Abstract
Parachlamydia acanthamoeba is an obligately intracellular bacterium that naturally infects free-living amoebae. It is a potential human pathogen and may survive in human macrophages. We studied P. acanthamoeba entry into, and multiplication within, human monocyte-derived macrophages. After 8 h of incubation, 80% of macrophages were infected with a mean of 3.8 P. acanthamoeba organisms per cell. Electron microscopy demonstrated that parachlamydiae were in an intracellular vacuole. After infection with living organisms, the number of parachlamydiae per macrophage increased 4 times from day 0 to day 4, whereas heat-inactivated parachlamydiae were eliminated during the same period. Quantitative PCR confirmed that P. acanthamoeba replicates within macrophages. Transcriptional activity of P. acanthamoeba was detected by reverse transcription-PCR targeting the gene encoding ADP-ATP translocase (tlc). P. acanthamoeba exerted a cytopathic effect on macrophages. When macrophages were infected with living bacteria, their number decreased significantly from day 0 to day 4 due to apoptosis, as shown by annexin-V binding and electron microscopy. This study shows that P. acanthamoeba enters and multiplies within human macrophages before inducing their apoptosis.
First described in 1997 (3, 4), Parachlamydiaceae are obligately intracellular bacteria that naturally infect free-living amoebae (18). Parachlamydiaceae are closely related to the Chlamydiaceae, sharing 80 to 90% sequence homology of rRNA genes (13, 18). They present a Chlamydia-like cycle of replication, with elementary and reticulate bodies and a third developmental stage, the crescent body (19). Parachlamydia acanthamoeba (3) and Neochlamydia hartmanellae (20) are the only described species of the clade (13).
Parachlamydiaceae are potential emerging pathogens (18). The first hint was the identification of this bacterium within an amoeba isolated from the source of an outbreak of humidifier-associated fever in the United States and the related serological study (4). In another serological study including 371 patients with community-acquired pneumonia, 2.2% were seropositive for Parachlamydia (26). Parachlamydia DNA was also detected by PCR in mononuclear cells of a patient with bronchitis (29) and in sputum (11) and bronchoalveolar lavage (10) samples from two other patients. More recent work suggested that Parachlamydia may be an agent of aspiration pneumonia (17) and an agent of community-acquired pneumonia in human immunodeficiency virus-infected patients with low CD4 cell counts (16).
There is growing evidence that amoebae play a role in the adaptation of microbes to macrophages (35). The adaptation of Legionella pneumophila to macrophages and its adaptation to amoebae exhibit similarities at both cellular (2, 5) and molecular (6, 9, 15, 32, 34) levels. Moreover, legionellae grown within amoebae exhibit enhanced growth in monocytes (7). Similarly, growth of Mycobacterium avium in amoebae resulted in enhanced entry into macrophages (8). The fact that nonencapsulated strains of Cryptococcus neoformans do not survive in amoebae, while virulent strains do, is another example of comparable adaptations to free-living amoebae and macrophages (33).
These findings suggest that the virulence traits important for entry and survival in macrophages may be those used to survive within environmental amoebae. Consequently, we wondered whether P. acanthamoeba organisms that survive the bactericidal effects of free-living amoebae may enter and multiply in human macrophages.
MATERIALS AND METHODS
P. acanthamoeba culture and purification.
P. acanthamoeba strain Hall coccus and Acanthamoeba polyphaga Linc-AP1 were kindly provided by T. J. Rowbotham (Public Health Laboratory, Leeds, United Kingdom). P. acanthamoeba was grown at 32°C within A. polyphaga in 150-cm2-surface cell culture flasks (Corning Glass Works, Corning, N.Y.) with 30 ml of peptone-yeast extract-glucose broth (19). After 6 days of incubation, cultures were harvested and the broth was centrifuged at 180 × g for 10 min to eliminate most amoebae. The supernatant was then centrifuged at 6,600 × g for 30 min. The bacterial pellet was suspended in 10 ml of Dulbecco's phosphate-buffered saline (PBS) (Gibco-BRL, Life Technologies, Paisley, Scotland), loaded onto 10% sucrose (Sigma) in PBS, and centrifuged at 5,800 × g for 30 min at 4°C. To improve purification, the pellet was suspended in PBS, loaded onto a discontinuous Gastrografine (Schering, Lys-Lez-Lannoy, France) gradient, and ultracentrifuged at 140,000 × g. Parachlamydiae, which clustered in a large lower band, were collected, centrifuged at 5,800 × g, and resuspended in PBS twice before freezing at −80°C. Parachlamydiae were titered by using a lysis test. Two hundred microliters of 5 × 105 A. polyphaga organisms/ml in Page's modified Neff's amoeba saline (PAS) were distributed in a 96-well Costar microplate (Corning). Parachlamydiae were serially diluted from 10−1 to 10−14 in another microplate. Ten microliters of each dilution was inoculated into six wells of the Acanthamoeba microplate, which was read daily to determine the highest dilution that led to amoebal lysis.
