Abstract
Genotypes 1 and 2 of Cryptosporidium parvum are the primary types associated with infections in humans, with type 1 being by far the predominant genotype. The frequency of mixed infection with both genotypes in humans is relatively rare, while type 1, which experimentally infects other mammals, has been found to naturally infect almost exclusively humans. One possible explanation for the absence of type 1 in other mammals and the low frequency of mixed infections in humans is the inability of type 1 to compete with type 2 in nature when both occur simultaneously. To investigate this, we challenged gnotobiotic piglets with equal number of oocysts of type 1 and type 2, given either simultaneously or with type 2 given 24 or 48 h after type 1. The genotype of the oocysts excreted in feces and the relative distribution of each of the genotypes throughout the gut at necropsy were determined. Regardless of the time interval between challenges with the two genotypes, type 2 invariably displaced type 1. The rate of displacement was rapid when both genotypes were given simultaneously, after which no traces of type 1 were detected in the feces or in gut sections by PCR. Infection with type 1 24 or 48 h before challenge with type 2, while permitting type 1 to become established, was still rapidly eliminated within 3 days after challenge with type 2. These observations have major implications regarding the relative perpetuation and survival of these two genotypes in mammals.
Cryptosporidium parvum, an enteric protozoan belonging to the class Apicomplexa, is associated with diarrheal disease in humans and animals (10, 11, 17, 32). The infection can be acquired through direct contact with infected humans or animals or through consumption of food or water contaminated with feces. Seepage of human and cattle effluents into drinking-water supplies probably constitutes the most important risk for human infection (7, 12, 16, 17). Cryptosporidiosis in humans induces a spectrum of disease ranging from asymptomatic to self-limiting diarrhea. Symptoms of profound dehydrating diarrhea, associated with abdominal cramps, headaches, nausea, anorexia, wasting, and lethargy, have been attributed to cryptosporidiosis. In individuals with immunodeficiencies, including AIDS, or children with malnutrition, the infection can lead to protracted, debilitating, and life-threatening diarrhea and wasting (9, 31, 56). It is not clear whether this wide spectrum of disease is due largely to host factors, parasite factors, or the nature of the interaction between them. While host factors, such as the immune status, clearly play a major role in terms of resolution and reinfection, the role of parasite factors is unclear (33, 44, 50, 51). Despite its medical significance, cryptosporidiosis remains largely untreatable, although paromomycin, nitazoxanide, and azithromycin have shown promise as effective drugs (2, 3, 20, 45).
There are at least two major C. parvum genotypes that are responsible for approximately 98% of the infections of humans. Type 1 has so far been detected primarily in humans, but type 1 infections of nonhuman primates (47, 59), lambs (14), and dugongs (28) have also been reported, while type 2 is found in most mammals, including humans (4, 23, 26, 37, 48, 58). In recent studies of sporadic and outbreak cases of cryptosporidiosis, one genotype usually predominated. Type 1 was the predominant genotype in the majority of studies, except for studies conducted in the United Kingdom, France, and The Netherlands, in which type 2 was the predominant genotype (13, 15, 18, 19, 22, 23, 24, 27, 37, 40, 43, 48, 52, 53, 60). Other Cryptosporidium species found in humans include Cryptosporidium meleagridis (13, 18, 29, 39, 52, 53), Cryptosporidium felis (18, 29, 38, 42, 52, 60), Cryptosporidium canis (38, 42, 60), Cryptosporidium muris (13, 18, 21, 52), Cryptosporidium pig genotype (61), and Cryptosporidium cervine genotype (36).
There have been only a few studies examining whether parasite factors contribute to the nature and extent of disease seen in both animals and humans (8, 24, 33, 41, 51, 60). One such study involved human volunteers who were exposed to different doses of several well-characterized type 2 isolates. The data clearly demonstrated that, indeed, a different pattern of susceptibility and different spectrum of illness are induced by different C. parvum isolates (8, 33, 34, 35, 50, 51). Studies with the same isolates, repeated in the gamma interferon knockout mouse model, also demonstrated diversity of response to infection (35; S. Rich, G. Widmer, and S. Tzipori, unpublished data). Currently, these mouse studies have been limited to only type 2 isolates, because type 1 isolates do not infect rodents (1, 14, 30, 40, 59).
Recent studies with gnotobiotic piglets from other groups (30, 41) and preliminary data from our laboratory (D. E. Akiyoshi and S. Tzipori, unpublished data) suggest that there are type-specific parasite factors that are associated with virulence, efficiency of transmission, and severity of the disease. Pereira et al. (41) and Morgan-Ryan et al. (30) reported that type 2-infected pigs developed more severe disease and had shorter prepatent and patent periods than did type 1-infected pigs. Our preliminary findings also supported the belief that type 1 infections are less severe than type 2 infections. Examination of the available data of human patients with cryptosporidiosis also suggested that there are such factors, because in a number of studies, the majority of cases were type 1 infections (24, 53, 60). Given the ubiquitous nature of type 2 and the apparent restricted host range of type 1, the data suggest that type 1 has developed a very effective method for transmission between humans. Interestingly, most of these studies reported no or only an extremely low number of cases where both type 1 and type 2 were found together in the same host (6, 24, 37, 43, 53). Preliminary observations from our laboratory suggested that, when the two types simultaneously infect the same host, type 2 invariably predominates, displacing type 1 within a short period of time. This has been observed in infected humans, calves, and gnotobiotic piglets (Akiyoshi and Tzipori, unpublished).
