Abstract
The rabbit model of tuberculosis has been used historically to differentiate between Mycobacterium tuberculosis and Mycobacterium bovis based on their relative virulence in this animal host. M. tuberculosis infection in market rabbits is cleared over time, whereas infection with M. bovis results in chronic, progressive, cavitary disease leading to death. Because of the innate resistance of commercial rabbits to M. tuberculosis, 320 to 1,890 log-phase, actively growing inhaled bacilli were required to form one grossly visible pulmonary tubercle at 5 weeks. The range of inhaled doses required to make one tubercle allows us to determine the relative pathogenicities of different strains. Fewer inhaled organisms of the M. tuberculosis Erdman strain were required than of M. tuberculosis H37Rv to produce a visible lesion at 5 weeks. Furthermore, with the Erdman strain, only 7 of 15 rabbits had healed lesions at 16 to 18 weeks; among the other animals, two had chronic, progressive cavitary disease, a phenotype usually seen only with M. bovis infection. Genotypic investigation of the Erdman strain with an H37Rv-based microarray identified gene differences in the RD6 region. Southern blot and PCR structural genetic analysis showed significant differences between M. tuberculosis strains in this region. Correlation of the relative pathogenicity, including disease severity, in the rabbit model with the strain genotype may help identify stage-specific M. tuberculosis genes important in human disease.
Tuberculosis continues to be a leading cause of infectious disease mortality. Because of the complex pathogenesis of the disease, the development of animal models for specific stages of disease such as latency and cavitation remains a priority. The murine model is the most widely used animal model because of its economy, the extensive characterization of inbred strains, and the commercial availability of immunologic reagents (31). The guinea pig is another well-researched model of tuberculosis and has the advantages of remarkable susceptibility to low doses of aerosolized Mycobacterium tuberculosis and the development of a strong delayed-type hypersensitivity response (11, 36, 37, 40).
The rabbit model offers certain advantages over both the murine and guinea pig models. First, when infected with M. tuberculosis or Mycobacterium bovis, rabbits have a spectrum of disease that represents many of the specific stages of human disease. In general, rabbits are able to contain disease caused by virulent M. tuberculosis. Over time, the number of pulmonary bacilli decline and the lesions regress (22). With M. bovis infection, rabbits form chronic fibrosing pulmonary cavities (8). Both of these are prominent features in human disease. Finally, rabbit lung granulomas, with their caseous centers, closely resemble the human granuloma. Lurie and colleagues have found a remarkable similarity between the spectrum of rabbit tuberculosis and that found in humans (23).
In the era before genomics and before the biochemical property differences were elucidated, the rabbit infection model was used to differentiate between M. bovis and M. tuberculosis because of the remarkable difference in virulence for these animals (39). More recently, rabbits have been used to examine the difference in relative pathogenicities between M. tuberculosis strains CDC1551 and H37Rv. Tubercles from animals infected with CDC1551 are smaller and contain fewer bacilli than those infected with H37Rv (3).
In two recent reports, genomic deletion analysis was used to propose a new evolutionary schema for the M. tuberculosis complex of organisms (6, 29). The conclusion reached by both groups of authors was that successive loss of DNA from M. tuberculosis gave rise to M. bovis as well as the other members of the M. tuberculosis complex. Other genetic approaches such as bacterial artificial chromosome analysis have also been used to identify differences among the members of the M. tuberculosis complex (4, 5, 15). More information on the relative pathogenicity in vivo needs to be correlated with the genomic content of individual strains.
This report describes a series of infections in rabbits with three different strains of M. tuberculosis: Erdman, H37Rv, and CDC1551. In general, with increasing dose of any strain, more inhaled bacteria are required to produce one grossly visible tubercle at 5 weeks. Of the three strains, the Erdman strain appears to be the most virulent for rabbits, requiring the lowest number of inhaled bacilli to make one tubercle. This strain also has the greatest spectrum of disease at 16 to 22 weeks after initial aerosol infection. The Erdman strain's ability to produce coalescing lesions with occasional cavities by fewer inhaled bacteria compared with H37Rv led us to analyze its genomic DNA. Genomic analysis of the Erdman strain showed differences in the RD6 deletion region that is rich in PPE genes and that has been shown previously to be absent in many strains of M. bovis and in a few strains of M. tuberculosis (6). Further analysis of this region revealed interesting differences between strains.
MATERIALS AND METHODS
Microorganisms.
We compared M. tuberculosis strains CDC1551 (Oshkosh or CSU93), H37Rv, and Erdman (a kind gift of the late Frank Collins) (3). Strains were grown in roller bottles to log phase in Middlebrook 7H9 medium supplemented with 10% albumin dextrose complex, 0.2% glycerol, and 0.05% Tween 80. The mycobacteria were then bead vortexed for 1 min and frozen in aliquots at −70°C. Titered aliquots were thawed at the time of use, diluted with phosphate-buffered saline, and bead vortexed. The titer of the inoculum was verified by plating on Middlebrook 7H10 agar supplemented with albumin dextrose complex and glycerol. For log-phase cultures, these frozen aliquots were thawed and grown to log phase in complete Middlebrook 7H9 medium. Clumps were allowed to settle for 2 to 3 h. Cultures were then suspended in a 10% dilution of oleic albumin complex (Becton Dickinson Bioscience, Sparks, Md.) in 0.9% NaCl to a specific optical density. Bacilli were visualized in a hemacytometer to confirm that large clumps were absent and to estimate the number of CFU per milliliter that was confirmed by culture.
Animals and infection.
