Abstract
Variation in the expression of the different Tpr proteins in the syphilis spirochete, Treponema pallidum subsp. pallidum, may have important implications in its ability to evade host immune detection and cause persistent infection. In the present study we examined the pattern of antibody responsiveness to different Tpr members during infection with three isolates of T. pallidum. There was variability in the specificities and temporal patterns of reactivity of the antibodies elicited against the individual Tpr proteins, suggesting that isolates may express different repertoires of Tpr proteins during infection.
Syphilis is a chronic disease caused by Treponema pallidum subsp. pallidum. During the early stages of infection, the host mounts a vigorous immune response that is able to clear the majority of the treponemes from early lesions but is unable to completely eradicate the infection. Phagocytosis of opsonized treponemes by macrophages is the primary mechanism by which the host immune system clears treponemes from early lesions (1, 13, 15). While T. pallidum has been shown to be susceptible to bactericidal and opsonic antibodies (2-5, 15), the inability to culture the organism, combined with the fragile nature of its outer membrane, have made the positive identification of outer surface molecules that may be antibody targets difficult and often inconclusive (8).
The T. pallidum repeat (tpr) gene family is a 12-member gene family, originally identified in the Nichols strain of T. pallidum, that encodes proteins which share amino acid homology with the major surface proteins of Treponema denticola but whose function is unknown (5-7, 10). The 12 members of the gene family are divided into subfamily I (TprC, D, F, and I), subfamily II (TprE, G, and J), and subfamily III (TprA, B, H, K, and L) based on regions of sequence conservation shared by members within the same subfamily (5). TprK and members of subfamily I have putative cleavable signal peptides. Previous studies have shown that TprK is a target of opsonic antibody and that immunization with recombinant TprK is partially protective to challenge with T. pallidum, possibly suggesting a surface location for TprK (5, 16), although this conclusion is controversial (12). More recently it has also been shown that multiple alleles of tprK exist within isolates of T. pallidum subsp. pallidum (6), and there is some heterogeneity in tprD (7) and tprJ (18; unpublished data) sequences among strains. The Chicago and Baltimore 73-1 (Bal 73-1) strains share the same tprC and tprD sequences as the Nichols strain. Preliminary examination of sequence variation in the tprE, F, G, and I genes among T. pallidum strains has revealed very minor heterogeneity relative to that observed with the tprD, J, and K genes (unpublished data). Information regarding the degree of sequence heterogeneity among strains in the tprA, B, H, and L genes is currently unavailable. A possible surface location for some tpr gene products and sequence variation in some tpr genes suggest that the Tpr family could contribute to immune evasion and persistence of T. pallidum. In the present study we sought to determine which of the Tpr members in two “street” isolates of T. pallidum, Chicago and Bal 73-1, are expressed during infection and are thus recognized by the immune system; this pattern of antibody responsiveness was compared to the antibody response elicited by the laboratory-adapted Nichols strain.
The Nichols strain of T. pallidum has been propagated in rabbits since its initial isolation from the cerebrospinal fluid of a patient with secondary syphilis in 1912 (17). Despite extensive propagation in rabbits, it remains pathogenic for humans as demonstrated by accidental laboratory infections (9, 19). The Bal 73-1 strain was isolated from a newborn child with congenital syphilis in 1969 (11), and the Chicago strain was isolated in 1951 from a primary chancre (19). The Bal 73-1 and Chicago strains were passed in rabbits within our lab three and nine times, respectively, prior to their use in these studies. We have no information concerning the number of passages for these strains prior to our obtaining the stocks (from Paul Hardy and Ellen Nell, John Hopkins University); however, this number is certainly much lower than for the Nichols strain.
