Abstract
Ehrlichia canis virB9 was cloned and expressed. The sequences of virB9 from six geographic locations were identical. virB9 was transcribed by E. canis in dogs, ticks, and cell culture. Infected dogs had antibodies to recombinant VirB9, indicating that VirB9 was produced by E. canis in dogs and was antigenic.
Ehrlichia canis, the etiologic agent of canine monocytic ehrlichiosis (CME), is a gram-negative obligate intracellular bacterium that replicates in monocytes and macrophages (21, 29). CME has been recognized as a significant canine disease that causes considerable levels of morbidity and mortality worldwide (13, 21). E. canis is primarily transmitted by the brown dog tick, Rhipicephalus sanguineus (16, 18, 29). Without antibiotic treatment, dogs infected with E. canis remain infected; in some cases, the infection can persist even after antibiotic treatment (15, 29). There is currently no vaccine available for CME. The type IV secretion system (TFSS) machinery is a multicomponent pore that allows the delivery of virulence factors from bacteria across the bacterial and host membranes into the cytoplasm of the host cell (3). The TFSS has been shown to be essential for the intracellular survival of several facultative intracellular bacteria, including Brucella, Bartonella, and Legionella (1, 19, 24, 26). In a previous study, virB/D operons encoding the TFSS machinery in the human monocytic ehrlichiosis agent Ehrlichia chaffeensis and in the human granulocytic ehrlichiosis agent Anaplasma phagocytophilum were identified (20). VirB9 is one of a few outer membrane components of the TFSS machinery. It has been shown that the virB9 gene of A. phagocytophilum was transcribed in blood specimens from human patients and in an experimentally infected mouse and horse (20). However, it is not known whether VirB9 is expressed and immunogenic in infected mammalian hosts, or whether virB9 is transcribed in ticks. In the present study, we cloned and expressed virB9 from E. canis and investigated these questions.
Cloning and sequencing of virB9.
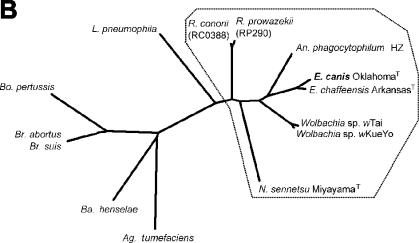
Primers EcavB9f1 and EcavB9r2 (Table 1) were designed to amplify the entire virB9 sequence from E. canis OklahomaT DNA extracted from purified bacteria as previously described (7, 23). PCR products were cloned into the pCRII TA cloning vector and sequenced (12). Analysis of the nucleic acid sequence of the 1,047-bp PCR product showed that it contained an open reading frame of 825 bp encoding a 275-amino-acid protein with a molecular mass of 31,948 Da. The deduced amino acid sequence of the E. canis OklahomaT VirB9 protein included a preferred cleavage site for signal peptidases (Ser-X-Ser) at amino acid positions 25 to 27, leading to a predicted N-terminal cleavable signal peptide of 27 amino acids. The CLUSTAL V method was used to align the nucleic acid sequence of the E. canis OklahomaT virB9 gene, including the signal sequence, with those of E. chaffeensis ArkansasT and A. phagocytophilum HZ. The results showed that the E. canis OklahomaT virB9 base sequence shared 78.0 and 46.5% identity with those of E. chaffeensis and A. phagocytophilum, respectively. The alignment of the predicted amino acid sequence of E. canis OklahomaT virB9 with those of Wolbachia sp. strain wTai, Wolbachia sp. strain wKueYo, and Neorickettsia sennetsu MiyayamaT showed identity levels ranging from 25.0 to 77.3%. A total of 47 amino acids (17% of total amino acids) were consistently conserved across all VirB9 proteins belong to the family Anaplasmataceae (Fig. 1A). A phylogenetic tree consisting of the amino acid sequences of VirB9 from 14 different gram-negative bacteria, including E. canis, was constructed based on the estimated evolutionary distances. VirB9 sequences from E. canis, E. chaffeensis, A. phagocytophilum, a Wolbachia sp., N. sennetsu, and two Rickettsia spp. formed a cluster, as shown in Fig. 1B. By using the Protean program (DNASTAR Inc., Madison, Wis.), nine surface-exposed regions containing 5 to 18 amino acids were identified by the Emini method on E. canis VirB9 that corresponded to the surface-exposed regions of E. chaffeensis VirB9 (data not shown). The surface-exposed regions of E. canis VirB9 had a high antigenic index by the method of Jamison-Wolf, similar to those of E. chaffeensis VirB9. E. canis VirB9 was predicted to contain seven T-cell motifs by the AMPHI method.