Macrophages.
Blood from healthy volunteers was drawn in tubes containing EDTA as an anticoagulant. Peripheral blood mononuclear cells were separated by centrifugation at 720 × g for 20 min on Ficoll (Eurobio, Les Ulis, France) and suspended in RPMI-HEPES supplemented with 200 mM l-glutamine (Gibco-BRL) and 10% fetal calf serum (Gibco-BRL). Then 106 peripheral blood mononuclear cells/ml were incubated for 1 h at 37°C in a four-well Lab-Tek Chamber Slide system (Nalge Nunc, Naperville, Ill.), in 12-well cell culture plates (Corning), or in 50-cm3 flasks (Corning) for 1 h at 37°C. After being washed, adherent cells were considered monocytes, because more than 95% of them were CD14 positive and exhibited phagocytic characteristics. Monocytes were further differentiated into macrophages by incubation at 37°C in the presence of fetal calf serum.
Infection procedure.
Macrophages were incubated with living or heat-inactivated (for 1 h at 100°C) bacteria in supplemented RPMI medium without antibiotics for 8 h at 37°C. Then cells were washed with RPMI-HEPES and further incubated for different periods at 37°C. Every day, cells were washed in PBS, fixed with 1% formaldehyde (Carlo Erba, Val de Reuil, France) for 10 min, and stained with Diff-Quik (Dade Behring, Düdingen, Switzerland). The number of bacteria per macrophage was determined by counting the parachlamydiae (magnification, ×1,000) within 100 macrophages. The infection index was calculated by multiplying the number of bacteria per macrophage by the number of macrophages per microscopic field. The proportion of infected macrophages was defined as the number of macrophages containing at least one Parachlamydia organisms divided by the total number of macrophages.
Quantitative PCR.
Macrophages were infected with inocula of 107, 106, 105, 104, or 103 parachlamydiae/ml. One hundred microliters of each harvested well was extracted by using the MagNA Pure LC DNA Isolation Kit III (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Quantitative PCR was performed using TaqMan technology. The master mixture was prepared from the TaqMan Universal Master Mix kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. Thus, 25 μl of reaction mixture comprised 200 nM forward primer (Adp81F, 5′-TAGTGATCTGCTACGGGATTT), 200 nM reverse primer (Adp84R, 5′-TTGGATTAGGATATTGCAATTT), 200 nM fluorescently labeled probe (6-FAM-5′-AACCTTGTAGAAGTAACCTGGAAGAACCAGC-3′-TAMRA, where 6-FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine), and 12.5 μl of 2× PCR Master Mix. Amplification was performed on the ABI 7700 sequence detection system (TaqMan system; Applied Biosystems), using 45 cycles, with a hybridization temperature of 52°C and an elongation temperature of 60°C. To prevent carryover, 200 μM uracil triphosphate was part of the master mixture and uracil-N-glycosylase was used systematically. DNAs extracted from parachlamydiae grown in amoebae and from sterile water were used as positive and negative controls, respectively. The positive control was diluted 10-fold to establish a calibration curve. The titer of each sample was determined by comparing its threshold cycle to that of the titered positive control.
Quantitative reverse transcription-PCR (RT-PCR).
One hundred microliters of each well was mixed with 200 μl of RNAprotect Bacteria reagent (Qiagen, Courtaboeuf, France). RNA was extracted by using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Quantitative RT-PCR was performed on the ABI 7700 sequence detection system by using the TaqMan One-Step RT-PCR Master Mix Reagents kit (Applied Biosystems) as recommended by the manufacturer and using the same primers and probe as those for PCR, at a concentration of 200 nM each. After a 30-min retrotranscription step at 48°C, 45 cycles of amplification were performed as described above. DNAs extracted from parachlamydiae grown in amoebae and from sterile water were used as positive and negative controls, respectively. The positive control was 10-fold diluted to establish a calibration curve. The number of copies of each sample was determined by comparing its threshold cycle to that of the positive control.