Using the gnotobiotic piglet model, which unlike the mouse is susceptible to both types, we have experimentally infected animals with a mixture of C. parvum type 1 and type 2 to more rigorously investigate their behavior within a single host. Several groups of piglets were inoculated with the type 2 isolate at different time intervals following challenge with type 1 to determine if this affected the rate of displacement of the type 1 isolate.
(This work was part of the thesis requirement for the degree of Bachelor of Science [Veterinary] from the University of Sydney for Siobhan Mor.)
MATERIALS AND METHODS
C. parvum isolates.
Type 1 isolate TU502 (1, 53), originally isolated from a child with diarrhea, and type 2 isolate GCH1 (57), isolated in 1991 from a patient with chronic diarrhea, were used for these studies.
Infection of gnotobiotic piglets.
Five groups of 1-day old piglets, delivered by cesarean section and maintained in microbiological isolators (54, 57), were orally inoculated with 100,000 oocysts of type 1 (group 1) or type 2 (group 2) or a mixture of 50,000 oocysts of each type (groups 3 to 5) (Table 1). As a reference point, day 0 is defined as the date of inoculation with type 1. For the mixed infections, the piglets were challenged with type 1 on day 0 and then were inoculated with type 2 on either day 0 (group 3), day 1 (group 4), or day 2 (group 5). The oocysts used for inoculum were purified from the gut contents of experimentally infected piglets and were confirmed to be the correct genotype. Starting on day 3, fecal samples were collected once or twice daily from the infected animals and were microscopically examined for oocysts. DNA was extracted from oocyst-positive fecal samples and PCR-restriction fragment length polymorphism (RFLP) analysis was performed to determine the genotype. The piglets were euthanized on days 4 to 6 postchallenge. Gut sections (n = 12 to 16) from each animal were taken from the stomach and the small and large intestines for genotype analysis and histology.
TABLE 1.
Number of piglets used in this study and the challenge schedule for C. parvum type 1 (TU502), type 2 (GCH1), or both types
| Group no./no. of piglets | Inoculum | Time of challengea (day) | Time of euthanasiab (day) | Representative pigletc |
|---|---|---|---|---|
| 1/4 | TU502 | 0 | 5 | Piglet 1 |
| 2/4 | GCH1 | 0 | 5 | Piglet 2 |
| 3/4 | TU502/GCH1 | 0 | 5 | Piglet 3 |
| 4/4 | TU502/GCH1 | 0 and 1d | 4 | Piglet 4 |
| 6 | Piglet 5 | |||
| 5/4 | TU502/GCH1 | 0 and 2d | 4 | Piglet 6 |
| 6 | Piglet 7 |
Where day 0 represents the time of challenge with type 1 (groups 1, 4, and 5), type 2 (group 2), or both genotypes (group 3).
Time of euthanasia after the first challenge.
Selected piglets from each group included in the genotype and histology studies reported in this paper.
Day of challenge with type 2.
DNA extraction and genotype analysis.
Intestinal mucosa were scraped from sequential gut sections collected from the piglets at necropsy. DNA was extracted from these intestinal scrapings and from fecal samples as previously described by Buckholt et al. (5). Genotype was determined by using Cryptosporidium outer wall protein (COWP) PCR-RFLP analysis as previously described (1).
Histology.
Intestinal sections, adjacent to those sections taken for DNA extraction, were also collected, fixed in 10% phosphate-buffered formalin, and processed for histology by using standard techniques (25, 46). Each section was examined under a light microscope for pathological changes and was scored according to the extent of parasites visible on the intestinal villi (see Fig. 3 legend).
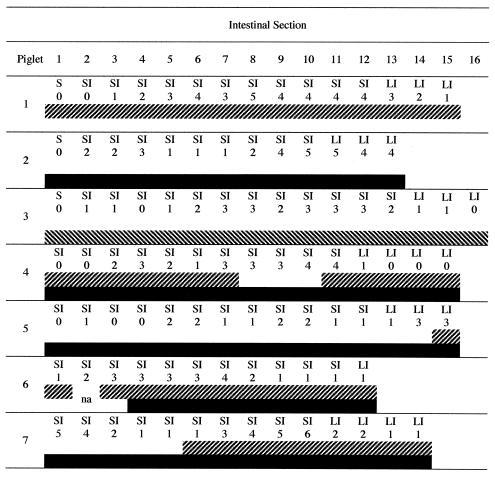
FIG. 3.
