Abstract
Pseudomonas aeruginosa is an important pathogen in immunocompromised patients and secretes a diverse set of virulence factors that aid colonization and influence host cell defenses. An important early step in the establishment of infection is the production of type III-secreted effectors translocated into host cells by the bacteria. We used cDNA microarrays to compare the transcriptomic response of lung epithelial cells to P. aeruginosa mutants defective in type IV pili, the type III secretion apparatus, or in the production of specific type III-secreted effectors. Of the 18,000 cDNA clones analyzed, 55 were induced or repressed after 4 h of infection and could be classified into four different expression patterns. These include (i) host genes that are induced or repressed in a type III secretion-independent manner (32 clones), (ii) host genes induced specifically by ExoU (20 clones), and (iii) host genes induced in an ExoU-independent but type III secretion dependent manner (3 clones). In particular, ExoU was essential for the expression of immediate-early response genes, including the transcription factor c-Fos. ExoU-dependent gene expression was mediated in part by early and transient activation of the AP1 transcription factor complex. In conclusion, the present study provides a detailed insight into the response of epithelial cells to infection and indicates the significant role played by the type III virulence mechanism in the initial host response.
Pseudomonas aeruginosa is one of the most virulent opportunistic pathogens in humans. In the setting of preexisting epithelial injury, particularly in immunocompromised patients, it leads to devastatingly acute infections, including pneumonia and sepsis (52). Up to 75% of patients in intensive care units are colonized with P. aeruginosa, and the mortality from pneumonia is 40%, even with antibiotic treatment (39).
P. aeruginosa boasts an impressive array of cell-associated and secreted virulence factors that contribute to its pathogenesis. Key among these is type IV pili, the major bacterial adhesin, and the type III secretion system with its secreted exotoxins. Type IV pili are polar fimbriae comprised of pilin, the product of the pilA gene (reviewed in reference 40). A unique feature of type IV pili is their ability to extend and retract (41). In addition to their roles in adhesion, type IV pili are also involved in biofilm formation (55) and twitching motility (63) and serve as bacteriophage receptors (49). They are required for full virulence in a mouse model of acute pneumonia (3, 11).
Upon host cell contact, the type III secretion system allows bacteria to directly inject toxins into the host cell, where they subvert host cell defense and signaling systems (24). Four type III-secreted effectors have been identified in P. aeruginosa, although few if any strains secrete all four of them (reviewed in reference 9). PA103, a human lung isolate, encodes and secretes two effectors, ExoU and ExoT. ExoU is a potent cytotoxin whose host cell targets and mechanism of action are not yet known (12, 13, 21). ExoT is a bifunctional protein, possessing an N-terminal GTPase-activating domain with GAP activity toward Rho, Rac, and Cdc42, and a C-terminal ADP-ribosyltransferase domain (16, 32, 35). The presence of a functional type III secretion system is associated with poor outcome in acute P. aeruginosa infections in humans (19, 51).
Evidence also suggests that type III effectors, in addition to their effects on host cellular enzymes, are likely to evoke rapid and specific changes in gene expression. Peripheral blood mononuclear cells respond to purified ExoS exposure by increasing the transcription of proinflammatory cytokines and chemokines (10, 34), and macrophages upregulate numerous genes in response to effectors of the homologous Yersinia enterocolitica type III secretion system (53). The lung epithelium, which represents an important barrier to infection and a primary target of type III-secreted effectors, responds to P. aeruginosa infection through the expression of many genes involved in host defense and immune cell activation (25). However, the role played by type III secretion in this response is not known. Here we use microarray analysis and well-characterized mutant strains of cytotoxic P. aeruginosa to investigate the response of lung epithelial cells to different type III effectors. More than 18,000 genetic elements were analyzed, and this resulted in the classification of expression patterns both dependent and independent of the type III toxin function. The type III-independent response consisted of genes involved in inflammatory responses and cytoprotective roles, whereas the majority of type III-dependent genes, induced through the action of either ExoU or other unidentified type III effectors, were transcriptional or cell signal regulators. We also show that the mechanism by which ExoU induced gene expression involves the early and transient activation of the AP1 transcription factor complex.
MATERIALS AND METHODS
Bacteria, cells, and infection protocol.
P. aeruginosa strain PA103, a cytotoxic lung isolate, and mutants derived from this strain that were used in the present study are described in Table 1. Two days prior to use, bacteria were inoculated from glycerol stocks onto Luria-Bertani (LB) agar plates and grown overnight at 37°C, and single colonies were subsequently incubated overnight in 2 ml of LB broth at 37°C without shaking. The 9HTEo− cells are an simian virus 40 T-antigen-transformed line derived from human tracheal epithelium and were kindly provided by D. Gruenert (University of Vermont). The cells were maintained in Dulbecco modified Eagle medium supplemented with glutamine (2 mM; Gibco), streptomycin (50 μg/ml; Gibco), penicillin (50 U/ml; Gibco), and Serum Supreme (10%; BioWhittaker). The cells were grown on plastic vessels precoated with a solution containing Vitrogen (0.2 mg/ml; Cohesion), NaOH (0.83 mM), and Ham F-12 medium (10%; Gibco). For infection studies, cells (between passages 18 to 22) were seeded at 150,000 cells/cm2, grown until completely confluent (usually 3 days), and then washed and incubated in serum- and antibiotic-free medium for a further 20 h. Freshly grown bacteria in LB broth were diluted into the serum- and antibiotic-free medium and added to the cells at a multiplicity of ca. 10 bacteria/cell. The amount of bacterial inoculum was enumerated by serial dilution and plating on LB agar plates.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristic(s)a | Reference |
|---|---|---|
| PA103 | Virulent lung isolate of P. aeruginosa; known type III secreted effector proteins are ExoT and ExoU | 1,38 |
| PA103 exoT | PA103 with an xylE aacC1 cassette replacing aa 36 to 348 of exoT; Gmr | 16 |
| PA103 exoU | PA103 with an in-frame deletion of aa 330 to 571 of exoU | 16 |
| PA103 exoT exoU | PA103 exoU with an xylE aacC1 cassette replacing aa 36 to 348 of exoT; Gmr | 16 |
| PA103 pscJ::Tn5 | Tn5 Gmr cassette inserted into pscJ; defective in type III secretion | 29 |
| PA103 pilA | PA103 with StuI/Bsu36I region of pilA replaced with a Gmr cassette | 4 |
aa, amino acids; Gmr, gentamycin resistance.
Care was taken to use completely confluent monolayers of 9HTEo− cells in the infection studies since they become polarized, form tight junctions (18), and resist the cytotoxic effects of PA103 up to 5 h after infection (B. McMorran, unpublished observations).