Specific pathogen-free New Zealand White rabbits (2.5 kg; female) were purchased from Covance Research Products, Inc. (Denver, Pa.), maintained in standard cages in biosafety level 3 conditions, and fed commercial rabbit chow and water ad libitum. All animals were maintained in accordance with protocols approved by the institutional animal care and use committees of The Johns Hopkins University, George Washington University, and the U.S. Army Medical Research Institute of Infectious Diseases (8). Just prior to infection, the volume of air that each rabbit breathed per minute was calculated from the per-minute ventilation, respiratory rate, and tidal volume of air inhaled by each rabbit (measured in a whole-body plethysmograph) according to Guyton's formula by using Pulmonary Mechanics computer model no. 6 software (Buxco Electronics, Sharon, Conn.) (17). Animals were infected by aerosol at the U.S. Army Medical Research Institute of Infectious Diseases by using a nose-only system where aerosols are generated in a class III biosafety glove box cabinet under negative pressure or in a completely contained biosafety level 4 air-locked area. The biosafety level 3 exposure chamber was a 16-liter Plexiglas box with one side containing a circular latex dam with a cutout into which the nose and mouth of the rabbit were inserted (41). The aerosol was generated with a Collison nebulizer containing bacilli suspended in the 10% dilution of oleic albumin complex. The aperture of the nebulizer disperses clumps of bacilli into units of one to three bacilli. Throughout the 10-min exposure period, a one-fourth-inch, stainless steel sampling port placed in the exposure chamber box at the level of the animal's nose was used to collect samples into 10% oleic albumin complex containing antifoam A (Sigma Chemical Co., St. Louis, Mo.) with an all-glass impinger (AGI-30; Ace Glass, Inc., Vineland, N.J.). Rabbits were placed in a loose-fitting canvas bag and then gently hand held. The oleic albumin complex solutions containing the aerosolized bacteria were cultured at various dilutions on 7H10 Middlebrook agar (Fisher Scientific). For each rabbit, the number of viable bacilli inhaled was calculated based on the volume of inhaled air during exposure and the number of CFU cultured from the impinger samples per milliliter. The animals were housed in biosafety level 3 facilities at the George Washington University Medical Center immediately following infection. Four weeks after infection, a 1:30 dilution of 4× Old Tuberculin concentrate (Wyeth Lederle, Pearl River, N.Y.) was injected intradermally, and 2 days later the resulting skin reaction was measured with calipers. Delayed-type hypersensitivity responses were quantified by measuring the skin fold thickness of the indurated skin minus the double skin fold thickness of normal unaffected skin and then multiplying by the width and length of the indurated area. At 5 weeks after infection, the rabbits were euthanized with intravenous pentobarbital (Euthasol; NLS Animal Health, Baltimore, Md.) and the number of grossly visible primary tubercles in the lungs was counted by Lurie's tubercle count method (12, 24). Other animals were sacrificed at later time points as noted in Results.
Microarrays.
A set of 70-bp oligonucleotides representing all 4,295 open reading frames of H37Rv (Qiagen Operon Technologies, Alameda, Calif.) were spotted in duplicate on glass slides coated with poly-l-lysine in a 384-well-format arrayer (GeneMachines, San Carlos, Calif.). Bacterial genomic DNA prepared by previously established methods (19) was labeled with either Cy3 or Cy5 probes (Amersham Pharmacia, Piscataway, N.J.) using the aminoallyl labeling method (34). The slides were scanned with an Axon scanner, and the Genepix (Union City, Calif.) array viewing software system was used to define and quantify spot intensities. Erdman genomic DNA was compared with that of CDC1551. Reverse labeling was performed to confirm the hybridization of individual spots. As each 70-mer was spotted in duplicate and reverse labeling was performed, four readings were available for each gene. A positive signal was defined as a greater than 20-fold difference in fluorescence hybridization signals between the two strains.
Southern blotting and PCR.
Reaction mixtures contained 0.5 mM concentrations of deoxynucleoside triphosphates, 5 μl of Expand High Fidelity 10× buffer, 1 μM primers, 0.75 μl of Expand HF polymerase, 500 μM magnesium chloride, and sterile distilled water in a final volume of 50 μl. Thermal cycling was performed in thin-walled tubes on a Mastercycler (Eppendorf, Westbury, N.Y.) with an initial denaturation step of 96°C for 10 min, followed by 30 cycles of 94°C for 15 s, 59°C for 30 s, and 72°C for 20 s. See Table 3 for the primers used to amplify intragenic regions of the genes in the RD6 region.
TABLE 3.