Anatomical site, histology scores, and genotyping results of intestinal samples obtained from piglets at necropsy: piglet 1 (type 1 only); piglet 2 (type 2 only), piglet 3 (type 1 and type 2 challenge on day 0), piglets 4 and 5 (mixed infection with type 2 challenge on day 1), and piglets 6 and 7 (mixed infections with type 2 challenge on day 2). Piglets 1 through 3 were euthanized on day 5, piglets 4 and 6 were euthanized on day 4, and piglets 5 and 7 were euthanized on day 6. Sections are numbered consecutively from stomach to distal spiral colon (left to right). Histology scoring is as follows: 0, no parasites detected on intestinal villi; 1, few parasites detected; 2, 25% of villus surface covered by parasites; 3, 50% of villus surface covered by parasites; 4, 75% of villus surface covered by parasites; and 5, 100% of villus surface covered by parasites. S, stomach; SI, small intestine; LI, large intestine; ▨ band, intestinal sample positive for type 1 by COWP PCR-RFLP; ▪ band, intestinal sample positive for type 2 by COWP PCR-RFLP; and na, genotyping results not available.
Mixed infection in cell culture.
Madin-Darby bovine kidney (MDBK) cells were grown to confluence in 96-well tissue culture plates. For the mixed-infection wells, oocysts of both types (50,000 each) were added together. As a control, MDBK cells were infected separately with oocysts (n = 100,000) of either C. parvum type 1 or type 2. Heat-inactivated oocysts (65°C for 15 min) of type 1 and type 2 (separately or together) were added to MDBK cells as additional controls. Cells were harvested 48 h postinfection, and DNA was extracted and genotyped as described above.
RESULTS
Infection of piglets.
In general, the extent of the clinical signs, including onset, extent of oocyst excretion, and duration of diarrhea and anorexia, weight loss, and dehydration, observed in piglets infected with type 1 were milder than those in pigs infected with either type 2 or a mixture of types 1 and 2. These observations are based on many earlier inoculation experiments with animals that were challenged over the last 6 years with either genotype or with a mixture of both (data not shown). In the experiments described here animals were challenged with relatively small doses of oocysts and were euthanized within 4 to 6 days after challenge; therefore, clinical signs were not fully developed. All animals, however, became infected and showed various degrees of diarrhea that depended on the inoculum and time of euthanasia. Oocyst excretion in the feces commenced within 3 or 4 days postinoculation, regardless of the inoculum given. Table 1 describes the groups of piglets and the number of animals included in each group.
Genotyping of feces.
As expected, detection of infection by COWP PCR-RFLP was more sensitive than that by microscopy. This was also true for the detection of parasite forms in a particular intestinal segment, compared with histological detection of infection on the mucosal surface. Figures 1 to 3 provide genotype analysis of a representative animal from each group. Piglet 1 was challenged with type 1 only (group 1), while piglet 2 was challenged with type 2 only (group 2). Piglet 3 was challenged with both type 1 and type 2 (group 3). These three animals were euthanized on day 5. Piglets 4 and 5 were challenged with both types, but type 2 was given 24 h after type 1 (group 4). Piglets 6 and 7 were also challenged with both types, but type 2 was given 48 h later than type 1 (group 5). Piglets 4 and 6 were euthanized on day 4, and piglets 5 and 7 were euthanized on day 6.
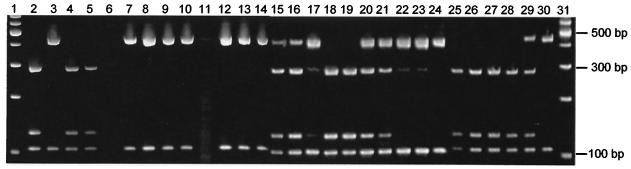
FIG. 1.
Genotype analysis of fecal samples from the gnotobiotic piglets. COWP PCR was performed, and the PCR products were digested with RsaI for 90 min at 37°C. The RsaI digests were electrophoresed on a 15% Tris-borate-EDTA acrylamide gel and were visualized by ethidium bromide staining. The samples are as follows: 100-bp ladder (lanes 1 and 31 [Promega Corp.]); type 1 control (lane 2); type 2 control (lane 3); piglet 1 (type 1), day 4 before noon (lane 4), day 4 after noon (lane 5), and day 5 before noon (lane 6); piglet 2 (type 2), day 3 (lane 7), day 4 before noon (lane 8), day 4 after noon (lane 9), and day 5 before noon (lane 10); piglet 3 (mixed), day 3 before noon (lane 11), day 4 before noon (lane 12), day 4 after noon (lane 13), and day 5 before noon (lane 14); piglet 4 (mixed, type 2 challenge on day 1, euthanized day 4), day 3 before noon (lane 15), day 3 after noon (lane 16), and day 4 before noon (lane 17); piglet 5 (mixed, type 2 challenge on day 1, euthanized day 6), day 3 before noon (lane 18), day 3 after noon (lane 19), day 4 before noon (lane 20), day 4 after noon (lane 21), day 5 before noon (lane 22), day 5 after noon (lane 23), and day 6 before noon (lane 24); piglet 6 (mixed, type 2 challenge on day 2, euthanized day 4), day 3 before noon (lane 25), and day 4 before noon (lane 26); piglet 7 (mixed, type 2 challenge day 2, euthanized day 6), day 3 before noon (lane 27), day 4 after noon (lane 28), day 5 before noon (lane 29), and day 6 before noon (lane 30). Digestion with RsaI yields fragments of 285, 125, 106, and 34 bp for type 1 and 410, 106, and 34 bp for type 2. Samples showing fragments of 410, 285, 125, 106, and 34 bp are indicative of a mixed infection with both type 1 and type 2 present.
