformans AOX1 also contains a potential cleavage site for mitochondrial presequences between Ser residues.
Disruption of alternative oxidase activity of C. neoformans.
We utilized the biolistic DNA method to disrupt AOX1 of C. neoformans by inserting a 1,900-bp URA5 fragment into the XhoI site of the AOX1 to create the aox1::URA5 disruption allele. We screened 50 transformants using colony PCR, and two homologous gene disruptants (aox1-1 and aox1-2) were found. The null mutant aox1-1 was used for all biological experiments in this study and is designated aox1. PCR analyses demonstrated that the expected 700-bp fragment of AOX1 was amplified from the wild-type strain (Fig. 2B, H99), while an expected 2,600-bp fragment containing the 1,900 bp of URA5 inserted within the 700 bp of AOX1 fragment was amplified from the aox1 mutants (Fig. 2B, lanes aox1-1 and aox1-2). To confirm the PCR results, Southern analysis was performed. DNA from the transformants was cut with XbaI, and it was expected that aox1 mutants would produce a 4.9-kb fragment in the case of disruption of AOX1 by the insertion of URA5; this was observed (Fig. 2C, lanes aox1-1 and aox1-2). The aox1 mutant had the AOX1 replaced back into it on a nourseothricin-selectable plasmid in three independent reconstituted strains (AOXRec1, -2, and -3); the PCR products obtained from them demonstrated both the disrupted native locus of the aox1 mutant and the presence of a wild-type copy of AOX1 (Fig. 2B, lanes AOXRec1, -2, and -3). The reconstituted strain, AOXRec1, was used for all biological experiments. The aox1 mutant formed capsule, melanin, and urease similar to those of the wild-type strain.
A functional alternative oxidase is present in C. neoformans.
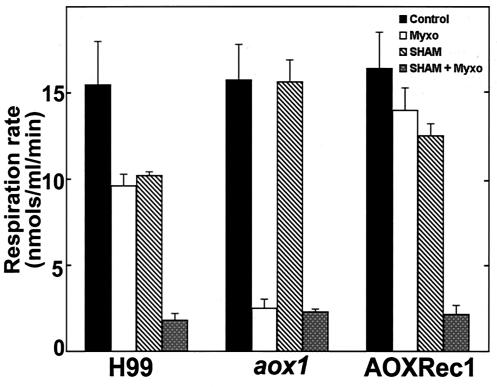
In order to investigate for the presence of a functional alternative oxidase pathway in C. neoformans, we utilized classic oxidative inhibitors that block either the alternative or cytochrome oxidase pathways. As shown in Fig. 1, SHAM inhibits the alternative oxidase pathway, while myxothiazol and potassium cyanide (KCN) are specific inhibitors for the cytochrome oxidase pathway. Our results from treatment of C. neoformans yeast cells with the inhibitors are presented in Fig. 3. The rate of oxygen consumption was measured to assess the effectiveness of these respiratory inhibitors on H99, aox1, and AOXRec1 strains. With application of myxothiazol, the rate of oxygen consumption as measured in nanomoles per milliliter per minute was inhibited by approximately 37% in H99 compared to that in the strain without inhibitors. When SHAM was used alone, it showed the same pattern of respiratory depression as myxothiazol. Furthermore, addition of both SHAM and myxothiazol to block both pathways almost completely inhibited respiration (Fig. 3). These findings suggest the existence of a functional alternative oxidase pathway in C. neoformans. Further support for two functional respiratory pathways was found when the application of myxothiazol severely reduced oxygen consumption in the aox1 mutant. It is noted that the reduction of oxygen consumption from myxothiazol treatment in the aox1 mutant is identical to the reduction in H99 cells treated with both myxothiazol and SHAM. When aox1 mutant was treated with SHAM alone, respiration remained unchanged. The fact that a functional alternative oxidase pathway in the aox1 mutant is absent or severely limited is confirmed by the results obtained with AOXRec1. This reconstituted strain followed the H99 pattern of oxygen consumption in response to the respiratory inhibitors.
FIG. 3.
C. neoformans has a functional cyanide-resistant respiratory pathway. Oxygen uptake was monitored in the presence of myxothiazol and SHAM. Data shown represent the means and standard errors of results from three independent experiments.