Detection of genes in M. bovis strains
| Gene | Primers used
|
Presence in strain:
|
|||
|---|---|---|---|---|---|
| Forward | Reverse | H37Rv | CDC1551 | Erdman | |
| PPE57 | GGCGCTGACTCATTGTTTTT | TGTGTCGTTATAGGCGTTCG | + | − | − |
| PPE58 | ACACTCCCGTCAGATTTTGC | AGATAGCGGTCCATTGCTTG | + | − | + |
| PPE59 | CAAATTCTGGAAACCGCCTA | GATAAACCGGGCGGTACTTT | + | + | + |
| Rv3427 | CACGCTGATGACCTCTACGA | ATTTGGTGGCTGGTGTTGAT | + | − | + |
| Rv3427 | CGTCGGAAAAACCCATGTAG | TAGAGGTCATCAGCGTGCAT | + | − | − |
| Rv3428 | CGCCAGAAAACCCAAAGATA | CAGCTGGTGAGTTCAAATGC | + | − | − |
Two micrograms of genomic DNA was digested with the specified restriction enzyme, electrophoresed on a 1% agarose gel, soaked in 0.25 M HCl at room temperature for 15 min, then soaked in 1.5 M NaCl-0.5 M NaOH for 1 h at room temperature, then soaked in 1.5 M NaCl-0.5 M Tris-HCl (pH 7.5) for 1 h at room temperature, and finally transferred to a nylon filter. A digoxigenin-labeled Rv3424c intragenic probe (forward, 5′-CGGAAAGCTGATCTGGTGAT-3′; reverse, 5′-GGATGTGGGGTGTTTTTGAG-3′) was made. Hybridizations were performed at 42°C overnight in a solution containing 30 ml of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10× Denhardt's reagent, 5 ml of 10% sodium dodecyl sulfate (SDS), 10 mg of boiled salmon sperm DNA/ml, and 50% formamide. Membranes were then washed in 2× SSC-0.1% SDS for 30 min at room temperature, washed in a roller bottle with 0.5× SSC-0.1% SDS for 1 h at 42°C, and then washed in 0.1 M maleic acid-0.15 M NaCl-0.3% Tween 20 (pH 7.5) for 5 min. Detection of the digoxigenin was achieved by using the Roche system (Mannheim, Germany).
Statistical analysis.
All comparisons of nonnormally distributed continuous data were analyzed with the Wilcoxon rank sum test using SAS version 8.2 (SAS Institute, Cary, N.C.).
RESULTS
Abilities of H37Rv, Erdman, and CDC1551 strains of M. tuberculosis to produce visible primary pulmonary tubercles in commercial New Zealand White rabbits.
The number of grossly visible primary pulmonary tubercles was counted 5 weeks after aerosol challenge with virulent M. tuberculosis. For the H37Rv strain, the number of inhaled log-phase bacteria required to produce one visible lesion ranged from 586 to 1,889 (Table 1). For the Erdman strain, the number of bacilli inhaled per tubercle ranged from 320 to 827, and for the CDC1551 strain, in the one experiment in which it was tested, the number of bacilli averaged 1,667 (Table 1) (3). The in vitro growth rates of all three strains were identical in broth media as measured by optical densities at 600 nm.
TABLE 1.
Results for rabbits given aerosol doses of different M. tuberculosis strains
| Expt no.a | Strainb | Mean impinger dose (CFU/ml) ± SD | Mean inhaled dose (CFU/ml) ± SE | Log10 inhaled bacilli | Mean no. of tubercles at 5 wk | Mean tubercle diameter (mm) ± SE | No. of bacilli inhaled/tubercle |
|---|---|---|---|---|---|---|---|
| 1 | H37Rv | 121,250 ± 16,530 | 393,330 ± 63,180 | 5.59 | 278 ± 65 | 2.3 ± 0.1 | 1,415 |
| 2 | H37Rv | 31,170 ± 3,820 | 60,050 ± 4,950 | 4.78 | 81 ± 21 | 1.6 ± 0.2 | 741 |
| 3 | H37Rv | 105,000 ± 6,190 | 410,000 ± 78,300 | 5.61 | 217 ± 53 | 1.6 ± 0.2 | 1,889 |
| 4 | H37Rv | 24,880 ± 2,490 | 38,880 ± 6,230 | 4.58 | 65 ± 10 | 2.2 ± 0.2 | 598 |
| 5a | H37Rv | 7,930 ± 1,340 | 3.90 | 8 ± 4 | 1.9 ± 0.2 | 991 | |
| 5b | CDC1551 | 600 | 8,500 ± 1,730 | 3.92 | 5.1 ± 1.5 | 1.2 ± 0.2 | 1,667 |
| 6 | CDC1551(f) | 14,100 ± 1,670 | 53,600 ± 6,570 | 4.73 | 3 ± 0.8 | 0.8 ± 0.1 | 17,867 |
| 7 | Erdman | 134,170 ± 16,700 | 309,300 ± 46,440 | 5.49 | 374 ± 97 | 1.93 ± 0.1 | 827 |
| 8a | Erdman | 12,170 ± 2,090 | 75,000 ± 20,980 | 4.87 | 210 ± 44 | 1.5 ± 0.1 | 357 |
| 8b | Erdman(f) | 329,440 ± 8,870 | 1,231,670 ± 196,940 | 6.09 | 353 ± 60 | 1.7 ± 0.1 | 3,489 |
| 9 | Erdman | 29,670 ± 1,930 | 63,700 ± 7,050 | 4.80 | 94 ± 19 | 1.5 ± 0.1 | 678 |
| 10 | Erdman | 34,330 ± 9,420 | 60,470 ± 20,560 | 4.78 | 189 ± 51 | 2.0 ± 0.4 | 320 |
| 11 | Erdman(f) | 52,330 ± 13,310 | 113,830 ± 32,690 | 5.06 | 135 ± 23 | 2.8 ± 0.3 | 843 |
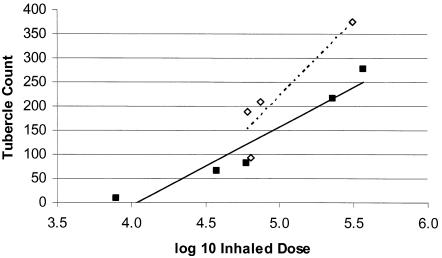
In general, with all strains tested, increasing inhalation dose tended to increase the number of bacteria required to produce one visible tubercle (Table 1) (10), We compared animals from experiments 2 and 4 with animals from experiments 8a, 9, and 10 that delivered approximately the same inhaled dose (log10 inhaled dose, ∼4.8). The Erdman strain produced a larger number of tubercles than did the same inhaled dose of strain H37Rv (averages of 163 ± 25 tubercles and 72 ± 12 tubercles, respectively; P = 0.011). Figure 1 shows the relationship between the number of tubercles and the log10 inhaled dose. All experiments with H37Rv and Erdman log-phase bacilli were used in this analysis. Inhaled frozen-thawed aerosols were excluded in the data for Fig. 1 as were experiments performed with CDC1551. The slope suggests a trend towards a widening of this virulence difference at higher doses (Fig. 1).