Macrophage viability and apoptosis.
Cells were harvested, diluted with an equal volume of 0.4% trypan blue (Sigma-Aldrich, Taufkirchen, Germany), and then transferred to a Kova counting slide (Hycor Biomedical, Garden Grove, Calif.). Numbers of unstained (living) and stained (dead) macrophages were determined in triplicate. Apoptosis was assessed by an annexin-V assay. Macrophages were infected with living and heat-inactivated bacteria, and uninfected macrophages were used as negative controls. The binding of Annexin-V-Fluos (Roche) was assessed at 0, 12, 24, 36, and 48 h postinfection. Cells were stained by addition of 20 μl of Annexin-V-Fluos (Roche) in 1 ml of a medium containing 10 mM HEPES-NaOH (pH 7.4), 140 nM NaCl, and 5 mM CaCl2 for 15 min. Since necrotic cells may also be stained with annexin according to the loss of membrane integrity, we then stained the cells simultaneously with annexin and propidium iodide (PI) by adding 20 μl of PI at 50 μg/ml (Sigma, Steinheim, Germany). The percentages of annexin- and annexin-PI-stained cells were determined with a fluorescence microscope (Axioscop; Zeiss, Jena, Germany).
Confocal microscopy.
Macrophages were infected with 107 parachlamydiae, fixed with 1% formaldehyde for 10 min, and permeabilized with 1% saponin (Research Organics, Cleveland, Ohio) for 30 min. Macrophages were first incubated with a homemade rabbit anti-Parachlamydia antibody serum at a 1:400 dilution for 30 min, followed by fluorescein-labeled anti-rabbit antibodies (AffiniPure goat anti-rabbit immunoglobulin G; Jackson ImmunoResearch Laboratories, West Grove, Pa.) diluted 1:200 in PBS with milk. Macrophages were then counterstained with Evans blue. Fluorescence was analyzed with a laser scanning confocal fluorescence microscope (Leica DMIRBE) equipped with a 100× oil immersion lens.
Electron microscopy.
Macrophages were infected with 107 parachlamydiae/ml for 8 h and harvested at 0, 12, or 24 h postinfection. After a wash, cells were fixed in 4% glutaraldehyde and prepared as described previously (19). Grids were examined with a Morgagni 268D electron microscope (Philips, Eindhoven, The Netherlands).
Statistical analysis.
The mean number of bacteria per macrophage and the mean number of macrophages were compared by using the Student t test (STATA software, version 7.0; Stata Corporation, College Station, Tex.).
RESULTS
Entry of P. acanthamoeba into macrophages.
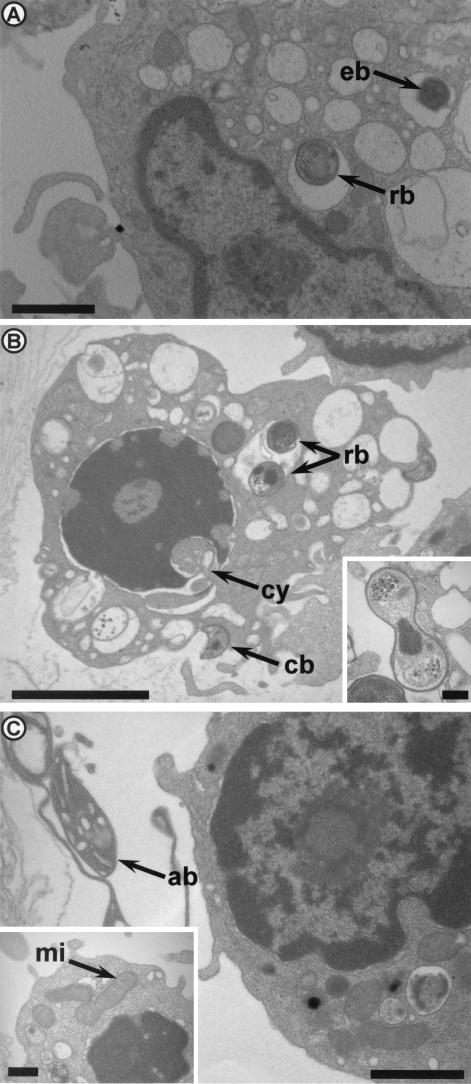
Monocyte-derived macrophages were incubated with 107 living or 107 heat-inactivated bacteria for 8 h. The percentage of infected macrophages and the number of bacteria per macrophage were determined. Eighty percent of macrophages contained bacteria (mean, 3.8 per macrophage) after incubation with living parachlamydiae. When macrophages were incubated with heat-inactivated bacteria, the percentage containing parachlamydiae was lower (63%), as was the number of bacteria per macrophage (2.3; P = 0.006). Confocal microscopy showed that parachlamydiae were intracellular, and electron microscopy demonstrated that they were located in an intracellular vacuole (Fig. 1 and 2). In addition, electron microscopy showed the presence of elementary bodies (one of the infective stages of Parachlamydia) and reticulate bodies (the dividing stage) within the vacuoles after 8 h of incubation (Fig. 2A). After 12 additional hours, crescent bodies and dividing bacteria were also seen (Fig. 2B); vacuoles containing more than six parachlamydiae were also observed.