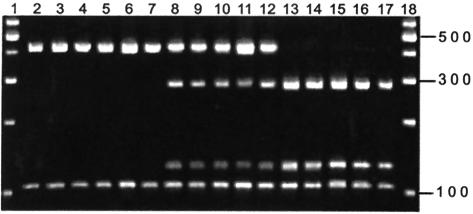
FIG. 4.
Cultured MDBK cells were infected with 50,000 oocysts/ml (type 2, types 1 and 2, or type 1). Cells were harvested as follows: type 2, 6 h (lane 3), 11 h (lane 4), 24 h (lane 5), and 48 h postinfection (lane 6) and heat inactivated (lane 7); types 1 and 2, 6 h (lane 8), 11 h (lane 9), 24 h (lane 10), 48 h (lane 11), and heat inactivated (lane 12); type 1, 6 h (lane 13), 11 h (lane 14), 24 h (lane 15), 48 h (lane 16), and heat inactivated (lane 17). COWP PCR-RFLP analysis was as previously described. Type 2 control (lane 2) and 100-bp ladder (Promega Corp.) (lane 1) are also shown. When both genotypes appear in the mixed-infection wells, there is a suggestion that type 2 is more predominant than when either genotype is alone, based on intensity of the PCR product. The heat-inactivated oocyst samples are representative of the oocyst populations that were added to the culture, in the absence of replication.
Figure 1 shows the genotype pattern of oocysts excreted in feces collected twice daily from piglets that were challenged with either type 1 only (piglet 1; lanes 4 to 6) or type 2 only (piglet 2; lanes 7 to 10) or were coinoculated with both types (piglet 3; lanes 11 to 14). Piglets inoculated with either type 2 or a mixture of the two genotypes started shedding on day 3, but shedding by the piglet inoculated with type 1 was delayed a day. The presence of type 1 was undetectable in the feces of the mixed-infection piglet (piglet 3), even on the first day of shedding. If type 2 was given 1 day after type 1, it rapidly displaced type 1 within 2 or 3 days (piglets 4 and 5; lanes 15 to 24). In piglet 5, this displacement was more gradual, with type 1 appearing first on day 3 after challenge and then gradually becoming displaced by type 2 on days 4 and 5 postchallenge with type 1 and disappearing entirely by day 6. The degree of displacement when the time interval between challenge with types 1 and 2 was increased to 2 days was even greater. No evidence of type 2 DNA was observed in either piglet (piglets 6 and 7; lanes 25 to 30) on day 3, but type 2 was detected on day 4 in piglet 6. In piglet 7, type 2 was detectable on day 5 and rapidly displaced type 1 by day 6, when type 1 was no longer detected.
Genotype analysis and histological observations of gut sections.