FIG. 1.
Relative virulence of our M. tuberculosis Erdman and H37Rv strains. Each point represents the mean tubercle count of six rabbits given the same aerosol dose. The slopes of the lines show the number of visible tubercles as a function of the number of bacilli inhaled. The Erdman strain (dashed line) produced more tubercles per bacilli inhaled than did the H37Rv strain (solid line).
Difference in the abilities to produce visible primary tubercles between log-phase and frozen-thawed M. tuberculosis.
The number of grossly visible tubercles at 5 weeks after infection seemed to depend on the growth state of the infecting bacilli. For example, despite an average inhaled dose of 54,000 frozen-thawed M. tuberculosis CDC1551, 1 of 12 rabbits had no grossly visible tubercles at necropsy 5 weeks later and the remaining 11 animals had an average of 3.3 tubercles. In contrast, in another experiment using log-phase M. tuberculosis CDC1551, an inhaled dose of only 8,500 bacilli produced a comparable number of tubercles (5.1 ± 3.7) and all animals had visible lesions (Table 1) (3). In other words, a large inhaled dose of frozen-thawed bacilli produced fewer tubercles than a much smaller inhaled dose of log-phase bacilli.
This was also true for the Erdman strain, the most virulent strain in our study (Table 2). In frozen-thawed M. tuberculosis Erdman aerosolization experiments, the mean number of bacilli required to produce one grossly visible tubercle ranged from 843 to 3,489 (Table 2). In contrast, in animals infected with log-phase bacilli, the average number of bacilli needed to produce one tubercle ranged from 419 to 827 depending on the aerosolized dose (Table 2) (Fig. 1). Furthermore, there was a statistically significant difference between the numbers of inhaled bacilli required to form one tubercle in both the higher (3,489 versus 827, P < 0.01) and lower (843 versus 419, P = 0.04) dose groups between frozen-thawed and log-phase bacilli in the inoculum.
TABLE 2.
Aerosol experiments with log-phase or frozen-thawed M. tuberculosis Erdman
| Expt no. | Type | No. of rabbits | Avg no. of inhaled bacilli (log10 inhaled) | Mean no. of tubercles ± SE | Mean tubercle diameter (mm) ± SE | No. of bacilli inhaled/tubercle | Response to old tuberculin (mm3) |
|---|---|---|---|---|---|---|---|
| 8b | Frozen | 6 | 1,231,670 (6.09) | 353 ± 60 | 1.74 ± 0.11 | 3,489 | 641 ± 180 |
| 11 | Frozen | 6 | 113,830 (5.06) | 135 ± 23 | 2.77 ± 0.28 | 843 | 740 ± 180 |
| 7 | Log | 6 | 309,330 (5.49) | 374 ± 97 | 1.93 ± 0.13 | 827 | 695 ± 222 |
| 8a and 9 | Log | 12 | 63,670 (4.80) | 152 ± 29 | 1.51 ± 0.08 | 419 | 1,133 ± 180 |
Since large clumps of bacilli were present in the frozen-thawed specimens before they were aerosolized, we conducted flow cytometric analyses of the impinger specimens to see whether the clumps remained after aerosolization and, therefore, could account for the lower tubercle-generating ability. Both log-phase and frozen impinger samples were found to be well dispersed with no detectable clumps when analyzed using dot plots of forward and side scatter (data not shown). The results of tuberculin skin tests of animals infected with log-phase and frozen-thawed M. tuberculosis Erdman strain were comparable, especially in those groups aerosolized with the most closely matched mean inhaled dose.
Results of the two frozen-thawed aerosol experiments and those of the three log-phase aerosol experiments were compared using the Wilcoxon rank sum test. There was a statistically significant 1-log difference in the inhaled dose between the two groups aerosolized with frozen-thawed bacilli (experiments 8b and 11) and the two groups aerosolized with log-phase, actively growing bacteria (experiments 7, 8a, and 9) (P < 0.01). In the animals in experiment 11 infected with 1-log-fewer frozen bacteria, the tubercle counts were significantly lower than those in rabbits in experiment 8b (P < 0.01), as expected, and the tubercle diameters were significantly larger (P < 0.01). For those animals infected with log-phase bacteria, however, the tubercle counts showed more variability, with insignificant differences in both tubercle counts and diameters despite the 1-log difference in inhaled dose.
M. tuberculosis Erdman spectrum of disease.
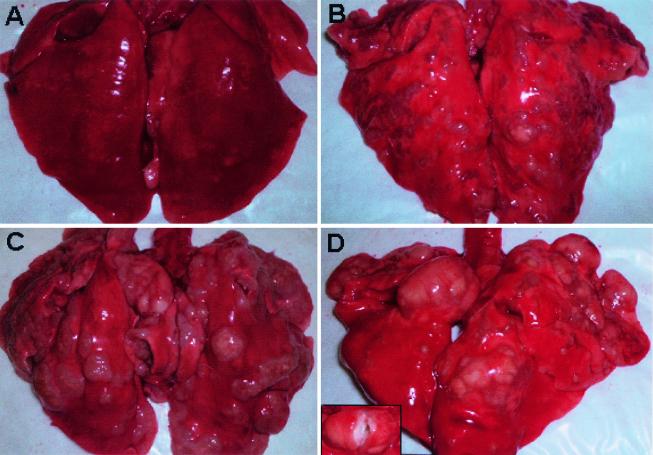
At 16 to 18 weeks after aerosol infection with frozen-thawed Erdman bacilli, the spectrum of disease varied greatly among the 15 rabbits necropsied. Seven (47%) had nearly or completely healed lesions, three had multiple, small caseous tubercles, and three had large coalescing lesions. Finally, two animals had large caseous lesions: one had early cavities filled with solid caseum and one had multiple large caseous lesions, some of which had frank liquefaction (Fig. 2).