FIG. 1.
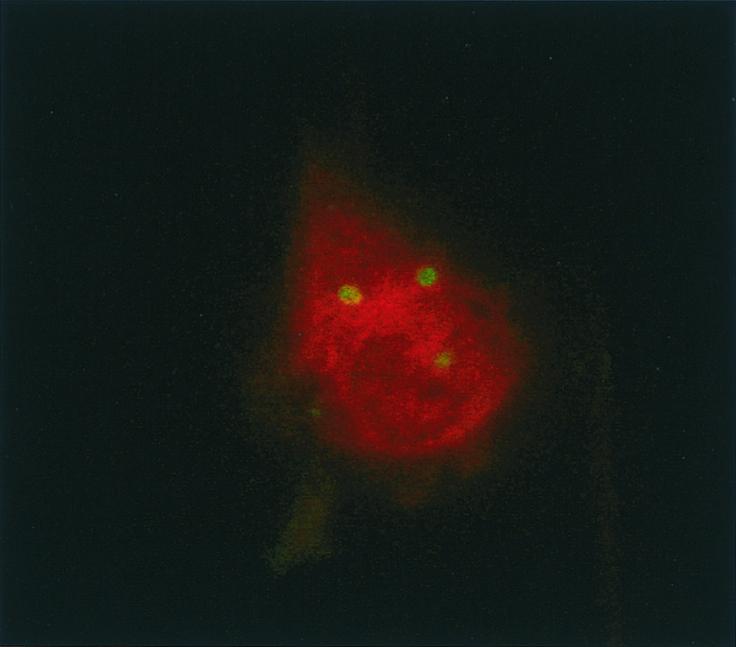
Confocal microscopy confirming the intracellular location of P. acanthamoeba. Infected macrophages were permeabilized with saponin and subjected to immunofluorescence with a 1:400-diluted rabbit anti-Parachlamydia polyclonal antibody. Magnification, ×1,000.
FIG. 2.
P. acanthamoeba within human macrophages, as observed by electron microscopy, after 8 (A), 20 (B), and 32 (C) h. (A) Human macrophage with two vacuoles containing one reticulate body (rb) and one elementary body (eb), respectively. Magnification, ×7,700. Bar, 1 μm. (B) Presence of two reticulate bodies within a vacuole and of crescent bodies (cb) outside a macrophage that is undergoing early apoptosis with chromatin condensation. Note also the fragmentation of the nuclear membrane, leading to cytoplasmic invagination (cy). Magnification, ×6,230. Bar, 2 μm. (Inset) Reticulate body undergoing binary fission. Magnification, ×30,800. Bar, 0.2 μm.(C) Human macrophage and typical apoptotic body (ab). Magnification, ×9,800. Bar, 1 μm. (Inset) Human macrophage undergoing apoptosis with chromatin condensation and dilated mitochondria (mi). Magnification, ×15,400. Bar, 0.5 μm.
Multiplication of P. acanthamoeba within macrophages.