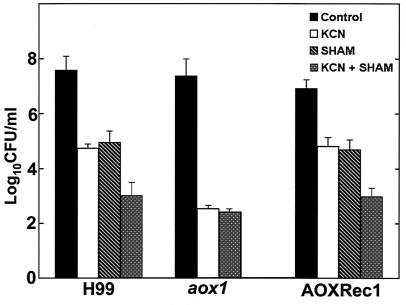
In the next set of experiments, the respiratory inhibitors' effect on respiration rates correlated with their impact on yeast growth in nutrient-rich media. H99, aox1, and AOXRec1 were grown on YPD medium in the presence of different inhibitors. H99 showed a reduction of about 50% in growth in the presence of KCN or SHAM alone. The growth rate was further reduced when KCN and SHAM were used together (Fig. 4). Furthermore, treatment with KCN led to a reduction of growth of the aox1 mutant to a level similar to that of H99 when treated with both respiratory inhibitors (KCN and SHAM). There was no further effect on aox1 mutant growth when the mutant was exposed to SHAM (Fig. 4). In fact, in the aox1 strain, the KCN effect was fungicidal, since the treatment with this inhibitor reduced the yeast cell counts below the initial inoculum concentration. The AOXRec1 strain followed the H99 pattern of growth in response to both inhibitors.
FIG. 4.
Growth patterns of C. neoformans (H99, aox1, and AOXRec1). A total of 104 CFU/ml were treated with 1 mM KCN and 2 mM SHAM at 30°C for 24 h, and cells were plated for number of surviving colonies. (SHAM alone was not tested with aox1.)
AOX1 transcription is up-regulated during temperature stress.
Fungal pathogens need to be able to survive and grow at mammalian body temperatures in order to successfully invade the human host (21). The apparent up-regulation of AOX1 at 37°C enabled us to clone this gene through subtractive hybridization of cDNA libraries. Therefore, we reconfirmed whether AOX1 expression was induced with high temperature exposure. H99 was grown under two different temperatures (30 and 37°C) for 4 or 24 h. There was no difference in growth rate of H99 at these temperatures. Transcript levels were examined by northern analysis, and the results are presented in Fig. 5. With exposure to an environmental temperature of 30°C at 4 h, AOX1 was expressed at a measurable level (Fig. 5, lane 2), but when yeast cells were further exposed to 30°C for 24 h, no change in AOX1 expression was detected (lane 4). In contrast, when RNA was isolated from H99 cells grown at 37°C for 4 h, AOX1 expression could clearly be seen as up-regulated (approximately 10-fold higher) compared to that in cells grown at 30°C (lane 3). The expression level increased to 20-fold when H99 cells were grown for 24 h at 37°C (lane 5). These results confirmed the initial screening of the differential cDNA library subtraction technique and suggest that AOX1 in C. neoformans might be important in responding to environmental temperature stresses. We then attempted to determine if this up-regulation of AOX1 by temperature was correlated with a temperature-sensitive growth phenotype. The growth rates of aox1 and H99 were the same at 37°C in YPD broth, and thus AOX1 induction by temperature was not correlated with a temperature-sensitive growth phenotype in nutrient-rich conditions.
FIG. 5.
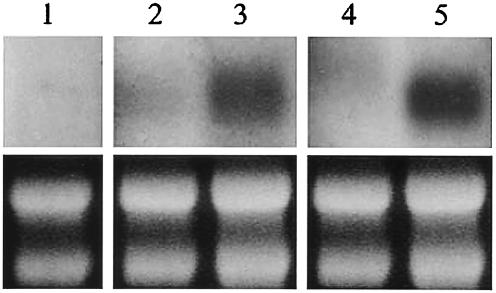
Northern analysis of AOX1 expression. Total RNA obtained from aox1 grown at 37°C for 4 h, H99 grown at 30°C for 4 h, H99 grown at 37°C for 4 h, H99 grown at 30°C for 24 h, and H99 grown at 37°C for 24 h (lanes 1 to 5, respectively) was probed with AOX1 (top). Below are photos of gels showing rRNA bands for measurement of RNA loading (bottom).
Impact of AOX1 on pathogenesis.