Three to six separate animals per strain were infected intratesticularly with 108 treponemes as previously described (14). Sera were collected from animals prior to infection and at days 10, 17, 30, 45, 60, and 90 postinfection (day 45 samples were not available from Nichols-infected animals). Enzyme-linked immunosorbent assays (ELISAs) were performed using recombinant peptides that were derived from the sequences of the tpr genes originally reported in the T. pallidum (Nichols) genome sequence and then produced in Escherichia coli (10). The peptides were histidine tagged and purified on nickel columns (5). TprC and D are identical in the Nichols strain, so a single peptide represented both; TprL could not be expressed. With the exception of TprF, each recombinant peptide represented the variable region, which contains sequence that differentiates the individual Tpr proteins; only small portions of the sequence that is conserved among members of the same subfamily were present in the peptides. The subfamily III recombinant proteins (TprA, B, H, and K) have virtually no overlap in constant regions, while TprC/D, Tpr I, TprE, TprG, and TprJ each contain 23 amino acids (aa) of the 5′ conserved region, and TprC/D and TprI also contain 21 aa of the 3′ conserved region. TprF, in contrast, contains 262 aa of the 5′ constant region of subfamily I in addition to 87 aa representing the variable region (Table 1). The wells of a 96-well ELISA plate (EIA II Plus Microplate; ICN Biomedicals) were coated with the appropriate Tpr antigen resuspended in phosphate-buffered saline (PBS) containing 0.1% sodium azide and 0.1% sodium dodecyl sulfate and used at a final concentration of 10 μg/ml at 50 μl/well. Plates were incubated at 37°C for 2 h, followed by a 4°C overnight incubation. The wells were then washed three times with PBS (used for this and all subsequent washing steps) and blocked by incubation for 1 h at room temperature with 200 μl of 3% nonfat milk-PBS/well. After washing the wells, 100 μl of the primary antibody solution (made from pools of infected or normal rabbit sera diluted 1:20 in 1% nonfat milk-PBS-0.05% Tween 20 [Sigma]) was added to each well, allowed to bind for 1 h at room temperature, and then washed three times. To reduce background due to rabbit antibodies cross-reactive with E. coli (in which our recombinant peptides were produced), all of the sera were preadsorbed with E. coli lysate prior to use in the ELISAs. Because no optical density (OD) values were above the linear range of the ELISA, only one concentration of serum was tested. Next, 100 μl of a 1:2,000 dilution of alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (H and L; Sigma) in 1% nonfat milk-PBS-0.05% Tween 20 was added to each well and allowed to bind at room temperature for 1 h before washing the wells three times. Fifty microliters of p-nitrophenylphosphate (Sigma) diluted to 1 mg/ml in development buffer (as specified by the manufacturer) was added to each well; after 45 min, readings were taken at 405 nm using a Multiskan MC plate reader (Titertek). The mean background OD405 readings (i.e., reading observed using preimmune rabbit sera) were subtracted from the values for each of the time points. Data presented represent the mean ± standard error of three replicate wells per condition.
TABLE 1.
Recombinant peptides
| Subfamily and peptide | Amino acid sequence limits | Size (aa) |
|---|---|---|
| Subfamily I | ||
| TprC/D | RLTLEP . . . . . . . . . YTHLLT | 200 |
| TprFa | YAGVLT . . . . . . . . . GTGGAC | 349 |
| TprI | RLTLEP . . . . . . . . . YTHLLT | 211 |
| Subfamily II | ||
| TprE | RLTLEP . . . . . . . . . QQTVAA | 191 |
| TprG | RLTLEP . . . . . . . . . DLIPKT | 219 |
| TprJ | RLTLEP . . . . . . . . . MRTEIT | 226 |
| Subfamily III | ||
| TprAb | MGLVVT . . . . . . . . . GCKITW | 368 |
| TprB | RLTLSP . . . . . . . . . SLSKLV | 194 |
| TprH | RITLTP . . . . . . . . . YTHLID | 180 |
| TprK | IEGYAE . . . . . . . . . DTSFLE | 315 |
The peptide for TprF contains 262 aa of the 5′ constant region of subfamily I in addition to 87 aa of the variable region.
TprA consists of two overlapping reading frames. Our peptide is encoded by the second reading frame.