Microarray hybridization and analysis.
Total RNA was prepared from infected and uninfected (control) cells by using the RNeasy RNA isolation kit (Qiagen). RNA samples (40 μg) were labeled with Cy5 or Cy3 dUTP, purified, and subjected to microarray hybridization as described previously (26). Equal amounts of control (noninfected, Cy3-labeled) and infected (Cy5-labeled) sample were hybridized to microarray chips (human 19K2.2; Ontario Cancer Institute) overnight at 45°C in humidified chambers. Slides were washed for 3 min in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.05% sodium dodecyl sulfate (SDS) and then twice for 3 min each time in 0.2× SSC prior to obtaining fluorescence images with a Genetic MicroSystems G418 scanner.
Spot intensities were quantified by using Imagene software (Biodiscovery). The proportion of “passed” spots (i.e., with detectable intensity) on each array was 60 to 70%, and no intrachip or spatial variations in hybridization signals were observed. Quantified data were normalized and analyzed by using the Genespring package (Silicon Genetics). For normalization the 40th percentile of all measurements was used as a positive control for each sample; each measurement for each gene was divided by this synthetic positive control, assuming that this was at least 0.01. The bottom tenth percentile was used as a test for correct background subtraction. To calculate the presented ratios, the measured intensity of each gene was divided by its control channel value. When the control channel intensity was <200, the datum point was considered unreliable and omitted. Raw data are viewable at http://microarray.imb.uq.edu.au/base/www/index.phtml (login: guest-wainwright; password: XuFEG1D2).
Clones corresponding to spots of interest were obtained from the Ontario Cancer Institute. Plasmids from pure cultures were sequenced with M13 primers (22), and identity was established by using the basic local alignment search tool (BLAST) and the NCBI GenBank database. Plasmids were digested to liberate insert without poly(A)+ tail, where possible, and separated on an agarose gel with the QiaQuick gel extraction kit (Qiagen) to be used as Northern probes.
Northern blotting.
Total RNA (15 μg) was separated on a 1% agarose-formaldehyde gel and transferred to Magna nylon membrane (Osmonics). Prehybridization (3 h) and hybridization (overnight) were performed in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt solution, 0.5% SDS, 1 μg of heat-denatured sheared salmon sperm DNA/ml (Sigma), and 50% formamide at 42°C. Probes were labeled by using [α-32P]CTP and a random priming (Megaprime; Amersham Pharmacia) and then purified on G-50 Sephadex. Hybridized blots were washed at high stringency (0.1× SSC-0.1% SDS at 42°C). Bands were detected and quantified by phosphorimaging with phosphor screens (Kodak) and a Storm scanner and ImageQuant software (both from Molecular Dynamics). A 600-bp fragment of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA was used a loading control in the quantification calculations.
Immunofluorescence staining.
Cells for staining studies were grown on Vitrogen-coated glass coverslips and, after infection, were washed in phosphate-buffered saline (with three 2-min washes) and fixed in cold methanol (30 min). Staining was performed on prepermeabilized cells (0.1% Triton X-100-phosphate-buffered saline for 10 s) in a solution containing 0.5% bovine serum albumin (Sigma). The primary antibodies and dilutions used were anti-cFos at 1/100 (clone 4-G, goat polyclonal immunoglobulin G [IgG]; Santa Cruz Biochemicals) and anti-P. aeruginosa at 1/400 (purified rabbit sera [a gift from C. Whitchurch, University of California at San Francisco]). Secondary antibodies, Cy3-labeled donkey anti-goat IgG, and Alexa Fluor 488-labeled chicken anti-rabbit IgG (Molecular Probes) were used at a 1/400 dilution. Fluorescence staining was visualized on an Olympus AX-70 microscope with a ×60 oil immersion objective lens. Images were captured from a CCD300-RC charge-coupled device camera (Dage-MTI) by using NIH Image 1.62 software and were merged and processed by using Adobe Photoshop 5.0.2.
Electrophoretic mobility shift assay (EMSA).
Preparation of the nuclear extracts, probe preparation and binding reactions were performed essentially as described previously (56). Extracts were prepared from ca. 1.2 × 107 9HTEo− cells. An AP1-specific double-stranded oligonucleotide (CGATTGACTCAGTACTGAGTCAATCG, with the consensus AP1-binding sites underlined) and a nonspecific oligonucleotide of similar size were used in the binding reactions. For the antibody inhibition experiments, 3 μg of anti-c-Fos or anti-β-galactosidase (goat polyclonal IgG; Cortex Biochem) antibody was included in the binding reaction. Protein-DNA complexes were separated on 6% nondenaturing polyacrylamide gels (acrylamide-bisacrylamide [29:1]) run at 100 V in 0.5× Tris-borate-EDTA buffer. The gels were dried and visualized by autoradiography.
RESULTS
Microarray analysis identifies genes regulated in 9HTEo− cells by P. aeruginosa infection.
To assess and compare the contributions of type III secretion to the transcriptional host cell response, exoT, exoU, exoT exoU, pscJ, and pilA isogenic mutants and the parental wild-type PA103 strains of P. aeruginosa were used (Table 1). The former three strains harbor deletions in the ExoU and/or ExoT genes and do not secrete the respective toxins, although they possess a functional type III secretion apparatus. The pscJ mutant harbors a transposon insertion in the pscJ gene and has a nonfunctional type III secretion apparatus (29). The pilA strain contains an antibiotic resistance cassette in the pilin gene and lacks type IV pili; it is also defective for type III-dependent effects (29; Jakobsen and J. Engel, unpublished results). Cells were infected for 4 h and subsequently subjected to microarray analysis. cDNA was prepared from cells infected with each strain (Cy5 labeled), pooled with cDNA from uninfected cells as a common reference (Cy3 labeled), and hybridized to microarrays containing over 18,000 cDNA clones (with ca. 25% redundancy) (Ontario Cancer Institute, Toronto, Ontario, Canada). The microarrays contained duplicate spots of each cDNA clone, providing two measurements of relative gene expression. In addition, duplicate hybridizations were performed by using independently grown and infected cell material (biological replicates) to allow for technical and biological variation between hybridizations. Raw duplicate data sets were merged into one experiment by using Genespring software (Silicon Genetics) and subjected to normalization and expression analysis as described in Materials and Methods.