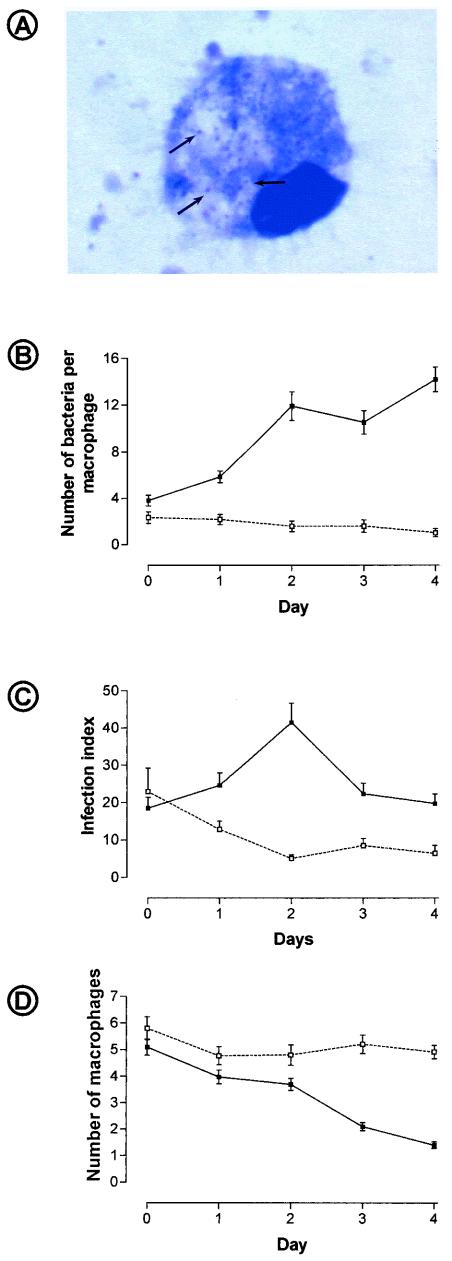
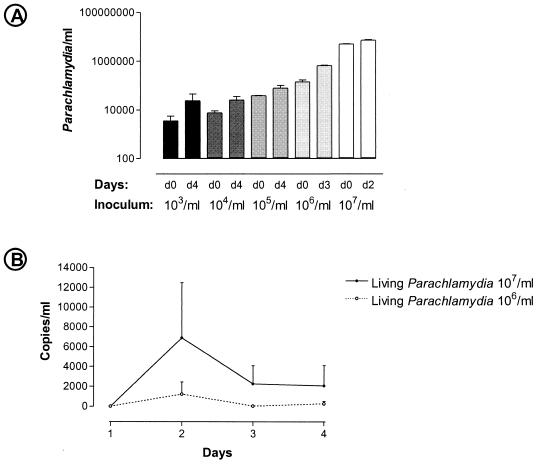
Living P. acanthamoeba organisms were shown to multiply in macrophages (Fig. 3A). When macrophages were infected with 107 living bacteria, the proportion of infected macrophages increased from 80% at day 0 to 96% at day 4, and the mean number of bacteria per macrophage increased 3 to 4 times (P < 0.001) (Fig. 3B). The infection index doubled from day 0 to day 2 (P = 0.001) (Fig. 3C) but subsequently decreased due to the diminution in the number of macrophages per field (Fig. 3D). When an inoculum of 107 heat-inactivated parachlamydiae was used as a negative control, the proportion of infected macrophages and the number of parachlamydiae per macrophage decreased as expected from day 0 to day 4 (P < 0.001) (Fig. 3B). Quantitative PCR confirmed that P. acanthamoeba initially grows within human macrophages, with a doubling time of about 2 days (Fig. 4A). A subsequent decrease in the number of parachlamydiae was observed after the 2nd or 3rd day following infection with a high inoculum of 107 or 106 parachlamydiae/ml, respectively. The number of parachlamydiae per milliliter increased until the 4th day after infection with a lower inoculum of 103 to 105 parachlamydiae/ml. The transcriptional activity of internalized P. acanthamoeba was confirmed by measuring the expression of the gene encoding ADP-ATP translocase (tlc), with peaks of expression at day 2 (Fig. 4B).
FIG. 3.
Growth of P. acanthamoeba within human macrophages evaluated on Diff-Quik-stained smears. (A) Diff-Quik-stained P. acan-thamoeba within human macrophages. Note the pyknotic nucleus sug-gesting apoptosis. Magnification, ×900. (B) Number of parachlamydiae per macrophage according to time. Solid and open symbols, living and killed parachlamydiae, respectively. (C) Infection index (number of bacteria per macrophage multiplied by the number of macrophages). Solid and open symbols, living and killed parachlamydiae, respectively. (D) Number of macrophages per microscopic field. Solid symbols, infected cells; open symbols, uninfected control.