TABLE 1.
virB9 primer pairs used in this study
| Primer | Sequence | Purpose |
|---|---|---|
| EcavB9f1 | 5′-GAATTTACCCATCCAAGTGGC-3′ | Cloning and sequencing |
| EcavB9r2 | 5′-CCAACAACGTTTACATTCTCTTC-3′ | |
| EcavB9f3 | 5′-TCGGGATCCGAGCATAGCTGATAA CCCTGTTAC-3′ | Cloning and expression |
| EcavB9r3 | 5′-AGTGCGGCCGCTTATAGTTTTTTGT TAGTTATTTTCACAG-3′ | |
| EcavB9of | 5′-CATTATCATTTCAATACGTAACTC-3′ | Cloning and sequencing |
| EcavB9or | 5′-TTTTGATTTTCTTCTGACATAGTG-3′ | |
| EcavB9f | 5′-GCACACTCCATAAGCATAGCTG-3′ | Expression analysis |
| EcavB9r | 5′-CTCTTAGGTCAGAGTACTCGTC-3′ |
FIG. 1.
(A) Deduced amino acid sequence alignment of VirB9 proteins from six members of the family Anaplasmataceae. Aligned identical amino acids are in white letters on a black background. Gaps in the alignment are indicated by dashes. The alignments were generated by the CLUSTAL V method in the MegAlign program (DNASTAR). Conserved sequences are indicated by arrows. The GenBank accession numbers of the VirB9 proteins of E. chaffeensis ArkansasT, A. phagocytophilum HZ, Wolbachia sp. strain wTai, and Wolbachia sp. strain wKueYo are AF392617, AF392618, AB045234, and AB045235, respectively. The sequence of N. sennetsu MiyayamaT is available at http://www.tigr.org. (B) Phylogram based on the deduced amino acid sequences of the virB9 genes of E. canis and similarities with selected gram-negative bacteria. The amino acid sequences were aligned by the CLUSTAL V method, and phylogenic analysis was performed with the use of the PHYLIP software package (version 3.5). The order Rickettsiales is boxed. The GenBank accession numbers of the virB9 sequences of the organisms used in this analysis are as follows: Agrobacterium tumefaciens Ti, J03320; Bartonella henselae, AF182718; Bordetella pertussis ptl, D47301; Brucella abortus, AF226278; Brucella suis, AF141604; Legionella pneumophila lvh, Y19029; Rickettsia prowazekii, AJ235271; Rickettsia conorii, NC_003103.
Sequence analysis of the virB9 gene from different geographic locations.
Full-length virB9 genes (959 bp) of E. canis were amplified by using the primer pair EcavB9of and EcavB9or (Table 1), from four dog blood 16S rRNA gene-based PCR-positive DNA samples derived from Arizona (strain Arizona-1), California, New Mexico (strain New Mexico-1), and Hawaii and from a DNA sample from the E. canis VDE strain culture isolated from a dog in Venezuela (26). The resulting PCR products were sequenced. These five DNA sequences were aligned with the Oklahoma sequence by the CLUSTAL V method, and all six were found to be 100% identical.
Analysis of virB9 expression.
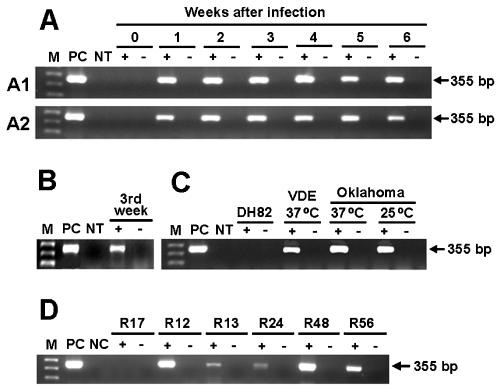
Two 1- to 2-year-old specific-pathogen-free female beagles were infected by intravenous inoculation with naturally infected samples of dog blood from Arizona (strain Arizona-2) and New Mexico (strain New Mexico-2), respectively. Two 1- to 2-year-old specific-pathogen-free female mixed-breed dogs, designated dogs B and C, were intravenously inoculated with E. canis OklahomaT cultivated in DH82 cells (27). Peripheral blood mononuclear cells (PBMCs) were isolated from dog blood samples, as described elsewhere (11). OklahomaT and VDE were cultured in DH82 cells. A pool of E. canis 16S rRNA-positive ticks was removed from each of five dogs in Arizona (a total of 11 ticks) (12). RNA was extracted from dog PBMCs, cell culture, and tick specimens and treated with DNase and reverse transcribed as previously described (10). The resulting cDNA template was amplified with the primers EcavB9f and EcavB9r (Table 1). As a negative control, an equivalent sample of RNA was subjected to the same procedure in the absence of reverse transcriptase, to exclude the possibility that the RNA preparation was contaminated with genomic DNA. virB9 expression was detected in two dogs experimentally infected with E. canis Arizona-2 and New Mexico-2 throughout the 6-week period after intravenous inoculation of infected dog blood (Fig. 2). virB9 expression was also detected 3 weeks after a dog was inoculated with the OklahomaT strain, although this was the only time point examined for that animal. virB9 was transcribed in the VDE and Oklahoma strains of E. canis in culture at 37°C. virB9 was also transcribed in the Oklahoma strain when it was incubated at 25°C, which approximates the natural tick temperature. virB9 expression was detected in all five groups of naturally infected ticks (Fig. 2).