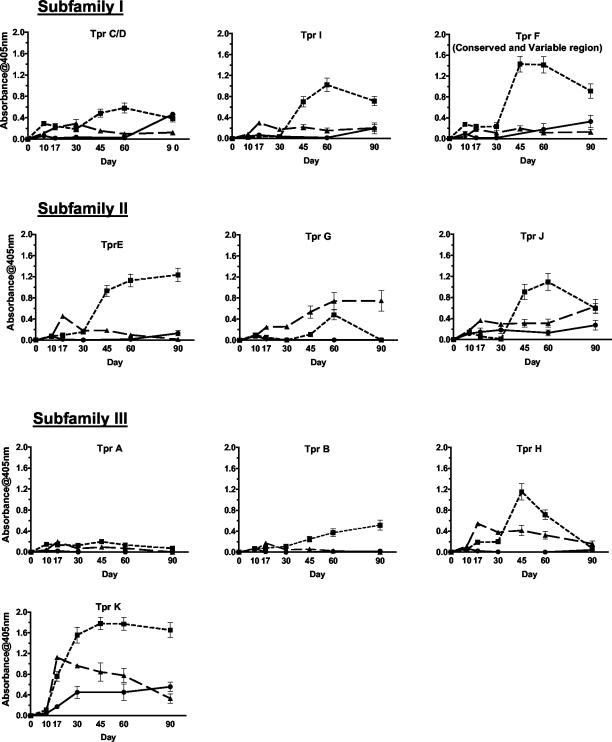
All rabbits demonstrated the strongest and earliest antibody reactivity to TprK (Fig. 1). In addition, Nichols-infected rabbits had low and late reactivity (days 60 to 90) to TprC/D, F, I, and J and little or no reactivity to A, B, E, G, and H (Fig. 1 and Table 2). The sera from animals infected with either the Bal 73-1 or Chicago isolates had similar levels of antibody reactivity as Nichols-infected animals against TprA and TprC/D, although reactivity to TprC/D was observed earlier than in Nichols-infected animals (Fig. 1). In contrast to the Nichols strain, Chicago-infected animals had strong antibody responses to TprE, F, G, H, I, and J as well as some reactivity to TprB. Bal 73-1-infected animals showed lower reactivity against these Tpr antigens, except TprG, which Bal 73-1 induced at the highest level of antibody reactivity among all three of the isolates tested (Fig. 1 and Table 2).
FIG. 1.
Development of antibody responses to Tpr peptides through day 90 postinfection. The mean OD405 readings and standard errors of the means are representative of three replicates of pooled sera from three to six animals per treponemal isolate. ▴, Bal 73-1 (n = 3); ▪, Chicago (n = 4 to 6); •, Nichols (n = 5).
TABLE 2.
Reactivities of antibodies in the sera of animals infected with T. pallidum isolates
| Isolate | Tpr proteina
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subfamily I
|
Subfamily II
|
Subfamily III
|
||||||||
| C/D | F | I | E | G | J | A | B | H | K | |
| Nichols | +/− | +/− | +/− | − | − | +/− | − | − | − | + |
| Chicago | +/− | ++ | ++ | ++ | +/− | ++ | − | +/− | ++ | ++ |
| Baltimore 73-1 | +/− | − | +/− | +/− | + | +/− | − | − | +/− | ++ |
Results are peak OD405 readings, as follows: − = <0.20; +/− = 0.20 to 0.59; + = 0.60 to 0.99; ++ = ≥1.00.
Differences in the time of appearance and patterns of antibody reactivity to the Tpr proteins in infected animals were features of the antibody responses elicited by infection with individual isolates. Chicago- and Nichols-infected animals generally showed a gradual increase in antibody reactivity to the Tpr proteins throughout the early stages of infection, with peak reactivity occurring at day 45 or 60 postinfection. A similar temporal pattern of antibody response was observed against TprG and J in Bal 73-1-infected animals; however, these animals showed a sharp early spike in reactivity to TprE, H, and K at day 17 or 30, which was followed by a general decline in antibody levels (Fig. 1).
In separate experiments the sera from the individual animals infected with Chicago or Bal 73-1 were tested against the Tpr peptides (data not shown). Differences in both the time of appearance and the level of reactive antibodies against some Tpr members were observed among sera from individual animals. These individual differences may be due to genetic diversity among the outbred rabbits. In a similarly diverse human population, variation in antibody responses might also be expected. Alternatively, different Tpr repertoires may be expressed in individual human or rabbit hosts, thus inducing different antibody specificities.
The independent recognition of some antigens in a subfamily, but not all antigens in the subfamily, demonstrates that the antibody reactivity measured in our assays is primarily directed to the variable regions of the recombinant peptides rather than the small constant regions present on most of the peptides. However, reactivity against our TprF peptide, which contains a 262-aa stretch of the 5′ constant region, may be directed either to the 5′ constant region of subfamily I or to the TprF variable regions; this distinction could not be determined in our studies. Furthermore, TprF is identical to TprI except for a stretch of 11 unique amino acids within its variable region (5). Thus, the strong level of antibody reactivity observed in Chicago-infected animals against our TprF peptide may actually be induced by expression of any of the subfamily I proteins, all of which contain this conserved region (Fig. 1). Further studies are under way to determine whether there are antibody responses specifically directed to the constant regions of the Tpr proteins.