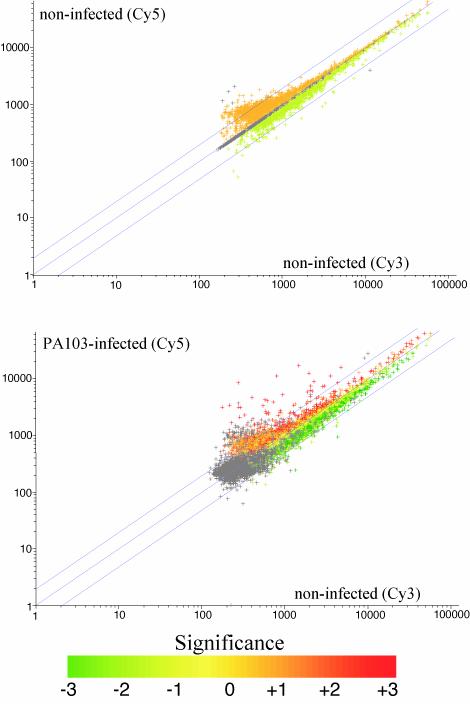
We first sought to identify genes that were regulated by infection by comparing expression levels of PA103 wild-type infected and noninfected cells. Representative scatter plots for noninfected/noninfected and parental/noninfected hybridizations are illustrated in Fig. 1. Extremely low variation is evident in the first plot, indicating that the Cy3 and Cy5 incorporation and hybridization efficiencies were similar. In the second plot, the majority of genes lie within a twofold ratio range and were considered unregulated by infection. Datum points located outside the twofold boundaries were taken to be significantly different from control expression and chosen for further analysis.
FIG. 1.
Scatterplot profiles of 9HTEo− cell genes after microarray hybridization and normalization. Each plot represents the Cy3 versus Cy5 expression intensity ratios of 9,243 genes derived from microarray hybridization experiments comparing noninfected and noninfected cells (top) or comparing noninfected and PA103-infected cells (bottom). Genes which lie outside the upper and lower twofold boundaries (blue lines) are significantly up- or downregulated in infected versus noninfected cells. The significance of these expression changes are indicated by graded coloring from one, two, or three standard deviations difference between infected and noninfected intensities. Gray color refers to genes lacking duplicate datum points.
A total of 9,243 cDNA elements were used in the analysis, and 55 clones, representing ca. 46 unique genes, were significantly and reproducibly up- or downregulated by P. aeruginosa infection compared to noninfected cells (Table 2). These genes represent the total transcriptional response to infection measured by the arrays. Each of the clones was independently sequenced to verify its identity on the array. Although this is a relatively small proportion of the total number analyzed, an initial examination indicated several upregulated genes previously shown as inducible by P. aeruginosa infection. These genes included monocyte chemotactic protein (MCP1/SCYA2), tristetraproline (TTP), ras homolog RhoB, viral oncogene homolog c-fos and urokinase type plasminogen activator receptor (PLAUR) genes (25). Raw expression intensity values were also very similar between the different samples, indicating that the changes were due to infection and not to variation in the reference signal. Northern blots performed on RNA from additional infection experiments were used to check the reproducibility of the array data and were in close agreement with expression patterns (see Fig. 3).
TABLE 2.
Relative expression ratios of 9HTEo− cell genes regulated by P. aeruginosa infection
| Class and gene designation | Full gene name | Accession no.a | Function | Relative expression ratio of P. aeruginosa strain:
|
Pb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PA103 | exoT | exoU | exoTU | pscJ | pilA | |||||
| Class A: type III independently activated genes | ||||||||||
| VEGFA | Vascular endothelial growth factor A | n91060 | Proinflammatory | 8.04 | 4.98 | 5.65 | 5.72 | 5.69 | 6.17 | 0.001 |
| VEGFA | Vascular endothelial growth factor A | aa126903 | Proinflammatory | 4.9 | 4.46 | 2.76 | 2.65 | 3.86 | 3.37 | 0.001 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | n24118 | Proinflammatory | 2.58 | 2.02 | 2.71 | 2.58 | 3.65 | 2.91 | 0.1 |
| ADM | Adrenomedullin | w19284 | Proinflammatory, antimicrobial | 6.43 | 2.15 | 9.62 | 4.97 | 5.76 | 5.43 | 0.1 |
| FSTL3 | Follistatin-like 3 | n75101 | Proinflammatory | 2.13 | 1.78 | 1.63 | 2.4 | 2.18 | 1.59 | 0.04 |
| IRS2 | Insulin receptor substrate-2 | aa035325 | Signal regulator | 3.41 | 1.47 | 2 | 2.31 | 2.57 | 2.05 | 0.01 |
| ANKRD3, DIK | Dual-specificity Ser/Thr/Tyr kinase 3 (dik gene) | w37332 | Signal regulator | 3.71 | 2.63 | 4.66 | 3.49 | 4.41 | 3.59 | 0.06 |
| STC1S | Stanniocalcin 1 | r48681 | Autocrine signal | 20.37 | 4.75 | 11.75 | 11.22 | 13.66 | 6.23 | 0.08 |
| SRA1 | Steroid receptor RNA activator | r50254 | Transcription regulator | 7.34 | 4.34 | 6.49 | 6.57 | 8.44 | 9.68 | 0.08 |
| NFIL3 | Nuclear factor regulated by IL-3 | w67706 | Transcription activator | 5.59 | 4.69 | 4.85 | 4.74 | 7.56 | 3.99 | 0.03 |
| MT1E | Metallothionein 1E | h93127 | Oxidant protection | 4.64 | 1.61 | 4.05 | 2.06 | 4.68 | 3.42 | 0.06 |
| MT1E | Metallothionein 1E | h52525 | Oxidant protection | 3.47 | 1.43 | 3.87 | 2.49 | 4.4 | 4.06 | 0.17 |
| MT1G | Metallothionein 1G | w78010 | Oxidant protection | 4.3 | 1.37 | 3.08 | 3.25 | 4.1 | 3.71 | 0.15 |
| MT1G | Metallothionein 1G | h57208 | Oxidant protection | 2.9 | 1.55 | 2.7 | 2.52 | 4.22 | 4.13 | 0.2 |
| MT2A | Metallothionein 2A | h91612 | Oxidant protection | 4.22 | 1.54 | 2.95 | 2.91 | 3.81 | 4.08 | 0.2 |