FIG. 2.
Lungs from outbred New Zealand White rabbits at 16 to 18 weeks after aerosol infection with M. tuberculosis Erdman strain. Panels show lungs completely healed of disease with no visible tubercles (A), multiple coalescing lesions scattered throughout the lungs (B), multiple large coalescing lesions, some with early cavity formation (C), and a large cavitary lesion (arrow) filled with liquefied caseum (inset) and multiple coalescing lesions (D).
Comparison of M. tuberculosis Erdman and H37Rv by microarray genotyping.
After eliminating differences relating to IS6110 copy number, we found that only one gene, Rv3428c, was absent in Erdman relative to H37Rv by our fluorescence intensity criteria and confirmed by reverse labeling. As would be expected with the use of an array based on the H37Rv sequence, we could not find genes that were present in Erdman but absent in H37Rv. Interestingly, the deleted gene from the Erdman strain was part of a five-gene deletion of the RD6 deletion region seen in many strains of M. bovis (6). Intragenic PCR of each of the predicted open reading frames in H37Rv (http://genolist.Pasteur.fr/TubercuList) showed the presence of Rv3424c, PPE57, PPE58, Rv3427, and PPE59 and the absence of Rv3428c in the Erdman strain (Table 3). All genes were present in H37Rv by PCR. As predicted by the genome sequence of CDC1551 (www.tigr.org), Rv3425 through Rv3428c were missing by PCR. Using primers that spanned regions from PPE57 to Rv3427 and PPE57 to PPE58, products were obtained in Erdman whose sequences were identical to those of H37Rv except for two base substitutions at positions 382 and 387 (both alanine-to-guanine substitutions). These resulted in a nonsynonymous substitution (threonine to alanine in the Erdman strain) and one synonymous substitution. PPE58 in Erdman contained four guanines in the sequence at position 373 leading to a stop codon after positoin 528 and resulting in a truncated protein. In H37Rv there are give guanines resulting in a 696-bp sequence.
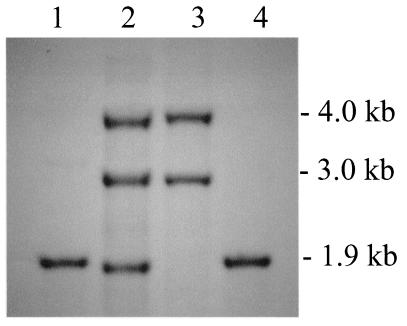
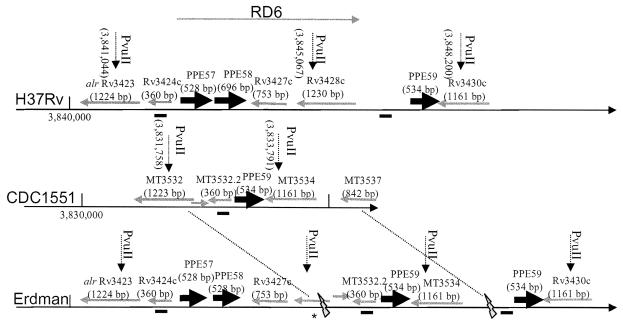
Southern blot data are presented in Fig. 3 and 4. Using three different restriction enzymes and a 244-bp Rv3424c intragenic probe, two bands were obtained for H37Rv. A BLAST search of the Rv3424c sequence against the H37Rv sequence showed an exact match not only for the annotated Rv3424c gene but also for a 208-bp region (with infrequent mismatches across the remaining 152 bp) located between Rv3428c and PPE59. The truncated copy of Rv3424c in H37Rv putatively codes for a 73-amino-acid protein that is not annotated in the genome. CDC1551 has only one copy, denoted MT3532.2, in the database. The Erdman strain has three copies of Rv3424c (one truncated as in H37Rv). Three PPE genes (PPE57 to PPE59) in the RD6 region of the H37Rv genome bear remarkable homology. PPE57 and PPE58 are absent in CDC1551, as was previously reported (14). The Erdman strain appears to contain segments in the RD6 region similar to both H37Rv and CDC1551. With both PvuII and BglII digests in the Erdman strain genomic DNA, Rv3424c hybridized to three fragments with the sizes predicted for H37Rv and CDC1551 (∼4.0, 2.9, and 1.9 kb and ∼5.5, 3.2, and 2.2 kb, respectively). Other genomic DNA digests with PmlI, PmlI/NheI, and KpnI were also performed and Southern blotted, and the results agreed with the putative map presented in Fig. 4. Further investigation of Rv3428c, an IS1532 transposon that may be truncated, is needed as we were unable to obtain PCR products of this region.
FIG. 3.
Southern blot analysis with PvuII-cut genomic DNA probed with a 244-bp fragment of Rv3424c. Lane 1, CDC1551; lane 2, Erdman; lane 3, H37Rv; lane 4, M. bovis BCG Pasteur.