These results demonstrate that different specificities of antibody responses are induced by infection with individual isolates of T. pallidum, suggesting that T. pallidum strains may express different repertoires of Tpr proteins. This may reflect intrinsic strain-to-strain differences in expression, or it may reflect the fact that the Nichols strain is highly rabbit adapted, with Tpr expression distinct from that in strains that have been passed less extensively in rabbits. A difference in the antibody responses to our Tpr peptides was also observed in a previous study using a limited number of human samples (unpublished results). Extrapolation of our findings to natural human infection suggests that humans infected with individual strains of T. pallidum may be exposed to varying Tpr repertoires, depending upon their infecting strain. This possibility may need to be considered when designing a Tpr-based diagnostic test or vaccine.
Acknowledgments
This work was supported by U.S. Public Health Service grants AI 34616 and AI 42143 (S.A.L.) and AI 43456 (W.V.V.).
We thank Heidi Pecoraro for manuscript preparation.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Baker-Zander, S. A., and S. A. Lukehart. 1992. Macrophage-mediated killing of opsonized Treponema pallidum. J. Infect. Dis. 165:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, N. H., and J. N. Miller. 1976. Humoral immunity in experimental syphilis. II. The relationship of neutralizing factors in immune serum to acquired resistance. J. Immunol. 117:197-207. [PubMed] [Google Scholar]
- 3.Blanco, D. R., J. N. Miller, and P. A. Hanff. 1984. Humoral immunity in experimental syphilis: the demonstration of IgG as a treponemicidal factor in immune rabbit serum. J. Immunol. 133:2693-2697. [PubMed] [Google Scholar]
- 4.Blanco, D. R., E. M. Walker, D. A. Haake, C. I. Champion, J. N. Miller, and M. A. Lovett. 1990. Complement activation limits the rate of in vitro treponemicidal activity and correlates with antibody-mediated aggregation of Treponema pallidum rare outer membrane protein. J. Immunol. 144:1914-1921. [PubMed] [Google Scholar]
- 5.Centurion-Lara, A., C. Castro, L. Barrett, C. Cameron, M. Mostowfi, W. C. Van Voorhis, and S. A. Lukehart. 1999. Treponema pallidum major sheath protein homologue TprK is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centurion-Lara, A., C. Gordones, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect. Immun. 68:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centurion-Lara, A., E. S. Sun, L. K. Barrett, C. Castro, S. A. Lukehart, and W. C. Van Voorhis. 2000. Multiple alleles of Treponema pallidum repeat gene D in Treponema pallidum isolates. J. Bacteriol. 182:2332-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, D. L., P. Chang, A. W. McDowall, and J. D. Radolf. 1992. The outer membrane, not a coat of host proteins, limits its antigenicity of virulent Treponema pallidum. Infect. Immun. 60:1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. J., R. C. Johnson, and M. Smith. 1976. Accidental laboratory infection with Treponema pallidum, Nichols strain. J. Am. Vener. Dis. Assoc. 3:76-78. [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutto, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 11.Hardy, J. B., P. H. Hardy, E. H. Oppenheimer, S. J. Ryan, Jr., and R. N. Sheff. 1970. Failure of penicillin in a newborn with congenital syphilis. JAMA 212:1345-1349. [PubMed] [Google Scholar]
- 12.Hazlett, K. R., T. J. Sellati, T. T. Nguyen, D. L. Cox, M. L. Clawson, M. J. Caimano, and J. D. Radolf. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukehart, S. A., S. A. Baker-Zander, R. M. C. Lloyd, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. II. Nature of cellular infiltrations and Treponema pallidum distribution in testicular infection. J. Immunol. 124:461-467. [PubMed] [Google Scholar]
- 14.Lukehart, S. A., S. A. Baker-Zander, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. I. In vitro response to mitogens and Treponema pallidum antigens. J. Immunol. 124:454-460. [PubMed] [Google Scholar]
- 15.Lukehart, S. A., and J. N. Miller. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121:2014-2024. [PubMed] [Google Scholar]
- 16.Morgan, C. A., B. J. Molini, S. A. Lukehart, and W. C. Van Voorhis. 2001. Segregation of B and T cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J. Immunol. 169:952-957. [DOI] [PubMed] [Google Scholar]
- 17.Nichols, H. J., and W. H. Hough. 1913. Demonstration of Spirochaeta pallida in the cerebrospinal fluid. JAMA 60:108-110. [Google Scholar]
- 18.Stamm, L. V., S. R. Greene, H. L. Bergen, J. M. Hardham, and N. Y. Barnes. 1998. Identification and sequence analysis of Treponema pallidum tprJ, a member of a polymorphic multigene family. FEMS Microbiol. Lett. 169:155-163. [DOI] [PubMed] [Google Scholar]
- 19.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.