FIG. 2.
virB9 expression by E. canis in naturally infected R. sanguineus tick groups from Arizona and in samples of dog blood. Total RNA was extracted and subjected to reverse transcription-PCR, as described in Materials and Methods. The amplified products were separated on agarose gels and visualized with ethidium bromide. Lane M, molecular size marker (1 Kb Plus DNA ladder; Invitrogen); lane PC, product amplified by PCR with DNA from E. canis culture as a template and respective primer pairs, used as a positive control for PCR and as a determinant of amplicon size on the gel; lane NT, no template (negative control). Samples were run in the presence (+) and absence (−) of reverse transcriptase, as a control for DNA contamination. DH82, uninfected DH82 cells. (A) Weekly expression levels of virB9 between day 0 and 6 weeks after infection of dog PBMCs (A1, Arizona-2; A2, New Mexico-2); (B) virB9 expression in dog C (OklahomaT, virB9 expression was analyzed only at the third week after infection in dog C); (C) virB9 expression in culture of E. canis VDE and OklahomaT in DH82 cells; (D) virB9 expression in naturally infected tick groups (R12, one engorged female; R13, two engorged females and one unengorged male; R24, one male and one semiengorged female; R48, two males and one semiengorged female; R56, two unengorged females; and R17 [E. canis 16S rRNA-negative ticks as a control], two engorged females and one male).
Expression of recombinant VirB9 (rVirB9) in Escherichia coli and Western blot analysis.
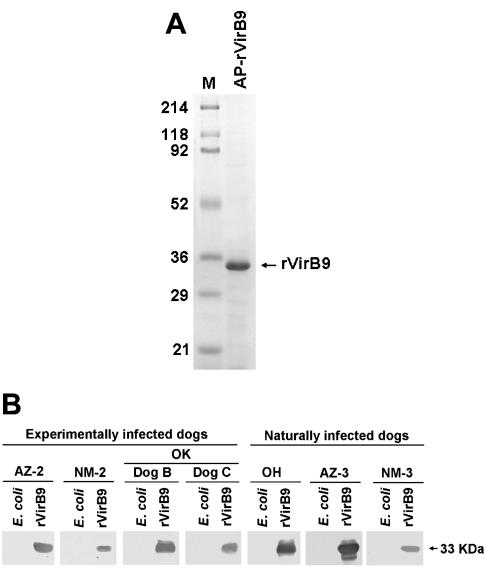
A pair of primers (EcavB9f3 and EcavB9r3) (Table 1) was designed to clone and express the VirB9 protein. The resulting PCR products were digested with BamHI and NotI, ligated into the BamHI and NotI sites of the pET33b vector (Novagen Inc., Madison, Wis.), and amplified in E. coli Novablue cells (Novagen). E. coli BL21(DE3)pLys was transformed with the recombinant plasmid for protein expression. rVirB9 was harvested after 4 h of induction at 27°C with isopropylthio-β-d-galactoside and affinity purified by His-Bind resin (Novagen) column chromatography. E. coli transformed with the E. canis virB9 expression plasmid (designated pET33becavirB9) produced a 287-amino-acid fusion protein (32,932 Da). The protein included 39 amino acids at the N terminus derived from pET33b. The expressed rVirB9 fusion protein was purified by affinity chromatography and detected as a single band of approximately 33 kDa on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Fig. 3A). Western blot analysis was performed using the samples from four experimentally infected dogs as described above, along with three other naturally infected dog plasma samples (Ohio, Arizona-3, and New Mexico-3), to detect antibodies to rVirB9 as previously described (22). All plasma samples from E. canis-infected dogs reacted with rVirB9, as determined by Western blot analysis. None of the samples tested reacted with the E. coli lysate, included as a negative control (Fig. 3B). Three E. canis indirect-fluorescent-antibody-negative plasma samples (from noninfected dogs) did not react with either rVirB9 or E. canis lysate (data not shown). These results showed that VirB9 is expressed by E. canis in infected dogs and that it is highly immunogenic.