FIG. 4.
Genetic map of the RD6 region in M. tuberculosis strains H37Rv, CDC1551, and Erdman. The hybridization site of the 244-bp Rv3424c intragenic probe is shown as a thick bold line underneath each linearized genome. The RD6 region is fully present in H37Rv and is shown as an arrow extending from Rv3425 to Rv3428c. PPE family genes are shown in grey. An area that is not fully characterized in Erdman is indicated by an asterisk.
DISCUSSION
In 1898, Theobald Smith wrote one of the first descriptions of rabbit infection with tuberculosis. He pointed out remarkable differences in the abilities of rabbits to control intravenous infections by M. bovis and M. tuberculosis (38). Only 1 of 10 animals died following intravenous inoculation of M. tuberculosis, whereas all 9 of 9 rabbits died with a similar dose of M. bovis. Further studies with rabbits confirmed these phenotypic differences (1, 9, 22). The relative virulence of many mutant strains has been measured by intravenously infecting immunodeficient mice and quantifying the time to death. Guinea pigs that are exquisitely susceptible to M. tuberculosis have been used in a similar way in time-to-death studies coupled with histopathology to assess the relative virulence of strains. An early definition of virulence was simply the ability of an organism to multiply in the lungs of mice (32). The bacillary burden of particular organs is not, however, the only measure of a particular strain's virulence. A strain's abilities to induce lung pathology and to cause mortality are also important measures of virulence (21, 30). The relative virulence of M. tuberculosis complex organisms has been described in mice, guinea pigs, and rabbits (2, 7, 11, 13, 32, 40). Others have looked at the cellular immune response to determine whether virulence can be characterized more by the host response than the properties of the organism (27).
For example, the HN878 strain from an epidemic in Houston, Tex., was shown to be hypervirulent in a mouse model of tuberculosis (27). Subsequently, this strain was tested in a rabbit meningitis model where organisms were injected directly into the cerebrospinal fluid. HN878 had the highest bacillary load, the greatest extrameningeal dissemination, and the most severe clinical symptoms. There was also evidence of high organism burdens and an early cavitary response in the lungs compared with strains H37Rv and CDC1551, indicating continuous extrameningeal seeding (L. Tsenova, A. Bergtold, B. Mangaliso, V. H. Freedman, and G. Kaplan, Abstr. 36th Tuberc. Leprosy Res. Conf., p. 134, 2001). Although genotypic differences in the organism may be important to the pathogenicity seen in both mice and rabbits, Tsenova and colleagues showed in the murine model that an earlier Th1 response may be the key to the host's ability to limit disease severity (26). CDC1551 induced a rapid, vigorous interleukin 12-driven Th1 cytokine response and was cleared, whereas HN878 had a reduced response and resulted in a more severe disease phenotype. Due to limited cytokine reagents in the rabbits, the specific evolution of Th1 cytokines could not be studied, but the HN878 strain produced a later overzealous plasma tumor necrosis factor response that appeared to correlate with the worsened clinical status.
The relative pathogenicity of a specific strain seems to be related to its genotype, the way in which it was cultivated, and the response of the host. Because it is relatively resistant to infection caused by M. tuberculosis and because it has a wide range of disease manifestations (bacillary clearance to cavitation), the rabbit presents a unique niche in the menu of animal testing systems. Ultimately, the ability to correlate the clinical manifestations of a human isolate with those in the rabbit would be a powerful tool to study both the host response and the strain genotype's contribution to the pathogenic outcome (28).
With the genome sequencing of strains in the M. tuberculosis complex, our understanding of fundamental genetic differences among the species has broadened. It is now clear that M. bovis differentiated phylogenetically from M. tuberculosis and that the deletions in the genome of M. tuberculosis led to an expanded host range for M. bovis (6, 29). With M. microti, however, other deletions led to its attenuation for most animal species (except for the vole) and to a loss of lung tropism (4, 25). Kato-Maeda et al. used high-density oligonucleotide arrays to understand genetic differences between strains of M. tuberculosis (20). Their findings concurred with those of others showing that many deletions are mediated by insertion sequences (16, 18). These authors also concluded that increasing the number of genes deleted may lead to an attenuation of virulence and a decreased ability to produce cavities.
The rabbit model of tuberculosis has historically been an important way to differentiate between M. tuberculosis and M. bovis. Fewer than 30 inhaled bacilli of M. bovis results in a progressive infection that eventually leads to the death of the rabbit (8, 9, 33). One to 2 orders of magnitude more inhaled bacilli of H37Rv M. tuberculosis are needed to create one visible pulmonary tubercle at 5 weeks (3, 10, 23, 24). However, the majority of these rabbits recovered, as is the case with humans.