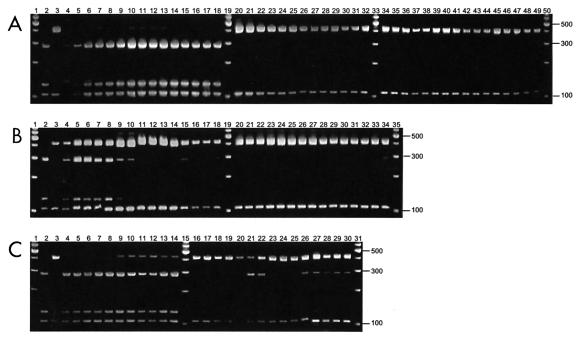
Figure 2 shows the distribution of the parasite DNA in the various gut segments taken at necropsy 4 to 6 days after challenge with type 1 as determined by COWP PCR-RFLP analysis. The genotype profiles at necropsy (day 5) of piglets infected with either type 1 (piglet 1; lanes 4 to 18), type 2 (piglet 2; lanes 20 to 32), or a mixture of both types given simultaneously (piglet 3; lanes 34 to 49) are shown in Fig. 2A. Piglets infected with only a single type displayed a profile specific to that genotype. However, when the two types were given at the same time, there was no trace of type 1 in any of the gut segments on day 5 (lanes 20 to 32). While all three piglets had parasites distributed throughout the entire intestinal tract, the apparent intensity of the banding profiles differed. The heaviest banding in the type 1 piglet was in the distal small intestine, compared to findings with the type 2 piglet, which had the highest intensity in the proximal small intestine. The mixed-infection piglet (piglet 3) had a fairly uniform distribution of type 2 throughout the entire intestinal tract. When the time interval was 1 day between challenges with the two types (Fig. 2B), the piglet euthanized on day 4 (piglet 4; lanes 4 to 18) had parasite DNA of type 2 throughout the gut, while type 1 was confined to the proximal gut, suggesting that the normal progression to the large intestine did not occur once type 2 became firmly established. The piglet euthanized on day 6 (piglet 5; lanes 20 to 34) showed no traces of type 1 in the first 14 sections and only a faint band in the last section. This indicated that it took only 5 days for type 2 to completely displace type 1 throughout the entire gut. With a 2-day interval between challenges with the two types, there were only trace amounts of type 2 in the distal small intestine and large intestine of piglet 6 (Fig. 2C). However, when the piglet was euthanized on day 6 (piglet 7; lanes 16 to 30), rather than on day 4 (piglet 6; lanes 4 to 14), the situation was different. Type 2 was able to predominate and effectively displaced most of the type 1 from the proximal small intestine, and only small amounts of type 1 were present in the mid- and distal intestines.
FIG. 2.
Genotype analysis of the intestinal sections from infected gnotobiotic piglets by COWP PCR-RFLP. Sections were obtained at necropsy and extend from the stomach (left) to the distal spiral colon (right). (A) Piglet 1 (type 1; lanes 4 to 18), piglet 2 (type 2; lanes 20 to 32), piglet 3 (mixed; lanes 34 to 49), and 100-bp ladder (lanes 1, 19, 33, and 50). (B) Piglet 4 (mixed, type 2 day 1, euthanized day 4; lanes 4 to 18), piglet 5 (mixed, type 2 day 1, euthanized day 6; lanes 20 to 34), and 100-bp ladder (lanes 1, 19, and 35). (C) Piglet 6 (mixed, type 2 day 2, euthanized day 4; lanes 4 to 14), piglet 7 (mixed, type 2 day 2, euthanized day 6; lanes 16 to 30), and 100-bp ladder (lanes 1, 15, and 31). Type 1 and type 2 controls are in lanes 2 and 3 of each panel, respectively.
Intestinal sections, collected from each infected piglet, were examined histologically, and the extent of infection was scored as summarized in Fig. 3 for seven piglets representing the five groups at the time of euthanasia. The number of parasites observed in a particular section did not necessarily correlate with the extent of the lesions, because some areas with relatively few parasites had severe pathological changes, while some highly colonized areas showed relatively insignificant changes. Several animals had focal lesions in the proximal small intestine; however, the most-severe lesions were consistently seen in the distal ileum. Lesions in the small intestine were comprised of villous atrophy and fusion, metaplasia of epithelial cells from columnar to low cuboidal, and inflammatory cell infiltration. Peyer's patches were reactive in all animals, as indicated by many germinal centers, and were associated with atrophy of overlying villi. In general, the piglets euthanized on day 4 had mild gut lesions compared to piglets euthanized on day 6, in whom moderate gut lesions were observed, which tended to be distributed in the mid- to terminal ileum but occurred more extensively in the large intestine. The extent of lesions was more severe in animals infected with type 2 or a mixture of types 1 and 2 than in those infected with type 1, reflecting again that type 1 induces less severe mucosal alterations than type 2, which confirms earlier observations.
Mixed infection in cell culture.
Results of the COWP PCR-RFLP analysis are shown in Fig. 4 for the mixed-infection experiment in vitro. Wells that contained either type 2 (lanes 3 to 7) or type 1 (lanes 13 to 17) infected the cells within, and the parasites had the expected genotype. The mixed-infection wells (lanes 8 to 12) showed both types present, suggesting that type 2 did not prevent infection by type 1 in vitro. However, there appeared to be a slight reduction in the intensity of the type 1 PCR product, compared to that of the type 2 product, as the time interval increased. Clearly, the dynamics of mixed infection in cell culture, which is confined to the asexual phase of the life cycle, is not reflective of the situation in the host.