FIG. 4.
(A) Number of parachlamydiae per milliliter, as determined by quantitative PCR demonstrating the growth of parachlamydiae within macrophages. Baseline and maximum values obtained with different inocula are shown on a logarithmic scale. (B) Expression of the ADP-ATP translocase gene (tlc), showing peak expression on day 2. The number of copies is quantified compared to tlc mRNA expression of a single viable P. acanthamoeba organism grown in amoebae.
Cytopathic effect of P. acanthamoeba on macrophages.
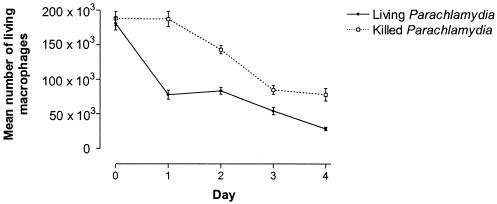
With an inoculum of 107 parachlamydiae/ml, the number of living macrophages that excluded trypan blue decreased in 4 days from 182 × 103 to 28 × 103 (P = 0.003) (Fig. 5). Heat-inactivated P. acanthamoeba also decreased the number of living macrophages, from 188 × 103 at day 0 to 76 × 103 at day 4, but at a lower rate than that for macrophages infected with living bacteria (P = 0.018).
FIG. 5.
Cytopathic effect of P. acanthamoeba on human macrophages. Graph shows the number of living macrophages (expressed as macrophages per well) as determined by staining of cells with trypan blue.
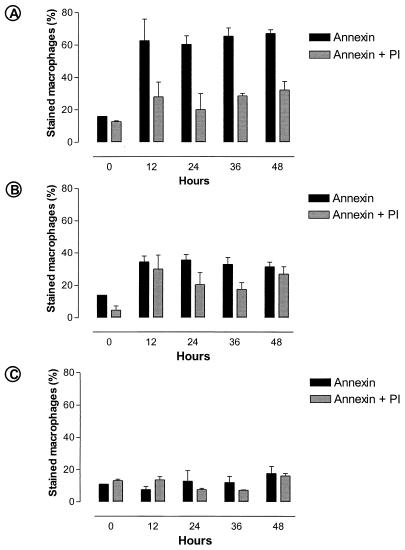
To test whether the cytopathic effect of P. acanthamoeba on macrophages was due to P. acanthamoeba-induced apoptosis, apoptosis was assessed by electron microscopy and the binding of annexin-V (36). Cytoplasmic vacuolation, fragmentation of the nuclear membrane, and chromatin condensation were observed 12 h after infection with living parachlamydiae (Fig. 2B). Apoptotic bodies and dilated mitochondria were also seen (Fig. 2C). The annexin assay confirmed that the morphological features of apoptosis were present early following P. acanthamoeba infection. Thus, 63% of the macrophages were stained with annexin after 12 h of infection with living P. acanthamoeba (Fig. 6A), whereas only 28% were simultaneously stained with annexin and PI. Heat-inactivated bacteria did not induce macrophage apoptosis, but like living P. acanthamoeba, they exerted a moderate, nonspecific cytotoxic effect associated with increased membrane permeability and simultaneous annexin and PI staining (Fig. 6B). This contrasted with the rate of 8% for annexin-stained cells in the negative control (P < 0.001) (Fig. 6C).
FIG. 6.
Proportions of annexin- and annexin-plus-PI-stained macrophages at different times, showing induction of apoptosis by living P. acanthamoeba (A) and the increased membrane permeability of macrophages infected with killed P. acanthamoeba (B) compared to that of uninfected cells (C).
DISCUSSION
This study shows that P. acanthamoeba enters and multiplies within human macrophages. This is the first demonstration that a strictly intracellular bacteria naturally infecting free-living amoebae may survive within human macrophages.
P. acanthamoeba was shown to enter into macrophages. The intracellular location of P. acanthamoeba within a vacuole was demonstrated by electron microscopy. Further work is needed to define the exact compartment of replication of P. acanthamoeba. The fact that both living and heat-inactivated P. acanthamoeba organisms may enter suggests that entry is a passive process occurring by a mechanism such as phagocytosis that does not require viable bacteria.