With our bacillary strains, we found that fewer inhaled bacilli of the Erdman strain than of the H37Rv strain were required to produce a visible lesion in commercial rabbits at 5 weeks. With H37Rv infection, most of the lesions healed in 4 to 6 months (1), whereas only half of the rabbits infected with the Erdman strain had healed lesions at this time. Several of the other rabbits had large, caseous, coalescing tubercles, consistent with progressive disease, with two of the rabbits having cavitary lesions. In an attempt to understand the genomic basis of this difference, we used an H37Rv-based microarray to determine whether the Erdman strain contained deletions relative to H37Rv. We found that Rv3428c in deletion region 6 (RD6) was absent in the Erdman strain. The RD6 region is known to be deleted in CDC1551 and in many strains of M. bovis (6). We did not find any of the other large deletion regions described to be associated with M. bovis despite the M. bovis-like pattern of disease that the Erdman strain produced in some of the rabbits. Microarray analysis using an H37Rv-based array would not detect regions deleted from M. tuberculosis after its divergence from M. bovis strains (e.g., TbD1) (6). Furthermore, rearrangements and point mutations, insertions, and small deletions could not be detected with this methodology (14). Because of these known limitations of microarray-based genomic analysis, we used traditional Southern blotting mapping techniques and PCR to substantiate the microarray data. We found that Erdman contained a segment identical to H37Rv interrupted by a segment identical to CDC1551. Therefore, the Erdman strain has four copies of PPE genes in this region and three copies of Rv3424c (one truncated). Furthermore, PPE57 appears to have a nonsynonymous substitution (T128A). It is noteworthy that a region with both transposons and multiple PPE genes should be so diverse among the three strains. The PPE gene family encoding acidic, glycine-rich proteins is thought to be expressed on the extracellular surface of M. tuberculosis. Others have speculated on the role of these proteins as potential antigens for host immunity, thereby implicating a role in virulence since PE and PPE genes have the highest rates of synonymous and nonsynonymous substitutions when M. tuberculosis whole genomes are compared (14). PE and PPE family proteins have also been shown to be immunogenic in a human peripheral blood mononuclear cell model and therefore may be useful in vaccine discovery (35).
Several interesting points about aerosol infection of rabbits emerge from our study. First, at higher inhaled doses of any M. tuberculosis strain, more bacilli are usually required to produce one visible lesion at 5 weeks. One possible explanation for this is that higher inhaled doses may provide a larger antigenic stimulus that can hasten acquired immunity and thereby prevent more microscopic tubercles from reaching visible size. Second, there is a significant difference between viable, frozen-thawed bacilli and log-phase, actively growing bacilli in the aerosol dose required to produce an equal number of grossly visible primary pulmonary tubercles. A greater number of frozen-thawed bacilli than log-phase bacilli are required. This may be because frozen-thawed bacilli are in lag phase and are therefore more easily destroyed by the highly activated alveolar macrophage population than are actively growing log-phase bacilli. Interestingly, the tuberculin skin test responses of the two groups of rabbits given the same inhaled dose of bacilli (frozen thawed or log phase) were not different, suggesting that once grossly visible tubercles were established in the host little difference in the two types of infecting bacilli was still present.
In conclusion, aerosol infection with M. tuberculosis allows us to determine the relative pathogenicities of different bacillary strains in the rabbit model of tuberculosis. With log-phase actively growing bacilli, the wide range of the infectious dose required to form one grossly visible pulmonary tubercle 5 weeks after infection allows us to assess the relative pathogenicities of various strains. The finding of a long-term outcome with increased propensity towards cavitary disease with some strains should be followed-up with in-depth studies of clinical strains to determine whether the rabbit outcomes mirror that of humans. Genetic analysis revealed heterogeneity in the RD6 region between Erdman and H37Rv which may account, in part, for the different patterns of disease resulting from modulation of the host response to infection that these strains produce in rabbits. Further studies of pathogenicity in the rabbit model produced by different strains within the M. tuberculosis complex along with their genotypes may elucidate specific genes that are critical in different stages of human disease.
Acknowledgments
This work was supported by the Sequella Global Tuberculosis Foundation, NIH grants K08 AI 01689-01 and 1R01 HL71554-01, and the Ellison Foundation.
We gratefully acknowledge the animal care provided by Walter Johnson and Mike Manion at George Washington University Medical Center.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Allison, M., P. Zappasodi, and M. B. Lurie. 1962. Host-parasite relationships in natively resistant and susceptible rabbits on quantitative inhalation of tubercle bacilli. Am. Rev. Respir. Dis. 85:553-569. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, V., E. H. Wiegeshaus, and D. W. Smith. 1992. Growth characteristics of recent sputum isolates of Mycobacterium tuberculosis in guinea pigs infected by the respiratory route. Infect. Immun. 60:4762-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishai, W. R., A. M. Dannenberg, N. Parrish, R. Ruiz, P. Chen, B. C. Zook, W. Johnson, J. W. Boles, and M. L. Pitt. 1999. Virulence of Mycobacterium tuberculosis CDC1551 and H37Rv in rabbits evaluated by Lurie's pulmonary tubercle count method. Infect. Immun. 67:4931-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, F. M., and M. M. Smith. 1969. A comparative study of the virulence of Mycobacterium tuberculosis measured in mice and guinea pigs. Am. Rev. Respir. Dis. 100:631-639. [DOI] [PubMed] [Google Scholar]
- 8.Converse, P. J., A. M. Dannenberg, Jr., J. E. Estep, K. Sugisaki, Y. Abe, B. H. Schofield, and M. L. Pitt. 1996. Cavitary tuberculosis produced in rabbits by aerosolized virulent tubercle bacilli. Infect. Immun. 64:4776-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Converse, P. J., A. M. Dannenberg, Jr., T. Shigenaga, D. N. McMurray, S. W. Phalen, J. L. Stanford, G. A. Rook, T. Koru-Sengul, H. Abbey, J. E. Estep, and M. L. Pitt. 1998. Pulmonary bovine-type tuberculosis in rabbits: bacillary virulence, inhaled dose effects, tuberculin sensitivity, and Mycobacterium vaccae immunotherapy. Clin. Diagn. Lab. Immunol. 5:871-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dannenberg, A. M., W. R. Bishai, N. Parrish, R. Ruiz, W. Johnson, B. C. Zook, J. W. Boles, and L. M. Pitt. 2000. Efficacies of BCG and vole bacillus (Mycobacterium microti) vaccines in preventing clinically apparent pulmonary tuberculosis in rabbits: a preliminary report. Vaccine 19:796-800. [DOI] [PubMed] [Google Scholar]
- 11.Dannenberg, A. M., Jr., and F. M. Collins. 2001. Progressive pulmonary tuberculosis is not due to increasing numbers of viable bacilli in rabbits, mice and guinea pigs, but is due to a continuous host response to mycobacterial products. Tuberculosis 81:229-242. [DOI] [PubMed] [Google Scholar]
- 12.Dannenberg, A. M., Jr. 1998. Lurie's tubercle-count method to test TB vaccine efficacy in rabbits. Front. Biosci. 3:27-33. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, P. L., and R. J. North. 1995. Virulence ranking of some Mycobacterium tuberculosis and Mycobacterium bovis strains according to their ability to multiply in the lungs, induce lung pathology, and cause mortality in mice. Infect. Immun. 63:3428-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 17.Guyton, A. C. 1947. Measurement of the respiratory volumes of laboratory animals. Am. J. Physiol. 150:70-77. [DOI] [PubMed] [Google Scholar]
- 18.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 20.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lurie, M. B. 1928. The fate of human and bovine tubercle bacilli in various organs of the rabbit. J. Exp. Med. 48:155-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lurie, M. B. 1964. Resistance to tuberculosis. Harvard University Press, Cambridge, Mass.