| Oatprp1 | Organic anion transporter member 12 | h84604 | Membrane transport | 3.05 | 2.33 | 1.85 | 5.51 | 2.7 | 3.12 | 0.2 |
| SLC2A1 | Solute carrier protein 2 | w58375 | Membrane transport | 2.96 | 3.66 | 2.35 | 3.18 | 2.93 | 2.66 | 0.02 |
| ADFP | Adipose differentiation-related protein (adipophilin) | w20444 | Membrane protein, inducible | 3.42 | 2.51 | 1.94 | 1.69 | 2.87 | 2.01 | 0.2 |
| U2AF65 | U2 small nuclear ribonucleoprotein auxiliary factor | aa053859 | mRNA processing | 2.73 | 1.64 | 1.94 | 1.89 | 1.42 | 1.45 | 0.01 |
| RNA pol I (16 kDa) | RNA polymerase I 16-kDa subunit | n71041 | mRNA processing | 2.28 | 1.72 | 1.58 | 1.83 | 2.01 | 1.63 | 0.2 |
| PCR17 | Rab GTPase-activating protein | aa047487 | Signal regulator | 2.25 | 0.94 | 2.07 | 2.53 | 1.91 | 1.7 | 0.2 |
| SYN2 | Syndecan 2 | aa156696 | Wound repair | 3.1 | 2.43 | 3.83 | 3.37 | 3.68 | 2.6 | 0.1 |
| EHD4 | EH domain containing 4 | n94535 | 2.28 | 1.48 | 2.52 | 1.29 | 2.15 | 2.53 | 0.2 | |
| EST | Chromosome 14q24.3, BAC 201F1 | h91088 | 3.52 | 3.34 | 3.21 | 2.9 | 3.09 | 2.89 | 0.01 | |
| EST | Hypothetical protein FLJ21616 | r98518 | 3.03 | 1.33 | 2.54 | 2.33 | 3.33 | 3.68 | 0.07 | |
| EST | No homology | n68160 | 3.01 | 2.68 | 2.69 | 3.33 | 1.94 | 1.96 | 0.17 | |
| EST | Hypothetical protein DKFZp566J091 | aa040473 | 2.91 | 1.9 | 2.09 | 2.15 | 2.19 | 2.26 | 0.02 | |
| Class B type III independently repressed genes | ||||||||||
| DUSP6 | Dual-specificity phosphatase 6 | t65557 | Signal regulator | 0.65 | 0.39 | 0.46 | 0.59 | 0.56 | 0.54 | 0.17 |
| TRXIP | Thioredoxin-interacting protein | n71361 | Oxidant protection | 0.24 | 0.26 | 0.15 | 0.13 | 0.15 | 0.15 | 0.001 |
| EST | KIAA1515 protein | r40307 | 0.24 | 0.15 | 0.079 | 0.073 | 0.13 | 0.08 | 0.1 | |
| EST | Chromosome 11q, clone:RP11-687M24 | h09816 | 0.35 | 0.7 | 0.49 | 0.66 | 0.53 | 0.64 | 0.003 | |
| EST | Hypothetical protein (HSPC210) | h72619 | 0.43 | 0.06 | 0.43 | 0.12 | 0.17 | 0.16 | 0.1 | |
| Class C: exoU-dependent activated genes | ||||||||||
| DUSP1 | Dual-specificity phosphatase 1 | r79387 | Signal regulator | 50.19 | 13.76 | 3.97 | 3.15 | 2.16 | 2.49 | 0.09 |
| DUSP1 | Dual-specificity phosphatase 1 | h87493 | Signal regulator | 11.43 | 11.52 | 3.86 | 6.25 | 2.18 | 2.61 | 0.005 |
| DUSP1 | Dual-specificity phosphatase 1 | h29136 | Signal regulator | 15.85 | 5.48 | 4.22 | 3.85 | 1.91 | 2.12 | 0.001 |
| DUSP1 | Dual-specificity phosphatase 1 | h01773 | Signal regulator | 14.36 | 8.64 | 4.45 | 6 | 2.02 | 2.89 | 0.03 |
| KNSL5 | Kinesin-like 5 | aa043507 | Signal regulator | 7.07 | 6.34 | 1.98 | 2.05 | 3.54 | 2.32 | 0.003 |
| RhoB | Transforming protein RhoB (ras homolog) | w67471 | Signal regulator | 7.3 | 0.668 | 2.85 | 2.06 | 2.46 | 1.41 | 0.1/PICK> |
| RhoB | Transforming protein RhoB (ras homolog) | r32081 | Signal regulator | 4.54 | 4.82 | 2.01 | 2.78 | 2.32 | 2.22 | 0.07 |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | aa019816 | Transcription activator | 20.8 | 3.1 | 2.72 | 4.01 | 4.3 | 1.62 | 0.1 |
| MYC | v-myc proto-oncogene protein | w87861 | Transcription activator | 10.37 | 5.66 | 3.42 | 2.44 | 5.95 | 3.69 | 0.1 |
| MYC | v-myc proto-oncogene protein | h43827 | Transcription activator | 7.73 | 10 | 2.99 | 3.28 | 2.63 | 3.26 | 0.02 |
| NR4A1/nur77 | Nuclear receptor subfamily 4, grpA, murine nur77 homolog | n64388 | Transcription activator | 5.37 | 1.6 | 1.26 | 2.02 | 1.56 | 1.04 | 0.2 |
| BPTF | Bromodomain transcription factor | h02279 | Transcription activator | 4.04 | 3.01 | 1.96 | 2.95 | 1.5 | 1.68 | 0.03 |
| TTP | Tristetraproline | aa054080 | Transcription activator mRNA stability | 3.7 | 1.51 | 2.02 | 2.78 | 1.18 | 1.6 | 0.1 |
| TCF8/ | Transcription factor 8 | aa150750 | Transcription repressor | 16.46 | 5.23 | 3 | 2.99 | 1.78 | 1.49 | 0.02 |
| TCF8 | Transcription factor 8 | r43502 | Transcription repressor | 4.44 | 3.17 | 2.07 | 2.43 | 1.34 | 1.61 | 0.03 |
| PLAUR | Urokinase type plasminogen activator receptor | t75241 | Protease regulator | 4.85 | 1.4 | 0.69 | 0.9 | 0.51 | 0.61 | 0.1 |
| EST | Chromosome 14, BAC R-156E22 | r85513 | 20.4 | 20.8 | 2.79 | 2.9 | 2.16 | 1.62 | 0.004 | |
| EST | Similar to hypothetical protein FLJ11328 | aa044730 | 6.3 | 6.98 | 2.22 | 2.46 | 2.9 | 2.41 | 0.1 | |
| EST | No homology | h87673 | 4.97 | 3.65 | 1.55 | 1.71 | 1.14 | 1.74 | 0.09 | |
| EST | Hypothetical protein FLJ22182 | aa156747 | 4.47 | 5.49 | 2.02 | 1.87 | 2.2 | 2 | 0.05 | |
| Class D: type III-dependent activated genes | ||||||||||
| MCP1/SCYA2 | Monocyte chemotactic protein 1 | aa024753 | Proinflammatory | 1.93 | 2.66 | 3.63 | 3.78 | 0.72 | 1.31 | 0.1 |
| MCP1/SCYA2 | Monocyte chemotactic protein 1 | r75975 | Proinflammatory | 1.12 | 2.32 | 2.44 | 2.88 | 0.7 | 0.82 | 0.1 |
| TIEG | TGF-early inducible gene | aa045730 | Transcription activator | 2.62 | 2.21 | 1.87 | 1.97 | 1.35 | 1.21 | 0.002 |
GenBank accession number.