Since this pathway has been used by pathogens in stress conditions, we tested whether AOX1 might impact the pathobiology of C. neoformans. We examined the effect of the aox1 mutant on animal models of infection. In the rabbit model of cryptococcal meningitis, equal numbers of yeast cells (108) from H99, aox1, and AOXRec1 strains were inoculated into the cisternae magnae of nine cortisone-treated rabbits, and the CSF was collected at days 4, 7, and 10 of infection. H99, aox1 mutant, and AOXRec1 strains persisted at similar CSF yeast concentrations during the first 10 days of infection. Rabbits infected with the aox1 mutant had a mean CSF yeast count (± standard error of the mean) of log 4.55 ± 1.04 CFU/ml of CSF, compared to log 5.49 ± 1.14 CFU/ml of CSF for rabbits infected with H99, at day 10 of infection. The values were not statistically different from each other (P = 0.17). At the earlier time periods (4 and 7 days of infection), H99, aox1, and AOXRec1 yeast counts were the same. Thus H99 and the aox1 mutant strain survived very well in the high temperature (39 to 40°C) of the rabbit CSF, and it is clear that AOX1 is not needed for the stresses of high-temperature growth. However, immunosuppressed rabbits have a severe compromise of their inflammatory response within the subarachnoid space, and thus there is little impact of intracellular yeast growth with its possible oxidative stresses. Therefore, we examined the yeast strains in a murine model in which there is more exposure to, and demands from, host cells to overcome the yeast.
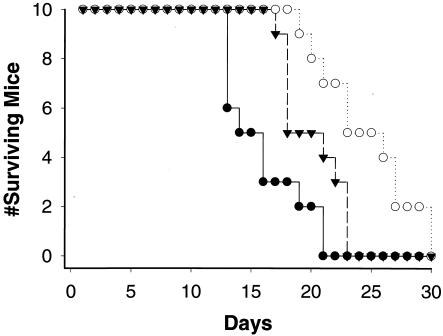
The pulmonary murine model of cryptococcosis was examined to appreciate (i) the impact of another animal species, (ii) a new site of infection, and (iii) added stresses of an active inflammatory response. To test the virulence of the aox1 mutant, mice were infected intranasally with yeast cells from strain H99, aox1, or AOXRec1, and survival was measured (Fig. 6). Mice infected with the aox1 mutant lived significantly longer than those infected with H99 cells. Mice infected with H99 had a mean survival of 14.9 days compared to a mean survival of 23.6 days in mice infected with the aox1 strain (P = 0.001). In confirmation that the difference in survival was specific to a mutation in AOX1, the AOXRec1 strain also resulted in significantly reduced survival compared to infection with the aox1 strain (P = 0.01).
FIG. 6.
Outcome of inhalational murine model. Shown is survival of mice inoculated with wild-type H99 (solid circles), AOXRec1 (solid inverted triangles), and the aox1 strain (open circles). Survival of mice infected with the aox1 mutant was significantly longer than that of mice infected with H99 (P = 0.001) and AOXRec1 (P = 0.01).
aox1 mutant is susceptible to growth inhibition in macrophages.
Since there was a difference in the outcome between the two animal models, we hypothesized that AOX1 might aid growth in certain unique environments or stresses, since the aox1 mutant was normal for known virulence factor phenotypes, namely, capsule production, melanin, synthesis, and urease formation. Recent studies have established that C. neoformans is a facultative intracellular pathogen and can survive the harsh environment of the macrophage phagolysosome (14, 39). It has been shown from histopathological studies that C. neoformans will encounter many more macrophages in the mouse than in the rabbit model (30). These facts suggested to us that it might be helpful to understand AOX1 impact on intracellular growth. Furthermore, to inhibit growth or kill invading microorganisms, phagocytic cells like macrophages generate reactive oxygen species (ROS), and the alternative oxidase in other systems has been shown to dampen the deleterious effects of endogenous ROS. We reasoned that the aox1 strain might be more susceptible to the harsh intracellular environment of the macrophages. The MH-S murine alveolar macrophage cell line was used to assay for its ability to inhibit or kill C. neoformans. The phagocytic indexes of the wild-type, aox1, and AOXRec1 strains were the same, demonstrating that the aox1 mutant had no defects in uptake by macrophages. The aox1 mutant showed significantly slower growth within the gamma interferon- and LPS-stimulated macrophages than did H99 (means, 2.70 × 103 ± 0.60 × 103 CFU versus 1.28 × 104 ± 0.20 × 104 CFU, respectively, at 24 h; P < 0.001). AOXRec1 also showed faster intracellular growth than the aox1 mutant, but it did not reach the H99 growth levels in the macrophages. All three strains grew equally well in unstimulated macrophages. In an attempt to identify the specific mechanism(s) for reduced intracellular growth, the impacts of oxidative and nitrosative stress were examined. Table 1 shows that the oxidative stress, and not the nitrosative stress, impaired the in vitro growth of the aox1 mutant.