- 24.Lurie, M. B., S. Abramson, and A. G. Heppleston. 1952. On the response of genetically resistant and susceptible rabbits to the quantitative inhalation of human-type tubercle bacilli and the nature of resistance to tuberculosis. J. Exp. Med. 95:119-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manabe, Y., C. Scott, and W. Bishai. 2002. Naturally attenuated, orally administered Mycobacterium microti is more effective than Mycobacterium bovis BCG as a tuberculosis vaccine. Infect. Immun. 70:1566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manca, C., L. Tsenova, C. E. Barry III, A. Bergtold, S. Freeman, P. A. Haslett, J. M. Musser, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740-6746. [PubMed] [Google Scholar]
- 27.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchison, D., L. Bhatia, S. Radhakrishna, J. Selkon, T. Subbaiah, and J. Wallace. 1961. The virulence in the guinea-pig of tubercle bacilli isolated before treatment from South Indian patients with pulmonary tuberculosis. Bull. W. H. O. 25:285-312. [PMC free article] [PubMed] [Google Scholar]
- 29.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 30.North, R. J., L. Ryan, R. LaCource, T. Mogues, and M. E. Goodrich. 1999. Growth rate of mycobacteria in mice as an unreliable indicator of mycobacterial virulence. Infect. Immun. 67:5483-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orme, I. M., A. D. Roberts, S. K. Furney, and P. S. Skinner. 1994. Animal and cell-culture models for the study of mycobacterial infections and treatment. Eur. J. Clin. Microbiol. Infect. Dis. 13:994-999. [DOI] [PubMed] [Google Scholar]
- 32.Pierce, C. H., R. J. Dubos, and W. B. Schaefer. 1953. Multiplication and survival of tubercle bacilli in the organs of mice. J. Exp. Med. 97:189-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratcliffe, H. L., and W. F. Wells. 1948. Tuberculosis of rabbits induced by droplet nuclei infection. J. Exp. Med. 87:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder, B. G., L. M. Peterson, and R. D. Fleischmann. 2002. Improved quantitation and reproducibility in Mycobacterium tuberculosis DNA microarrays. J. Mol. Microbiol. Biotechnol. 4:123-126. [PubMed] [Google Scholar]
- 35.Skeiky, Y. A., P. J. Ovendale, S. Jen, M. R. Alderson, D. C. Dillon, S. Smith, C. B. Wilson, I. M. Orme, S. G. Reed, and A. Campos-Neto. 2000. T cell expression cloning of a Mycobacterium tuberculosis gene encoding a protective antigen associated with the early control of infection. J. Immunol. 165:7140-7149. [DOI] [PubMed] [Google Scholar]
- 36.Smith, D. W., D. N. McMurray, E. H. Wiegeshaus, A. A. Grover, and G. E. Harding. 1970. Host-parasite relationships in experimental airborne tuberculosis. IV. Early events in the course of infection in vaccinated and nonvaccinated guinea pigs. Am. Rev. Respir. Dis. 102:937-949. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. W., and E. H. Wiegeshaus. 1989. What animal models can teach us about the pathogenesis of tuberculosis in humans. Rev. Infect. Dis. 11:S385-S393. [DOI] [PubMed] [Google Scholar]
- 38.Smith, T. 1898. A comparative study of bovine tubercle bacilli and of human bacilli from sputum. J. Exp. Med. 3:451-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltys, M. A., C. A. St. Hill, and I. Ansell. 1952. Pathogenicity of tubercle bacilli in experimental animals, p. 47-58. In Tubercle bacillus and laboratory methods in tuberculosis. E. & S. Livingstone Ltd., London, England.
- 40.Wiegeshaus, E. H., D. N. McMurray, A. A. Grover, G. E. Harding, and D. W. Smith. 1970. Host-parasite relationships in experimental airborne tuberculosis. 3. Relevance of microbial enumeration to acquired resistance in guinea pigs. Am. Rev. Respir. Dis. 102:422-429. [DOI] [PubMed] [Google Scholar]
- 41.Wilhelmsen, C. L., and M. L. Pitt. 1996. Lesions of acute inhaled lethal ricin intoxication in rhesus monkeys. Vet. Pathol. 33:296-302. [DOI] [PubMed] [Google Scholar]