As determined by Student t test analysis.
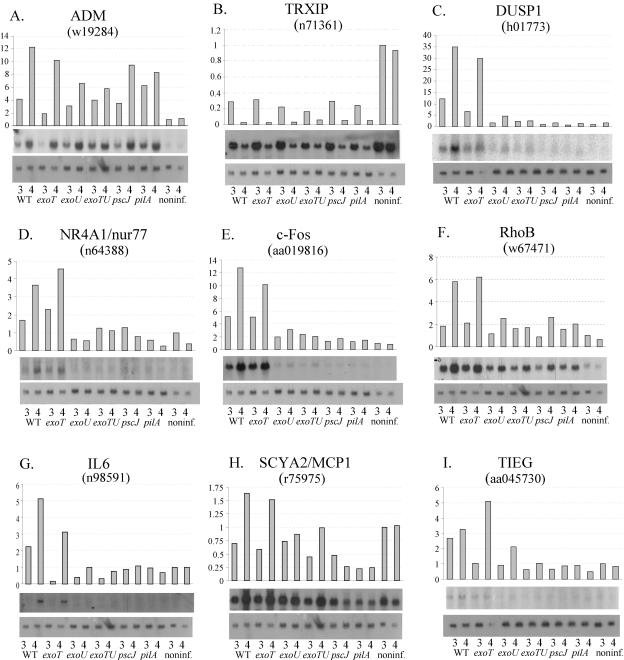
FIG. 3.
Northern blot analysis of P. aeruginosa PA103 induced genes. Each panel depicts the quantified expression level relative to GAPDH expression (top), Northern blot autoradiograms of the specific gene (center), and the GAPDH loading control (bottom). Cells were infected with each indicated strain for 3 and 4 h. ADM and TRXIP are examples of class A and B genes, respectively. DUSP1, NR4A1, Fos, RhoB, and IL-6 are examples of class C genes, and SCYA2 and TIEG are examples of class D genes. GenBank accession numbers of clones from which the Northern probes were constructed are also noted.
Identification of genes regulated dependently and independently by type III-secreted toxins.
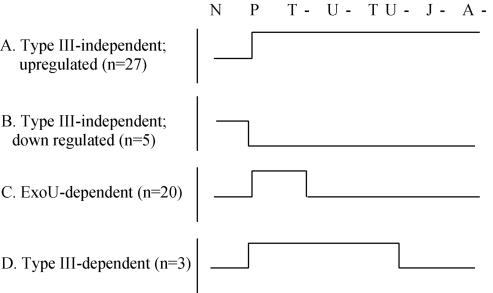
Cluster analyses allowed the infection-regulated genes to be categorized into one of four expression pattern classes (Fig. 2): (i) upregulated by infection with all six strains (class A); (ii) downregulated by all six strains (class B); (iii) upregulated only by the parental and exoT mutant strains (class C); and (iv) upregulated only by the parental and exoT, exoU, and exoT exoU mutant strains (class D). Since the first two classes of genes were regulated equivalently by all of the strains including parental, we termed this group type III-independent genes (Table 2). Class C genes were only induced by strains able to secrete ExoU, so this group was termed ExoU dependent (Table 2). Class D genes were induced by mutants with specific deletions in ExoU and ExoT, as well as the parental strain, but were not regulated by the mutants completely defective in type III secretion. Therefore, this group was termed type III secretion dependent (Table 2).
FIG. 2.
Expression patterns of 9HTEo− cell genes regulated after P. aeruginosa infection. A total of 55 genes that showed >2-fold difference between noninfected cells (N) and cells infected by P. aeruginosa PA103 parental (P), exoT (T-), exoU (U-). exoT exoT (TU-), pscJ (J-), and pilA (A-) were clustered into four different expression profile classes (A to D). The number of genes in each group is indicated in parentheses.
Approximately 23 unique genes (represented by 27 clones) were induced (class A) and 5 were repressed (class B) in a type III secretion-independent manner. The products of these genes were classified according to their major cellular functions. The majority were involved in either the control of immune and host defense mechanisms (n = 5) or oxidant protection (n = 4). Molecules with roles in transcriptional regulation (n = 2), cell signaling (n = 4), and solute transportation (n = 2) were also represented, along with five uncharacterized expressed sequence tags (ESTs). Northern blots on a selection of these genes confirmed that these expression pattern classes were reproducible. Adrenomedulin (ADM) was strongly induced as early as 3 h after infection and to equivalent levels by all strains (Fig. 3A), a finding consistent with the array results in Table 2. Genes downregulated by infection were also confirmed in Northern blotting experiments. As an example, expression of thioredoxin-interacting protein (TRXIP) was very high in untreated cells but strongly repressed between 3 and 4 h after infection (Fig. 3B).
Approximately 14 genes (represented by 20 cDNA clones) fell into the class C (ExoU-dependent expression pattern; Table 2). Genes in this group were consistently expressed at least twofold higher in the ExoU-sufficient parental and exoT strains compared to the ExoU-deficient mutants (i.e., the exoU, exoT exoU, pscJ, and pilA mutants). In many cases, induction by the latter four strains was completely absent. The vast majority of characterized ExoU-dependent genes are involved in transcriptional and cell signal regulation (n = 9). A further four clones represent unidentified ESTs. ExoU-dependent regulation of several of these genes was confirmed by Northern blotting. Transcripts encoding dual-specificity phosphatase 1-mitogen-activated protein (MAP) kinase phosphatase 1 (DUSP1/MKP1), nuclear orphan receptor 4A1 (NR4A1/nur77), and c-fos were all induced within 3 h of infection only by wild-type and exoT mutant strains (Table 2 and Fig. 3C to E). The expression pattern of RhoB suggested partial dependence on ExoU since all six strains induced the gene, but in the wild-type bacteria and ExoT mutant, induction was at least three times higher (Fig. 3F). We also found that interleukin-6 (IL-6), a well-characterized inflammatory cytokine not represented on the array, was regulated in an ExoU-dependent manner (Fig. 3G). No ExoU-dependent genes were downregulated by infection.
The type III-dependent group (class D) contained two unique genes, MCP1/SCYA2 and transforming growth factor β (TGF-β) early inducible growth factor (TIEG), which encode known host inflammatory response proteins. Northern blotting analysis of these two genes confirmed that they were regulated in the pattern inferred from the array analysis (Table 2 and Fig. 3H and I).