DISCUSSION
The present study with gnotobiotic piglets has confirmed our earlier observations that, when C. parvum type 1 and type 2 concurrently infect the same host, type 2 predominates and rapidly displaces type 1 in the gastrointestinal tract. This phenomenon has now been observed with several of our laboratory and field C. parvum type 1 isolates and in several hosts, including calves and humans (data not shown). PCR-RFLP analysis of both fecal samples and mucosal scrapings from the piglets clearly showed the disappearance of type 1 when given either concurrently or 1 or 2 days in advance of type 2. Challenging piglets with type 1, 1 or 2 days before type 2, only marginally prolonged the survival of type 1 in piglets. This suggests that type 2, in addition to becoming established more rapidly, may exhibit some inhibitory effect over the growth of type 1. This predominance of type 2 in mixed infections of piglets does not appear to be due to a higher rate of development of the different parasitic life cycle forms (55). However, the rate of infection was different, with generally more type 2 than type 1 parasite forms seen in a given villus from an infected piglet. We can only speculate as to the cause of this phenomenon at this time. It is also not clear if these properties are shared by a few, most, or all isolates of type 1. To address this question, additional studies on isolates obtained from geographically and genetically diverse sources will be required. However, indirect evidence suggesting that displacement of type 1 by type 2 is a general phenomenon can be drawn from clinical observations in which only a few patients had mixed infections of type 1 and type 2 (23, 37, 43, 53). The paucity of reported mixed infections in humans may reflect the low sensitivity of the assays used to genotype the samples. Evidence from studies in this laboratory indicates that a miniscule number of type 1 samples are often found to coexist with type 2 (49), which again may have significant epidemiological implications.
The detection of parasite DNA by COWP PCR-RFLP in feces and gut sections was more sensitive than the detection of oocysts or endogenous parasite forms by microscopy. The high degree of sensitivity of PCR-RFLP was reflected in the amplification of C. parvum DNA when no parasites were detected histologically. The distribution of parasites and lesions in the gut followed a similar pattern and depended largely on the interval between challenge and euthanasia. The distribution patterns of the parasites in the type 1, type 2, and mixed-infection piglets on day 4 differed slightly between the type 1 piglets and the other two piglet groups. Although both types were distributed throughout the gut, the greatest number of type 1 parasites was in the distal small intestine and large intestine, while the highest concentration of type 2 parasites was in the proximal small intestine and decreased toward the large intestine. The observed distribution patterns for types 1 and 2 in piglets infected with a mixture were similar to the parasite patterns reported by Pereira et al. (41) and Morgan-Ryan et al. (30) in piglets infected with a single type. We observed that piglets challenged with type 2 or a mixture of types 1 and 2 exhibited a greater intensity of infection and mucosal lesions. Pereira et al. (41) and Morgan-Ryan et al. (30) similarly reported moderate-to-severe mucosal attenuation in type 2-infected pigs and mild-to-moderate lesions in the type 1-infected pigs. While type 1 appears to induce milder symptoms in piglets (30, 41; Akiyoshi and Tzipori, unpublished) and in calves (Akiyoshi and Tzipori, unpublished), there is clinical evidence that in humans there are no appreciable differences in the manifestation of symptoms or severity between the two genotypes (53), which may reflect the existence of isolate variation in terms of infectivity and virulence, as was shown to be the case in studies with several type 2 C. parvum isolates in human volunteers (34, 35, 51). This is also supported by the findings of two studies of sporadic cases of cryptosporidiosis in which humans infected with type 1 were reported to have had longer patent periods and shed greater number of oocysts than did humans infected with type 2 (24, 60).
The growth of the parasites in cell culture, which was limited to 48 h and to the asexual phase of the life cycle, may be the reason for the failure to reproduce the predominance of type 2 over type 1 in this system. Further studies with different time intervals, or different cell lines, may have produced different results. Scanning and transmission electron microscopy studies currently in progress in this laboratory suggest that there is an appreciable delay in the ability of type 1 sporozoites to attach and become internalized by cells, compared to the ability of type 2. This delay may be due to differences in efficiency of the cellular signal transduction, or ligand-cell receptor affinity, which impact parasite attachment and invasion. The studies with piglets did not indicate that there were apparent differences in terms of parasite turnover, again possibly suggesting differences in efficiency.
Given these results, and the lack of evidence that the reverse is true for humans (Tzipori and Akiyoshi, unpublished), the mechanism of transmission and of survival of type 1 among humans, and other mammals in particular, remains a mystery. Clearly, type 1 has evolved an adept and clever mechanism of survival, because it is the predominant type in several studies of sporadic and outbreak infections in humans in environments where type 2 is ubiquitous. The predominance of type 2 and the associated apparent loss of type 1 in mixed infections among humans, as well as animals, add to the intrigue. The identification of mixed populations in which type 1 appears to coexist in miniscule fractions (0.01%) of the total population of type 2, on the other hand, adds to this complexity (49). Clearly, additional studies are needed to elucidate the mechanisms of survival of type 1 as a subpopulation, mode of transmission, and distribution among various mammalian populations. Only then can we begin to understand the contribution of both host and parasite factors to cryptosporidiosis.
Acknowledgments
D. E. Akiyoshi and S. Mor contributed equally to the manuscript.
This study was supported by National Institutes of Health grants NO1 AI-25466 and RO1-AI-50471 and Food and Drug Administration grant FD-U-001621.