TABLE 1.
Quantitative yeast counts after 48-h exposure to stressors
| Stressora | Count (CFU/ml) for strain:
|
||
|---|---|---|---|
| H99 | aox1 | AOXRec1 | |
| None (medium alone) | (1.97 ± 0.16) × 106 | (2.10 ± 0.09) × 106 | (1.92 ± 0.09) × 106 |
| DETA | (1.90 ± 0.18) × 106 | (1.95 ± 0.26) × 106 | (1.61 ± 0.32) × 106 |
| T-B00H | (1.80 ± 0.26) × 106 | (1.90 ± 0.7) × 105b | (1.68 ± 0.20) × 106 |
DETA, 2,2′-(hydroxynitrosohydrozono) bis-ethanamine; T-B00H, tert-butyl hydroperoxide.
Significantly reduced growth compared to that of H99 and AOXRec1 (P = 0.04).
DISCUSSION
With the use of a variety of techniques to examine global gene expression in pathogens, the study of gene expression profiles coordinated with phenotype analysis of null mutants has become a common strategy to understand microbial pathogenesis. Our group has used a differential display PCR method to identify C. neoformans unique transcripts at the site of infection (33). For instance, a transcript of the isocitrate lyase gene (ICL1) was found to be induced in C. neoformans cells during meningitis. However, the analysis of the mutant carrying the site-directed null mutation showed that this induction of gene expression was not necessary for the mutant’s ability to establish infection. This finding illustrates the importance of using global gene expression screens to ask questions about the needs of a pathogen, but these expression profiles require studies with specific null mutants to define the functional importance of these regulated genes and their networks for pathogenesis.
In this study, we used another differential global molecular screen, cDNA library subtraction, to identify C. neoformans genes regulated in response to environmental temperature changes. We reasoned that genes responding to the environmental cue of temperature might also be important to high-temperature growth and thus be a fundamental component of the virulence composite of this pathogen. The ability to grow at high temperatures (37 to 39°C) separates C. neoformans from all other nonpathogenic cryptococci, and the ability to survive at mammalian body temperatures is a crucial feature which separates some 200 fungal species causing clinical disease from the over 20,000 known fungal species in the biomass. Others have shown that C. neoformans has the ability to alter gene expression with exposure to high temperatures. It appears that many of these genes that are induced under high temperature are related to heat shock proteins and the translational machinery for the yeast (36). We similarly have found potential stress-related genes to be up-regulated by exposure to high temperature, and one of these genes, AOX1, attracted our interest because of its unique potential impact on oxidative stress response and/or energy production. We were able to confirm by our global screen with specific Northern analysis that AOX1 is highly induced in its expression in C. neoformans by high temperature. However, despite the importance to virulence of the temperature-sensitive phenotype for several pathways, including the calcineurin (28), RAS (44), and trehalose (E. A. Wills, G. M. Cox, and J. R. Perfect, Abstr. 5th Int. Conf. Cryptococcus Cryptococcosis, abstr. PO17, 2002) pathways, in C. neoformans, the alternative oxidase pathway was not associated with a high-temperature-growth phenotype. In contrast to AOX1, the calcineurin gene of C. neoformans is essential for high-temperature growth (28), but its expression is not regulated by temperature (unpublished data). These results illustrate the disconnect between gene expression and phenotype in C. neoformans for temperature responses and growth. Furthermore, the mechanism for attenuated virulence in the aox1 mutant is not mediated through its growth response to or viability under high-temperature stresses.
The C. neoformans Aox1 protein appears to have all conserved motifs of other Aox1 proteins, including the site for metal binding. Inhibitor studies have shown that Aox1 is also involved with a functional alternative oxidase pathway in C. neoformans which is similar to those of other fungal species and plants. However, in some fungal species, including C. albicans, the alternative oxidase is encoded by a small nuclear gene family whose members are regulated in a developmental and tissue-specific manner (23). In our Southern analysis, screening of the genome databases of C. neoformans, and functional analysis of the null mutant, there is strong evidence that C. neoformans possesses only one alternative oxidase gene.