ExoU-dependent induction of AP1 transcription factor activity.
A number of ExoU-dependent genes identified on the arrays are classified as immediate-early (IE) inducible genes (including c-Fos, c-Myc, DUSP1/MKP1, and RhoB), meaning that their regulation is subject to rapid changes in intracellular signaling and transcriptional activity rather than requiring de novo protein synthesis for their expression. Few of the type III-independent genes belong to this class, suggesting that ExoU was targeting a rapid response cell signaling pathway(s) during infection. Oncogene c-fos is one of the best-characterized IE genes. It is activated by a variety of stress-inducing stimuli and regulated via many signaling mechanisms including MAP kinase cascades. The c-Fos protein forms heterodimeric complexes with members of Jun protein family called AP1, which functions as a transcriptional activator of genes involved in cell protection and survival (65). Interestingly, other ExoU-dependent genes identified in the arrays were coregulated with c-fos in response to other stimuli, including c-myc (58) and DUSP1/MKP1 (62). In addition, NR4A1/nur77, SCYA2/MCP1, IL-6, and PLAUR are known to be activated by AP1 (7, 44, 46, 57). Therefore, c-Fos and AP1 may represent key mediators of the ExoU-stimulated response, and we decided to further characterize the mechanism of induction.
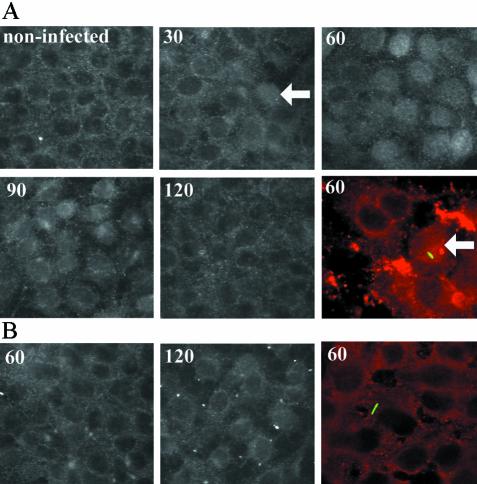
At the level of protein production, we observed no de novo increases in c-Fos protein during infection (data not shown). However, immunofluorescence studies revealed changes in the cellular location of c-Fos. The protein resides in the cytoplasm of quiescent cells but, upon stimulation, translocates to the nucleus, where it forms transcription factor complexes (50). In uninfected cells, significant levels of endogenous protein were localized to the cytoplasm (Fig. 4). Exposure to P. aeruginosa resulted in a rapid translocation of c-Fos to the nucleus (within 30 to 60 min), followed by the return to a cytoplasmic location by 120 min. This phenomenon was ExoU dependent since no nuclear translocation was observed in cells exposed to the ExoU-deficient mutant. No further changes in protein location were observed after extended lengths of infection (up to 4 h) with either parental or mutant strains (data not shown). Using an anti-P. aeruginosa antibody, bacilli were observed interacting with cells at similar, but relatively low frequencies for both strains (estimated at 5 to 10% cells bound by bacteria), a finding consistent with observations in other polarized epithelial cell infection models (36). Although both bound and unbound cells contained nuclear c-Fos after exposure to the wild-type strain, no cells treated with the mutant strain (bound or unbound) were activated (Fig. 4), further suggesting that translocation required the function of ExoU.
FIG. 4.
Immunolocalization of c-Fos expression. Black and white panels represent images of cells immunostained with anti-c-Fos antibody after exposure to P. aeruginosa strains PA103 (A) or PA103 exoU (B) for different lengths of time as indicated (in minutes). Color panels represent images of cells doubly stained with anti-c-Fos antibody (red channel) and anti-P. aeruginosa antibody (green channel). Examples of nuclear localized c-Fos are indicated with arrows. Magnification, ×600.
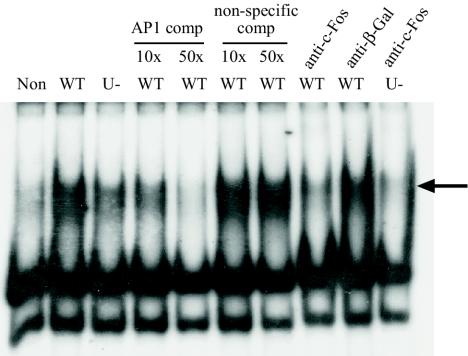
The c-Fos protein functions as a DNA-binding transcription factor in a heterodimeric complex called AP1, the activity of which can be detected by using labeled oligonucleotides containing the AP1 recognition site in an EMSA. Active AP1 complex was not present in the nuclei of uninfected cells but was detected within 40 min of infection with the parental PA103 strain (Fig. 5). This coincided with the time at which c-Fos protein relocates from the cytoplasm to the nucleus. No AP1 activity was detected in cytoplasmic extracts (data not shown), and the AP1 complex contained c-Fos protein since anti-c-Fos antibody specifically abolished the AP1 band and caused a weak supershifted band (visible on extended exposure). AP1 activity was also detected after infection by ExoU-deficient bacteria. However, consistent with the other regulatory studies the band intensity was significantly less than wild-type levels, indicating the requirement of AP1 in the regulation of ExoU-dependent gene expression.
FIG. 5.
AP1 EMSA. Nuclear extracts from noninfected cells (Non) and cells infected for 40 min with PA103 (WT) or PA103 exoU (U-) were incubated with radiolabeled AP1 oligonucleotide. An intense shifted band (arrow) indicates the presence of active DNA-binding AP1 complex. Competition experiments with 10- and 50-fold excess unlabeled (AP1 comp) AP1 oligonucleotide abolished the band, whereas nonspecific oligonucleotide (non-specific comp) did not, indicating the specificity of the DNA-protein complex. Inclusion of anti-c-Fos antibody, but not anti-β-galactosidase (anti-β-Gal) antibody, inhibited band formation, indicating that c-Fos protein was present in the active complex.
DISCUSSION
Type III secretion allows for rapid contact and communication between bacteria and host cells. For many gram-negative species, the role of secreted type III effectors is to evade host defense mechanisms. In the case of P. aeruginosa, the effectors characterized to date appear to disable host cell actin cytoskeleton and cell-cell junctions, inhibit phagocytosis, and cause cell necrosis, thereby (presumably) disrupting the epithelial cell barrier and preventing an effective immune response. While much is understood about the functions of these effectors, little is known about the host cell response they may evoke. We sought to address this through the use of microarray studies that allowed the simultaneous measure of expression of thousands of different genes in conjunction with a controlled in vitro infection system and well-defined strains of P. aeruginosa. Our studies indicate the important role of type III secretion.