The presence of dividing reticulate bodies, the increased number of bacteria per vacuole, the increased number of bacteria per macrophage, and the increased proportion of infected macrophages suggested the multiplication of P. acanthamoeba. Quantitative PCR and the significant increase in the infection index confirmed that P. acanthamoeba replicates within monocyte-derived macrophages.
Transcriptional activity of P. acanthamoeba was detected by RT-PCR targeting the gene encoding ADP-ATP translocase (tlc). The ADP-ATP translocase is a unique energy parasitism enzyme present in Chlamydiales and Rickettsiales (38) that exchanges ADP from the bacteria with ATP present in the cytoplasm of the host cell (37). The important expression of that gene on day 2 suggests that reticulate bodies may use the ATP of the infected macrophage. The reduced expression of the gene after 3 days may reflect the lower availability of cytosolic ATP within heavily infected and apoptotic macrophages, the decreased infection burden, and/or another developmental stage.
P. acanthamoeba should thus be added to the growing list of intracellular microorganisms that are able to replicate in both amoebae and macrophages, which already includes L. pneumophila (31), Listeria monocytogenes (25), Chlamydophila pneumoniae (12), Coxiella burnetii (23), Francisella tularensis (1), M. avium (8), Afipia spp. (24), Burkholderia pseudomallei (21), virulent Pseudomonas aeruginosa (30), and Cryptococcus neoformans (33).
Our data also showed that P. acanthamoeba is cytopathic for human macrophages. This finding contrasts with the absence of cytopathogenicity observed with the replication of Chlamydophila pneumoniae within macrophages (22). The cytopathic effect of P. acanthamoeba takes place, at least partially, through the apoptosis of infected cells. Indeed, we showed that P. acanthamoeba induced cytoplasmic vacuolation, condensation of the chromatin, fragmentation of the nuclear membrane, mitochondrial dilatation, apoptotic bodies, and translocation of phosphatidylcholine from the inner to the outer leaflet of the cell membrane of infected cells. Apoptotic cells sometimes become nonadherent, and consequently, it is possible that some apoptotic cells were washed during the staining procedure. This may explain the observed absence of increase in the rate of annexin-stained cells after 12 h postinfection (Fig. 6A). Apoptosis of infected cells may prevent P. acanthamoeba from using the macrophage as a replication niche for a prolonged time, and consequently, we wondered whether P. acanthamoeba may enter and multiply in other human cells, such as those of the respiratory epithelium. Although it is paradoxical that an intracellular bacterium induces the apoptosis of its host cell, this phenomenon has been observed for L. pneumophila early after macrophage infection (14) and for Chlamydia psittaci after about 1 day of infection (28). In addition, some intracellular bacteria, such as Shigella flexneri, escape from macrophages by inducing apoptosis. This allows infection of other target cells (27, 39). Apoptosis of infected cells was induced only by living P. acanthamoeba. However, we showed that both heat-inactivated and living P. acanthamoeba organisms exerted a moderate, nonspecific cytotoxic effect.
In conclusion, this study shows that P. acanthamoeba entered and multiplied within human macrophages, inducing their apoptosis. Its growth within human macrophages suggests that other microorganisms resistant to environmental amoebae may be naturally resistant to macrophages as well.
Acknowledgments
We thank the Swiss National Science Foundation for funding the postodoctoral fellowship of Gilbert Greub in the Unité des Rickettsies, Marseille, France.