The importance of cyanide-insensitive respiration (alternative oxidative pathway) in the biology of plants was first observed in the early 1900s. It was initially found to help produce heat for thermogenic plants to assist in their pollination by volatizing compounds to attract insects (27). However, recent studies have focused on the alternative oxidase pathway as being important to specific cellular functions. There have been multiple studies in which alternative oxidase activity has been induced in relationship to a variety of cellular stresses, including low temperature (40, 41), wounding (15), protein (47) and amino acid (2) synthesis inhibitors, stress during inhibition of the cytochrome pathway (40), pathogen attack (22, 35), aging (4, 5), and superoxide anion exposure (26). Most of the recent stress work has focused on alternative oxidase's ability to prevent the production or damage of ROS. It can limit mitochondrial reactive oxygen formation (32, 43) and keeps these potential toxins in the cell low (23). However, the pathway has received little interest in its potential for energy production compared to the classic oxidative phosphorylation pathway.
The generation of ROS can be a determinant of cell survival and growth in plants (24), in fungi such as Podospora anserine (4, 5), and in the worm Caenorhabditis elegans (3). There are direct links to a functioning alternative oxidase system and protection from ROS to preserve growth and prolong viability of the organism (13, 29, 31). It is clear from a variety of studies that both host oxygen- and nitrogen-reactive species are deleterious to C. neoformans and that this yeast must have protective mechanisms to meet the challenge of ROS. For instance, we have found that in C. neoformans, the SOD1 gene (superoxide dismutase), which converts superoxide radicals into hydrogen peroxide and oxygen, has been shown to contribute to the yeast's ability to grow intracellularly and impacts the yeast's efficiency in producing disease (9). However, the sod1 mutant can still produce a lethal infection, and thus other antioxidant systems may be available to this yeast. Similar findings have been found with a nitrosative challenge against C. neoformans in that fhb1 (flavohemoglobin) null mutants are more susceptible to nitric oxide and are attenuated but not completely avirulent in the animal model (M. de Jesus-Berrios, Abstr. 5th Int. Conf. Cryptococcus Cryptococcosis, abstr. P4.4, 2002).
Our findings with aox1 mutants appear to mimic the outcome of the sod1 mutant. Both mutants had no distinct virulence defect in the rabbit meningitis model, which measures primarily extracellular in vivo yeast growth, since corticosteroid treatment of rabbits produces a profound CSF leukopenia (9). On the other hand, in the murine inhalational model, yeast cells must interact with macrophages and their potential for producing oxidative damage. Furthermore, we found in this study that the aox1 mutant did have slower growth only when macrophages were stimulated by interferon and LPS, which induces oxidative products. With the use of an agent that produces direct oxidative damage (tert-butyl hydroperoxide), the aox1 mutant was found to be more susceptible to growth inhibition than H99 and the reconstituted strain. It is suggested from our experiments that aox1 does participate in protection of the yeast from oxidative stresses and may help the yeast with intracellular growth in activated macrophages. On the other hand, with our assay to produce direct exposure to nitric oxide through 2,2′-(hydroxynitrosohydrozono) bis-ethanamine, we did not detect any impact on the growth of aox1. This is an interesting and not predicted finding, since in other systems, such as plants, nitric oxide does activate transcription of an alternative oxidase (16).
The alternative oxidase pathway has been shown to interact with the classic oxidative pathway to protect against ROS, produce intracellular signaling during stress, or generate energy in some biological systems (24, 26, 37). We did not specifically test the impact of the energy production aspects of the alternative oxidase pathway in C. neoformans, but our results suggest that it would play a very minor role, since the yeast was able to grow very well within the rabbit CSF. It is not clear what will be the exact linkage between the cytochrome and alternative pathways in C. neoformans. However, results in this study emphasize that the ability to have at least one of these respiratory pathways be functional is important to the viability of this yeast. When the aox1 mutant had its cytochrome pathway blocked with KCN, there was a complete block of respiration, and despite a nutrient-rich environment with YPD, the yeasts began to die. This finding may have implications for drug targets. For instance, while a block of the alternative oxidase pathway has only temporary impact on the cell under stress and by itself would not be a good antifungal target, a combined inhibition of both respiratory pathways may completely shut down yeast viability under stress conditions. This dual block may impact both the yeast's ability to respond to certain intracellular stresses and its ability to generate large quantities of energy needed to establish infection in a hostile environment. Although the cytochrome oxidase system is conserved between yeasts and mammals, the alternative oxidase system and its interaction with the cytochrome system is not. Strategies to uniquely interrupt these two pathways together in this fungus would likely produce a fungicidal response.