FIG. 3.
(A) SDS-polyacrylamide gel electrophoresis analysis of purified E. canis OklahomaT rVirB9. Affinity-purified (AP) rVirB9 (2 μg) was subjected to SDS-polyacrylamide gel electrophoresis followed by Coomassie blue staining. M, molecular size marker. The numbers on the left are molecular masses, in kilodaltons. (B) Western immunoblot analysis to detect E. canis VirB9-specific antibody in plasma samples from infected dogs. pET33b-transformed E. coli was used as a negative control. rVirB9, affinity-purified recombinant fusion protein of E. canis. Antigens (15 μg of E. coli and 2 μg of rVirB9) were subjected to Western blot analysis with serum derived from the blood of various infected dogs (OK, Oklahoma; OH, Ohio; AZ, Arizona; NM, New Mexico). The arrow on the right indicates the apparent molecular mass of rVirB9, based on broad-range prestained standards (Bio-Rad Laboratories, Richmond, Calif.).
VirB9 is a channel protein and forms heterodimers with VirB7 that localize at the outer membrane and play a critical role in stabilizing other VirB proteins during assembly of the TFSS machinery (5). Previously, eight virB and virD genes were detected in the human ehrlichiosis agents E. chaffeensis and A. phagocytophilum. All eight virB and virD genes were found to be expressed by E. chaffeensis and A. phagocytophilum in culture (20). Five of these genes (virB8, virB9, virB10, virB11, and virD4) were polycistronically transcribed and controlled through at least two tandem promoters located upstream of virB8. Thus, it is likely that virB9 is transcribed by E. canis as a part of the virB/D operon in cell culture. Transcription of the virB operon in Agrobacterium tumefaciens has been reported to be regulated in vitro by various environmental factors, including temperature (2, 4). Expression of VirB proteins in Brucella suis was greater in cultures grown at 37 than at 20°C (4). We found that both the Oklahoma and VDE strains of E. canis expressed virB9 in DH82 canine macrophage cell cultures at 37°C and that the Oklahoma strain also expressed virB9 at 25°C. Since the cultures were not synchronized and reverse transcription-PCR was not quantitative, we did not compare the relative virB9 expression levels in this study. It has been clearly established that the TFSS is essential for the survival and replication of intracellular pathogens, such as Brucella, Bartonella, and Legionella pneumophila, in that the TFSS prevents bacterial inclusions from trafficking to lysosomes (1, 5, 6, 9, 14, 19). E. canis is an intracellular pathogen that resides in the membrane-bound inclusion that does not fuse with lysosomes in monocytes and macrophages. The function of E. canis virB9 may be related to facilitating intracellular survival and replication, similar to its function in other intracellular pathogens. Previous studies investigating the role of the TFSS in L. pneumophila showed that the organism has two types of TFSS. The Lvh system functions as a DNA conjugation system but does not contribute to pathogenesis, whereas the Dot/Icm system acts in protein delivery and is required for virulence (5, 17). Curiously, virB9 sequences of E. canis as well as other members of the order Rickettsiales are more closely related to the ortholog sequence in the Lvh system than to that in the Dot/Icm system. In the present study, we found that the virB9 sequences of E. canis strains derived from six geographic regions were identical, showing that virB9 is more highly conserved than either p30-10, which encodes one of the major outer membrane proteins (12), or the 16S rRNA genes of E. canis (27, 28). For vaccine development, the ideal antigenic epitopes should be conserved to protect against all strains (8, 25, 29). E. canis VirB9 seems to be both highly antigenic and expressed in both mammalian and tick hosts. Based on these facts, we speculate that inhibition of VirB9 function may inhibit the intracellular survival of E. canis and thus may serve as a useful vaccine candidate for canine ehrlichiosis.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the E. canis virB9 genes are as follows: OklahomaT, AY205339; Arizona-1, AF546158; California, AY205340; Hawaii, AY205341; New Mexico-1, AY205342; and VDE, AY205343.
Acknowledgments
This research was supported by grants R01AI47407 from the National Institutes of Health. The genome of N. sennetsu was sequenced at The Institute for Genomic Research, and the sequences are available at http://www.tigr.org. The sequencing project was supported by National Institutes of Health grant R01 AI47885 to Y.R.