We thank Julia Dilo and Sue Chapman for technical support.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Akiyoshi, D. E., X. Feng, M. A. Buckholt, G. Widmer, and S. Tzipori. 2002. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect. Immun. 70:5670-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allam, A. F., and A. Y. Shehab. 2002. Efficacy of azithromycin, praziquantel and mirazid in treatment of cryptosporidiosis in school children. J. Egypt. Soc. Parasitol. 32:969-978. [PubMed] [Google Scholar]
- 3.Amadi, B., M. Mwiya, J. Musuku, A. Watuka, S. Sianongo, A. Ayoub, and P. Kelly. 2002. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomized controlled trial. Lancet 360:1375-1380. [DOI] [PubMed] [Google Scholar]
- 4.Awad-El-Kariem, F. M., H. A. Robinson, F. Petry, V. McDonald, D. Evans, and D. Casemore. 1998. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol. Res. 84:97-101. [DOI] [PubMed] [Google Scholar]
- 5.Buckholt, M. A., J. H. Lee, and S. Tzipori. 2002. Prevalence of Enterocytozoon bieneusi: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carraway, M., S. Tzipori, and G. Widmer. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65:3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casemore, D. P., S. E. Wright, and R. L. Coop. 1997. Cryptosporidiosis: human and animal epidemiology, p. 65-92. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 8.Chappell, C. L., P. C. Okhuysen, C. R. Sterling, and H. L. DuPont. 1996. Cryptosporidium parvum: intensity of infection and oocysts excretion patterns in healthy volunteers. J. Infect. Dis. 173:232-236. [DOI] [PubMed] [Google Scholar]
- 9.Colford, J. M., Jr., I. B. Tager, A. M. Hirozawa, G. F. Lemp, T. Aragon, and C. Petersen. 1996. Cryptosporidiosis among patients infected with human immunodeficiency virus. Factors related to symptomatic infection and survival. Am. J. Epidemiol. 144:807-816. [DOI] [PubMed] [Google Scholar]
- 10.Dillingham, R. A., A. A. Lima, and R. L. Guerrant. 2002. Cryptosporidiosis: epidemiology and impact. Microbes Infect. 4:1059-1066. [DOI] [PubMed] [Google Scholar]
- 11.Fayer, R., C. A. Speer, and J. P. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 12.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., J. Greensill, R. W. Ashford, L. E. Cuevas, C. M. Parry, N. A. Cunliffe, N. J. Beeching, and C. A. Hart. 2003. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J. Clin. Microbiol. 41:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giles, M., K. A. Webster, J. A. Marshall, J. Catchpole, and T. M. Goddard. 2001. Experimental infection of a lamb with Cryptosporidium parvum genotype 1. Vet. Rec. 149:523-525. [DOI] [PubMed] [Google Scholar]
- 15.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths, J. K. 1998. Human cryptosporidiosis: epidemiology, transmission, clinical disease treatment, and diagnosis. Adv. Parasitol. 40:38-87. [DOI] [PubMed] [Google Scholar]
- 17.Guerrant, R. L. 1997. Cryptosporidiosis: an emerging, highly infectious threat. Emerg. Infect. Dis. 3:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe, A. D., S. Forster, S. Morton, R. Marshall, K. S. Osborn, P. Wright, and P. R. Hunter. 2002. Cryptosporidium oocysts in a water supply associated with a cryptosporidiosis outbreak. Emerg. Infect. Dis. 8:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadappu, K. K., M. V. Nagaraja, P. V. Rao, and B. A. Shastry. 2002. Azithromycin as treatment for cryptosporidiosis in human immunodeficiency virus disease. J. Postgrad. Med. 48:179-181. [PubMed] [Google Scholar]
- 21.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohno. 2000. Short report: possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 22.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp4040/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlin, J., S. Pedraza-Diaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mor, S. 2001. Aspects of biology relating to type 1 and type 2 Cryptosporidium parvum. B.S. (Veterinary) thesis. University of Sydney, Sydney, Australia.