We thank J. S. Dumler for review of the manuscript.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., N. Springer, W. Schönhuber, W. Ludwig, E. N. Schmid, K. D. Müller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 349:925-926. [DOI] [PubMed] [Google Scholar]
- 5.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo, S. L., L. Yan, M. Littman, M. M. Samrakandi, and J. D. Cirillo. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667-1677. [DOI] [PubMed] [Google Scholar]
- 10.Corsaro, D., D. Venditti, A. Le Faou, P. Guglielmetti, and M. Valassina. 2001. A new chlamydia-like 16S rDNA sequence from a clinical sample. Microbiology 147:515-516. [DOI] [PubMed] [Google Scholar]
- 11.Corsaro, D., D. Venditti, and M. Valassina. 2002. New parachlamydial 16S rDNA phylotypes detected in human clinical samples. Res. Microbiol. 153:563-567. [DOI] [PubMed] [Google Scholar]
- 12.Essig, A., M. Heinemann, U. Simnacher, and R. Marre. 1997. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl. Environ. Microbiol. 63:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 14.Gao, L. Y., and Y. Abu Kwaik. 1999. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect. Immun. 67:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greub, G., P. Berger, L. Papazian, and D. Raoult. 2003. Molecular hint suggesting that Parachlamydiaceae are agents of pneumonia. Emerg. Infect. Dis. 9:755-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greub, G., I. Boyadjiev, B. La Scola, D. Raoult, and C. Martin. 2003. Serological hint suggesting that Parachlamydiaceae are agents of pneumonia in polytraumatized intensive-care patients. Ann. N. Y. Acad. Sci. 990:311-319. [DOI] [PubMed] [Google Scholar]
- 18.Greub, G., and D. Raoult. 2002. Parachlamydiaceae, potential emerging pathogens. Emerg. Infect. Dis. 8:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greub, G., and D. Raoult. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68:3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn, M., M. Wagner, K. D. Müller, E. N. Schmid, T. R. Fritsche, K. H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 21.Inglis, T. J., P. Rigby, T. A. Robertson, N. S. Dutton, M. Henderson, and B. J. Chang. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect. Immun. 68:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaukoranta-Tolvanen, S. S. E., A. M. Teppo, K. Laitinen, P. Saikku, K. Linnavuori, and M. Leinonen. 1996. Growth of Chlamydia pneumoniae in cultured human peripheral blood mononuclear cells and induction of a cytokine response. Microb. Pathog. 21:215-221. [DOI] [PubMed] [Google Scholar]
- 23.La Scola, B., and D. Raoult. 2001. Survival of Coxiella burnetii within free-living amoeba Acanthamoeba castellanii. Clin. Microbiol. Infect. 7:75-79. [DOI] [PubMed] [Google Scholar]
- 24.Lührmann, A., K. Streker, A. Schüttfort, and A. Haas. 2001. Afipia felis induces uptake by macrophages directly into a nonendocytic compartment. Proc. Natl. Acad. Sci. USA 98:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ly, T. M. C., and H. E. Muller. 1990. Interactions of Listeria monocytogenes, Listeria seeligeri, and Listeria innocua with protozoans. J. Gen. Appl. Microbiol. 36:143-150. [Google Scholar]
- 26.Marrie, T. J., D. Raoult, B. La Scola, R. J. Birtles, and E. de Carolis. 2001. Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia. Emerg. Infect. Dis. 7:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarre, W. W., and A. Zychlinsky. 2000. Pathogen-induced apoptosis of macrophages: a common end for different pathogenic strategies. Cell Microbiol. 2:265-273. [DOI] [PubMed] [Google Scholar]
- 28.Ojcius, D. M., P. Souque, J. L. Perfettini, and A. Dautry-Varsat. 1998. Apoptosis of epithelial cells and macrophages due to infection with the obligate intracellular pathogen Chlamydia psittaci. J. Immunol. 161:4220-4226. [PubMed] [Google Scholar]
- 29.Ossewaarde, J. M., and A. Meijer. 1999. Molecular evidence for the existence of additional members of the order Chlamydiales. Microbiology 145:411-417. [DOI] [PubMed] [Google Scholar]
- 30.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone, B. J., A. Brier, and Y. Abu Kwaik. 1999. The Legionella pneumophila prp locus; required during infection of macrophages and amoebae. Microb. Pathog. 27:369-376. [DOI] [PubMed] [Google Scholar]
- 35.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 36.Vermes, I., C. Haanen, H. Steffens-Nakken, and C. Reutelingsperger. 1995. A novel assay for apoptosis: flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J. Immunol. Methods 184:39-51. [DOI] [PubMed] [Google Scholar]
- 37.Winkler, H. H., and H. E. Neuhaus. 1999. Non-mitochondrial ATP transport. Trends Biochem. Sci. 24:64-68. [DOI] [PubMed] [Google Scholar]
- 38.Wolf, Y. I., L. Aravind, and E. V. Koonin. 1999. Rickettsiae and Chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet. 15:173-175. [DOI] [PubMed] [Google Scholar]
- 39.Zychlinsky, A., M. C. Prevost, and P. J. Sansonetti. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature 358:167-169. [DOI] [PubMed] [Google Scholar]