Acknowledgments
This work is supported by an NIH grant (R01-A128388) and as part of the Duke University Mycology Research Unit (P01-AI44975). Gary M. Cox was a recipient of the Burroughs Wellcome Fund New Investigator Award in molecular pathogenic mycology.
We thank Ann Umbach for technical assistance with measuring respiration rate.
Editor: T. R. Kozel
REFERENCES
- 1.Albury, M. S., C. Affourtit, and A. L. Moore. 1998. A highly conserved glutamate residue (Glu-270) is essential for plant alternative oxidase activity. J. Biol. Chem. 273:30301-30305. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, S., R. Bligny, D. A. Day, J. Whelan, and R. Douce. 1997. Induction of alternative oxidase synthesis by herbicides inhibiting branched-chain amino acid synthesis. Plant J. 11:649-657. [Google Scholar]
- 3.Barja, G. 1999. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J. Bioenerg. Biomembr. 31:347-366. [DOI] [PubMed] [Google Scholar]
- 4.Borghouts, C., C. Q. Scheckhuber, O. Stephan, and H. D. Osiewacz. 2002. Copper homeostasis and aging in the fungal model system Podospora anserina: differential expression of PACtr3 encoding a copper transporter. Int. J. Biochem. Cell Biol. 34:1355-1371. [DOI] [PubMed] [Google Scholar]
- 5.Borghouts, C., A. Werner, T. Elthon, and H. D. Osiewacz. 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell. Biol. 21:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans, p. 1-541. ASM Press, Washington, D.C.
- 7.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, Cap64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, M., and G. C. Hill. 1996. Cloning, sequencing, and functional activity of the Trypanosoma brucei brucei alternative oxidase. Mol. Biochem. Parasitol. 83:125-129. [DOI] [PubMed] [Google Scholar]
- 9.Cox, G. M., T. S. Harrison, C. P. Taborda, H. C. McDade, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 11.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinant, M., D. Baurain, and R. F. Matagne. 1998. Characterization of a cDNA encoding the mitochondrial alternative oxidase (AOX) in Chlamydomonas reinhardtii and assays of AOX inactivation by the antisense strategy, p. 441-444. In I. M. Moller, P. Giardestrom, K. Glimelius, and E. Glaser (ed.), Plant mitochondria: from gene to function. Backhuys Publishers, Leiden, The Netherlands.
- 13.Doke, N., Y. Miura, L. M. Sanchez, and K. Kawakite. 1994. Involvement of superoxide in signal transduction: responses to attack by pathogens, physical and chemical shocks, and UV radiation, p. 177-199. In C. H. Foyer and P. M. Mullineaux (ed.), Causes of photooxidative stress and amelioration of defense systems in plants. CRC Press, Boca Raton, Fla.
- 14.Feldmesser, M., S. C. Tucker, and A. Casadevall. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273-278. [DOI] [PubMed] [Google Scholar]
- 15.Hiser, C., and L. McIntosh. 1990. Alternative oxidase of potato is an integral membrane protein synthesized de novo during aging of tuber slices. Plant Physiol. 93:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, X., V. Von Rad, and J. Durner. 2002. Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in arabidopsis suspension cells. Planta 215:914-923. [DOI] [PubMed] [Google Scholar]
- 17.Jarmuszkiewicz, W., C. M. Sluse-Goffart, L. Hryniewiecka, J. Michejda, and F. Sluse. 1998. Electron partitioning between the two branching quinol-oxidizing pathways in Acanthamoeba castelianii mitochondria during steady-state 3 respiration. J. Biol. Chem. 273:10174-10180. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, C. H., J. T. Prigge, A. D. Warren, and J. E. McEwen. 2003. Characterization of an alternative oxidase activity of Histoplasma capsulatum. Yeast 20:381-388. [DOI] [PubMed] [Google Scholar]
- 19.Joseph-Horne, T., D. Hollomon, and P. M. Wood. 2001. Fungal respiration: a fusion of standard and alternative components. Biochem. Biophys. Acta 1504:175-195. [DOI] [PubMed] [Google Scholar]
- 20.Kirimura, K., M. Yoda, and S. Usami. 1999. Cloning and expression of the cDNA encoding an alternative oxidase gene from Aspergillus niger WU-2223L. Curr. Genet. 34:472-477. [DOI] [PubMed] [Google Scholar]
- 21.Kwon-Chung, K. J., J. E. Bennett, and J. C. Rhodes. 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek 48:25-38. [DOI] [PubMed] [Google Scholar]