- 26.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. A. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 27.Morgan, U. M., L. Pallant, B. W. Dwyer, D. A. Forbes, G. Rich, and R. C. Thompson. 1998. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J. Clin. Microbiol. 36:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan, U. M., L. Xiao, B. D. Hill, P. O'Donoghue, J. Limor, A. Lal, and R. C. A. Thompson. 2000. Detection of the Cryptosporidium parvum “human” genotype in a dugong (Dugong dugon). J. Parasitol. 86:1352-1354. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. A. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijawi, I. Sulaiman, R. Fayer, R. C. A. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 31.Navin, T. R., R. Weber, D. J. Vugia, D. Rimland, J. M. Roberts, D. G. Addiss, G. S. Visvesvara, S. P. Wahlquist, S. E. Hogan, L. E. Gallagher, D. D. Juranek, D. A. Schwartz, C. M. Wilcox, J. M. Stewart, D. E. Thompson III, and R. T. Bryan. 1999. Declining CD4+ T-lymphocyte counts are associated with increased risk of enteric parasitosis and chronic diarrhea: results of a 3-year longitudinal study. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:154-159. [DOI] [PubMed] [Google Scholar]
- 32.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 33.Okhuysen, P. C., and C. L. Chappell. 2002. Cryptosporidium virulence determinants—are we there yet? Int. J. Parasitol. 32:517-525. [DOI] [PubMed] [Google Scholar]
- 34.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 35.Okhuysen, P. C., C. L. Chappell, M. M. Marshall, G. Widmer, and S. Tzipori. 2002. Infectivity of a Cryptosporidium parvum isolate of cervine origin for healthy adults and gamma interferon knockout mice. J. Infect. Dis. 185:1320-1325. [DOI] [PubMed] [Google Scholar]
- 36.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, S., S. Pedraza-Diaz, J. McLauchlin, and D. P. Casemore. 1998. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Commun. Dis. Public Health 1:231-233. [PubMed] [Google Scholar]
- 38.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type’ from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 39.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 40.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. L. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira, S. J., N. E. Ramirez, L. Xiao, and L. A. Ward. 2002. Pathogenesis of human and bovine Cryptosporidium parvum in gnotobiotic piglets. J. Infect. Dis. 186:715-718. [DOI] [PubMed] [Google Scholar]
- 42.Pieniazek, N. J., F. J. Bornay-Llinare, S. B. Slemenda, A. J. da Silva, L. N. S. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quiroz, E. S., C. Bern, J. R. MacArthur, L. Xiao, M. Fletcher, M. J. Arrowood, D. K. Shay, M. E. Levy, R. I. Glass, and A. Lal. 2000. An outbreak of cryptosporidiosis linked to a foodhandler. J. Infect. Dis. 181:695-700. [DOI] [PubMed] [Google Scholar]
- 44.Riggs, M. W. 2002. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 4:1067-1080. [DOI] [PubMed] [Google Scholar]
- 45.Rossignol, J. F., A. Ayoub, and M. S. Ayers. 2001. Treatment of diarrhea caused by Cryptosporidium: a prospective randomized double-blind placebo controlled study of nitazoxanide. J. Infect. Dis. 184:103-106. [DOI] [PubMed] [Google Scholar]
- 46.Sheehan, D., and B. Hrapchak. 1987. Theory and practice of histotechnology, 2nd ed. Battelle Press, Columbus, Ohio.
- 47.Spano, F., L. Putignani, A. Cristanti, P. Sallicandro, U. M. Morgan, S. M. LeBlancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanriverdi, S., M. O. Arslan, D. E. Akiyoshi, S. Tzipori, and G. Widmer. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 50.Teunis, P. F., C. L. Chappell, and P. C. Okhuysen. 2002. Cryptosporidium dose-response studies: variation between hosts. Risk Anal. 22:475-485. [DOI] [PubMed] [Google Scholar]
- 51.Teunis, P. F., C. L. Chappell, and P. C. Okhuysen. 2002. Cryptosporidium dose-response studies: variation between isolates. Risk Anal. 22:175-183. [DOI] [PubMed] [Google Scholar]
- 52.Tiangtip, R., and S. Jongwutiwes. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health 7:357-364. [DOI] [PubMed] [Google Scholar]
- 53.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am. J. Trop. Med. Hyg., in press. [PubMed]
- 54.Tzipori, S. 1998. Cryptosporidiosis: laboratory investigations and chemotherapy. Adv. Parasitol. 40:5-36. [DOI] [PubMed] [Google Scholar]
- 55.Tzipori, S., and H. Ward. 2002. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 4:1047-1058. [DOI] [PubMed] [Google Scholar]
- 56.Tzipori, S., and J. K. Griffiths. 1998. Natural history and biology of Cryptosporidium parvum. Adv. Parasitol. 40:5-35. [DOI] [PubMed] [Google Scholar]
- 57.Tzipori, S., W. Rand, J. Griffiths, G. Widmer, and J. Crabb. 1994. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulins. Clin. Diagn. Lab. Immunol. 1:450-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Widmer, G. 1998. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv. Parasitol. 40:223-239. [DOI] [PubMed] [Google Scholar]
- 59.Widmer, G., D. E. Akiyoshi, M. A. Buckholt, X. Feng, S. M. Rich, K. M. Deary, C. A. Bowman, P. Xu, Y. Wang, G. A. Buck, and S. Tzipori. 2000. Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol. Biochem. Parasitol. 108:187-197. [DOI] [PubMed] [Google Scholar]
- 60.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gillman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 61.Xiao, L., C. Bern, M. Arrowood, I. Sulaiman, L. Zhou, V. Kawai, A. Vivar, A. A. Lal, and R. H. Gilman. 2002. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 185:1846-1847. [DOI] [PubMed] [Google Scholar]