We detected a modest number of genes regulated in 9HTEo− cells by infection with P. aeruginosa. By comparison, macrophages may alter the expression of 10 to 20% of the total transcriptome in response to defined pathogen components such as lipopolysaccharide, and the response is idiosyncratic to particular pathogens (2). Other microarray studies have observed similarly small changes in gene expression in cultured lung epithelial cells infected with a different strain of P. aeruginosa (25) or in cytokine-stimulated lung epithelial cells (6). A comparison between our data and these earlier studies reveal similarly regulated genes, indicating that such microarray strategies are reproducible and comparable to findings of other labs. Using an alternative lung epithelial cell line (A549) and a different P. aeruginosa strain (PAK), which lacks flagella and secretes ExoS and ExoT but lacks ExoU, Ichikawa et al. (25) identified 22 genes that were upregulated after 3 h of P. aeruginosa exposure. Of the 15 represented on the microarrays used here, 5 were similarly induced. Other studies have addressed the response of epithelial cells to inflammatory stimuli such as tumor necrosis factor alpha and IL-1β exposure, and several genes are shared by these cells, including follistatin, stanniocalcin, TTP, MCP1/SCYA2 and vascular endothelial growth factor (VEGF) (6). This suggests the activation mechanism during P. aeruginosa exposure may involve the same signaling pathways as cytokines.
A major finding from the array study was the distinction between epithelial cell genes responsive to type III effectors and those regulated (presumably) by other bacterial factors. The latter set of genes fell into two expression classes: upregulated (class A) or downregulated (class B) by infection. It is interesting that there were no differences in the host cell transcriptional response to the type III secretion-defective mutant and the nonpiliated mutant. This observation underscores three important points. First, the lack of host cell transcriptional response to the pilA mutant is not simply a consequence of its decreased binding compared to the pscJ mutant. Second, this finding suggests that the type IV pilus does not itself modulate the host cell transcriptional response, although it may affect host cell signal transduction. Finally, while some of the mutant strains tested are more efficiently internalized than others (i.e., the exoU exoT and pscJ mutants), no transcriptosome pattern specific to bacterial internalization could be identified.
Many of the genes responsive to factors other than the type III-secreted effectors are clearly candidates for roles in the early phase of inflammation induced by the organism. ADM is a peptide hormone that acts at sites of trauma to dilate the endothelium and permit the passage of inflammatory cells. It also possesses antimicrobial activity and has been identified in epithelial cells in response to other infectious stimuli (30, 60). VEGF has a range of biological activities, including endothelial cell migration, proliferation, and increasing vascular permeability (5, 17), and its production has recently been associated with epithelial infections (42, 45). Many other genes products upregulated by infection have roles in the regulation of infection/inflammation/stress response pathways. Follistatins regulate cytokine production in the acute phase response (8), RhoB is a negative regulator of NF-κB signaling (15), and insulin receptor substrate 2 (IRS2) mediates signaling from cytokine receptors (61). All of the known downregulated genes also have roles that fit with regulation of cellular response to infection. DUSP6/MKP3 is a specific inhibitor of the MAP kinase and extracellular regulated kinase (66), and MAP kinases are activated by P. aeruginosa infection (48). Particularly interesting was the downregulation of TRXIP. This molecule mediates the cell response to oxidative stress by regulating the expression and activity of thioredoxin (28). The fact that three metallothionein genes were also upregulated on the arrays suggests that redox maintenance and regulation in epithelial cells is an important process in the host response to infection, perhaps protecting the cell from the toxic free radicals produced by P. aeruginosa.
Approximately 50% of the genes regulated by P. aeruginosa infection were not affected by the type III secretion mutants. This suggests that the secreted effectors influence gene activation pathways that are distinct from other virulence factors. The largest group of such genes were induced only by strains producing ExoU. ExoU-deficient mutants are known to exert reduced cytotoxicity on cells (13, 21), but studies to date have so far failed to determine the exact mechanism of ExoU function (12). Our studies indicate that ExoU activity may result in AP1 transcription factor activation. Nuclear translocation of c-Fos and concomitant AP1 activation occurred rapidly (within 30 to 40 min) after exposure to the bacteria and required ExoU. Whereas c-Fos translocation occurred only in monolayers exposed to ExoU-sufficient strains, P. aeruginosa-bound cells, as well as unbound cells, were stimulated. Either binding is relatively weak at this time point, or biological activation by ExoU may involve cell-cell communication mechanisms.
A number of ExoU-dependent genes identified by array and expression studies are known to be AP1 responsive, including NR4A1/nur77, SCYA2/MCP1, and IL-6 (44, 46, 57). What are the possible signaling pathways that may link ExoU and AP1? Recent studies have shown that P. aeruginosa activates Ca2+-dependent MAP kinase signaling in a pilus-dependent manner (48). Several lines of evidence from our studies suggest that MAP kinase signaling is also involved in mediating ExoU-dependent expression. Some of the genes are MAP kinase responsive in other systems, including MCP1/SCYA2 (64), PLAUR (34), and c-fos and c-myc (59). DUSP1/MKP1 is also upregulated during MAP kinase activation and plays a negative feedback role by directly inactivating multiple MAP kinases (33). AP1 activity is regulated in part by phosphorylation by the MAP kinase c-Jun N-terminal kinase (59). Therefore, ExoU may target components of a MAP kinase cascade that stimulate pathway activation or relieve its inhibition. Despite a lack of obvious cytotoxicity in the infected 9HTEo− cells during the infection time frame, the expression of several of the ExoU-dependent genes was consistent with an apoptotic and/or necrotic phenotype. NR4A1/nur77 has a proapoptotic role in T-cell development (43) and is upregulated after cytokine-induced growth arrest in melanoma cells (47). AP1 activity is involved in several apoptotic mechanisms (31). However, other ExoU-dependent gene products, such as DUSP1/MKP1 (14) and IRS2 (54, 67), have roles in protecting cells from apoptosis. Extended infection with P. aeruginosa (i.e., >24 h) does lead to cell death in 9HTEo− cells (E. O. Costelloe, unpublished data), and it is possible that these genes are induced in an attempt to delay the onset of apoptosis or necrosis.
The discovery of genes upregulated in an ExoU- and ExoT-independent, but type III-dependent manner (class D) suggests there are other secreted effectors in strain PA103 that can influence gene expression or that the type III secretion apparatus itself can activate host cell responses. Other studies have also raised this possibility (20). For example, the exoU exoT double mutant induces apoptosis in bone marrow-derived macrophages (Jakobsen and Engel, unpublished) and cytotoxicity when incubated with HeLa cells (16). However, the class D genes are likely to be markers of the host defense response rather than cytotoxicity in the 9HTEo− cells since no cytopathic effects were observed. MCP1/SCYA2 is a widely used marker of inflammatory action, and the product plays an important role in immune regulation by recruiting inflammatory cells to the site of infection (37). TIEG activation is indicative of TGF-β function (27), which is important in the repair of wounded epithelial cells (23). Therefore, the identification of the molecule(s) responsible for this activity may provide new targets for the prevention of cell damage and antinflammatory therapies in P. aeruginosa infection.