We appreciate Russell Greene, Deborah Cook, Scott C. Duston, and Maury Brown for sending infected dog blood specimens, and Robert Hamlin for arranging the transfer of two dogs.
Editor: J. T. Barbieri
REFERENCES
- 1.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 2.Baron, C., N. Domke, M. Beinhofer, and S. Hapfelmeier. 2001. Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 183:6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Lavigne, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90:341-348. [DOI] [PubMed] [Google Scholar]
- 4.Boschiroli, M. L., S. Ouhrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevielle, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, J. E., Y. Rikihisa, S. Ewing, and D. B. Fishbein. 1991. Serologic diagnosis of human ehrlichiosis using two Ehrlichia canis isolates. J. Infect. Dis. 163:564-567. [DOI] [PubMed] [Google Scholar]
- 8.De Groot, A. S., A. Bosma, N. Chinai, J. Frost, B. M. Jesdale, M. A. Gonzalez, W. Martin, and C. Saint-Aubin. 2001. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine 19:4385-4395. [DOI] [PubMed] [Google Scholar]
- 9.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 10.Donatien, A., and F. Lestoquard. 1935. Existence and algerie d'une rickettsia du chien. Bull. Soc. Pathol. Exot. 28:418-419. [Google Scholar]
- 11.Felek, S., A. Unver, R. W. Stich, and Y. Rikihisa. 2001. Sensitive detection of Ehrlichia chaffeensis in cell culture, blood, and tick specimens by reverse transcription-PCR. J. Clin. Microbiol. 39:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felek, S., R. Greene, and Y. Rikihisa. 2003. Transcriptional analysis of p30 major outer membrane gene of Ehrlichia canis in naturally infected ticks and sequence analysis of p30-10 of E. canis from diverse geographic regions. J. Clin. Microbiol. 41:886-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrus, S., T. Waner, H. Bark, F. Jongejan, and W. C. A. Cornelissen. 1999. Recent advances in determining the pathogenesis of canine monocytic ehrlichiosis. J. Clin. Microbiol. 37:2745-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqbal, Z., and Y. Rikihisa. 1994. Reisolation of Ehrlichia canis from blood and tissues of dogs after treatment with doxycycline. J. Clin. Microbiol. 32:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, E. M., S. A. Ewing, R. W. Barker, J. C. Fox, D. W. Crow, and K. M. Kocan. 1998. Experimental transmission of Ehrlichia canis (Rickettsiales: Ehrlichiae) by Dermacentor variabilis (Acari: Ixodidae). Vet. Parasitol. 74:277-288. [DOI] [PubMed] [Google Scholar]
- 17.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 18.Neer, T. M. 1998. Canine monocytic and granulocytic ehrlichiosis, p. 139-154. In C. E. Greene (ed.), Infectious diseases of dog and cat, 2nd ed. The W. B. Saunders Co., Philadelphia, Pa.
- 19.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70:2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rikihisa, Y. 1991. The tribe Ehrlichiae and ehrlichial diseases. Clin. Microbiol. Rev. 4:286-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rikihisa, Y., S. A. Ewing, and J. C. Fox. 1994. Western blot analysis of Ehrlichia chaffeensis, E. canis, or E. ewingii infection of dogs and human. J. Clin. Microbiol. 32:2107-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikihisa, Y., S. A. Ewing, J. C. Fox, A. G. Siregar, F. H. Pasaribu, and M. B. Malole. 1992. Analysis of Ehrlichia canis and a canine granulocytic Ehrlichia infection. J. Clin. Microbiol. 30:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy, C. R., K. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 25.Saunders, N. J., and E. R. Moxon. 1998. Implications of sequencing bacterial genomes for pathogenesis and vaccine development. Curr. Opin. Biotechnol. 9:618-623. [DOI] [PubMed] [Google Scholar]
- 26.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 27.Unver, A., M. Perez, N. Orellana, H. Huang, and Y. Rikihisa. 2001. Molecular and antigenic comparison of Ehrlichia canis isolates from dogs, ticks, and a human in Venezuela. J. Clin. Microbiol. 39:2788-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unver, A., Y. Rikihisa, M. Kawahara, and S. Yamamoto. Analysis of 16S rRNA gene sequences of Ehrlichia canis, Anaplasma platys, and Wolbachia species from the blood of dogs in Japan. Proc. N. Y. Acad. Sci. 990:692-698. [DOI] [PubMed]
- 29.Woody, B. J., and J. D. Hoskins. 1991. Ehrlichial diseases of dogs. Vet. Clin. N. Am. Small Anim. Pract. 21:75-98. [DOI] [PubMed] [Google Scholar]