- 22.Lennon, A. M., U. H. Neuenschwander, M. Ribas-Carbo, L. Giles, J. A. Ryals, and J. N. Seidow. 1997. The effects of salicylic acid and tobacco mosaic virus infection on the alternative oxidase of tobacco. Plant Physiol. 115:783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell, D. P., Y. Wang, and L. McIntosh. 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96:8271-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell, D. P., R. Nickels, and L. McIntosh. 2002. Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. Plant J. 29:269-279. [DOI] [PubMed] [Google Scholar]
- 25.McDade, H. C., and G. M. Cox. 2002. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 26.Minagawa, N., S. Koga, M. Nakano, S. Sakajo, and A. Yoshimoto. 1992. Possible involvement of superoxide anion in the induction of cyanide-resistant respiration in Hansenula anomala. FEBS Lett. 302:217-219. [DOI] [PubMed] [Google Scholar]
- 27.Moore, A. L., and J. N. Siedow. 1991. The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria. Biochim. Biophys. Acta 1059:121-140. [DOI] [PubMed] [Google Scholar]
- 28.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson, P. D., and J. E. Varner. 1993. Hydrogen peroxide and lignification. Plant J. 4:887-892. [Google Scholar]
- 30.Perfect, J. R., S. D. R. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177-194. [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad, T. K., M. D. Anderson, B. A. Martin, and C. R. Stewart. 1994. Evidence for chilling-induced oxidative stress in maize seedlings and regulatory role for hydrogen peroxide. Plant Cell 6:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purvis, A. C. 1997. Role of the alternative oxidase in limiting superoxide production by plant mitochondria. Physiol. Plant. 100:165-170. [Google Scholar]
- 33.Rude, T. H., D. L. Toffaletti, G. M. Cox, and J. R. Perfect. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect. Immun. 70:5684-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakajo, S., N. Minagawa, and A. Yoshimoto. 1993. Characterization of the alternative oxidase protein in the yeast Hansenula anomala. FEBS Lett. 318:310-312. [DOI] [PubMed] [Google Scholar]
- 35.Simons, B. H., F. F. Millenaar, L. Mulder, L. C. Van Loon, and H. Lambers. 1999. Enhanced expression and activation of the alternative oxidase during infection of arabidopsis with Pseudomonas syringae pv. tomato. Plant Physiol. 120:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steen, B. R., T. Lian, S. Zuyderduyn, W. K. MacDonald, M. Marra, S. J. M. Jones, and J. W. Kronstad. 2002. Temperature-related transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 12:1386-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, H., T. Kumagai, A. Goto, and T. Sugiura. 1998. Increase in intracellular hydrogen peroxide and up-regulation of a nuclear respiratory gene evoked by impairment of mitochondrial electron transfer in human cells. Biochem. Biophys. Res. Commun. 249:542-549. [DOI] [PubMed] [Google Scholar]
- 38.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker, S. C., and A. Casadevall. 2002. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. USA 99:3165-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanlerberghe, G. C., and L. McIntosh. 1992. Coordinate regulation of cytochrome and alternative pathway respiration in tobacco. Plant Physiol. 100:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanlerberghe, G. C., and L. McIntosh. 1992. Lower temperature increases alternative pathway capacity and alternative oxidase protein in tobacco. Plant Physiol. 100:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanlerberghe, G. C., and L. McIntosh. 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:703-734. [DOI] [PubMed] [Google Scholar]
- 43.Wagner, A. M., and A. L. Moore. 1997. Structure and function of the plant alternative oxidase: its putative role in the oxygen defense mechanism. Biosci. Rep. 17:319-333. [DOI] [PubMed] [Google Scholar]
- 44.Waugh, S. M., C. B. Nichols, C. M. Decesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. RAS1 and RAS2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 45.Williamson, P. 1997. Laccase and melanin in the pathogenesis of Cryptococ-cus neoformans. Front. Biosci. 2:99-107. [DOI] [PubMed] [Google Scholar]
- 46.Wills, E. A., I. S. Roberts, M. Del Poeta, J. Rivera, A. Casadevall, and G. M. Cox. 2001. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 40:610-620. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Q., L. Mischis, and J. T. Wiskich. 1996. Respiratory responses of pea and wheat seedlings to chloramphenicol treatment. Aust. J. Plant Physiol. 23:583-592. [Google Scholar]