ExoT activity induces marked changes in the cytoskeletal structure and the internalization capacity of cultured HeLa cells and macrophages (16). However, in the 9HTEo− cells no effects attributable to ExoT were observed at the morphological or gene expression level (i.e., in comparing the ExoT-sufficient strain PA103 exoU with the ExoT-deficient PA103 exoT exoU, pscJ, and pilA strains). Although the reasons for this are not clear, it is possible that more polarized cells resist the effects of ExoT, since others have noted no phenotypic effects in confluent MDCK epithelial cells infected with an alternate PA103 exoU strain (13).
In conclusion, we have provided a here detailed account of the responses evoked by epithelial cells upon contact with P. aeruginosa; this is the first such study addressing the importance of type III-mediated toxin secretion in epithelial cell gene regulation. The host cell response can be dissected into toxin-dependent and -independent pathways, with MAP kinase cascades and AP1 activation being important components in the former. Further characterization of the pathways mediating type III toxin actions will help us to understand the potential benefits or problems associated with drugs that may be developed to inhibit type III secretion.
Acknowledgments
The assistance of A. Cross and S. Grimmond in the collection and analysis of the microarray data is appreciated. We also thank M. Sweet for help with the EMSA.
This study was supported by a grant from the Australian National Health and Medical Research Council. The IMB incorporates the Centre for Functional and Applied Genomics, a Special Research Centre of the Australian Research Council. J.E. was supported by grants from the National Institutes of Health (AI 42806 and HL55980).
Editor: V. J. DiRita
REFERENCES
- 1.Apodaca, G., M. Bomsel, R. Lindstedt, J. Engel, D. Frank, K. E. Mostov, and J. Wiener-Kronish. 1995. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect. Immun. 63:1541-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldrick, J. C., A. A. Alizadeh, M. Diehn, S. Dudoit, C. L. Liu, C. E. Belcher, D. Botstein, L. M. Staudt, P. O. Brown, and D. A. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comolli, J. C., L. L. Waite, K. E. Mostov, and J. N. Engel. 1999. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect. Immun. 67:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly, D. T., J. V. Olander, D. Heuvelman, R. Nelson, R. Monsell, N. Siegel, B. L. Haymore, R. Leimgruber, and J. Feder. 1989. Human vascular permeability factor. Isolation from U937 cells. J. Biol. Chem. 264:20017-20024. [PubMed] [Google Scholar]
- 6.Cooper, P., S. Potter, B. Mueck, S. Yousefi, and G. Jarai. 2001. Identification of genes induced by inflammatory cytokines in airway epithelium. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L841-L852. [DOI] [PubMed] [Google Scholar]
- 7.Dang, J., D. Boyd, H. Wang, H. Allgayer, W. F. Doe, and Y. Wang. 1999. A region between −141 and −61 bp containing a proximal AP-1 is essential for constitutive expression of urokinase-type plasminogen activator receptor. Eur. J. Biochem. 264:92-99. [DOI] [PubMed] [Google Scholar]
- 8.de Kretser, D. M., M. P. Hedger, and D. J. Phillips. 1999. Activin A and follistatin: their role in the acute phase reaction and inflammation. J. Endocrinol. 161:195-198. [DOI] [PubMed] [Google Scholar]
- 9.Engel, J. Molecular pathogenesis of acute Pseudomonas aeruginosa infections. In A. Hauser and J. Rello (ed.), Severe infections caused by Pseudomonas aeruginosa, in press. Kluwer/Plenum Press, New York, N.Y.
- 10.Epelman, S., T. F. Bruno, G. G. Neely, D. E. Woods, and C. H. Mody. 2000. Pseudomonas aeruginosa exoenzyme S induces transcriptional expression of proinflammatory cytokines and chemokines. Infect. Immun. 68:4811-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farinha, M. A., B. D. Conway, L. M. Glasier, N. W. Ellert, R. T. Irvin, R. Sherburne, and W. Paranchych. 1994. Alteration of the pilin adhesin of Pseudomonas aeruginosa PAO results in normal pilus biogenesis but a loss of adherence to human pneumocyte cells and decreased virulence in mice. Infect. Immun. 62:4118-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck-Barbancon, V., and D. W. Frank. 2001. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 183:4330-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 14.Franklin, C. C., S. Srikanth, and A. S. Kraft. 1998. Conditional expression of mitogen-activated protein kinase phosphatase-1, MKP-1, is cytoprotective against UV-induced apoptosis. Proc. Natl. Acad. Sci. USA 95:3014-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritz, G., and B. Kaina. 2001. Ras-related GTPase Rhob represses NF-κB signaling. J. Biol. Chem. 276:3115-3122. [DOI] [PubMed] [Google Scholar]
- 16.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gospodarowicz, D., J. A. Abraham, and J. Schilling. 1989. Isolation and characterization of a vascular endothelial cell mitogen produced by pituitary-derived folliculo stellate cells. Proc. Natl. Acad. Sci. USA 86:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruenert, D. C., C. B. Basbaum, M. J. Welsh, M. Li, W. E. Finkbeiner, and J. A. Nadel. 1988. Characterization of human tracheal epithelial cells transformed by an origin-defective simian virus 40. Proc. Natl. Acad. Sci. USA 85:5951-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 20.Hauser, A. R., and J. N. Engel. 1999. Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67:5530-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 22.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-556. [DOI] [PubMed] [Google Scholar]
- 23.Howat, W. J., S. T. Holgate, and P. M. Lackie. 2002. TGF-beta isoform release and activation during in vitro bronchial epithelial wound repair. Am. J. Physiol. Lung Cell Mol. Physiol. 282:L115-L123. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. van't Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingram, W. J., C. A. Wicking, S. M. Grimmond, A. R. Forrest, and B. J. Wainwright. 2002. Novel genes regulated by Sonic Hedgehog in pluripotent mesenchymal cells. Oncogene 21:8196-8205. [DOI] [PubMed] [Google Scholar]
- 27.Johnsen, S. A., M. Subramaniam, T. Katagiri, R. Janknecht, and T. C. Spelsberg. 2002. Transcriptional regulation of Smad2 is required for enhancement of TGFβ/Smad signaling by TGFβ inducible early gene. J. Cell Biochem. 87:233-241. [DOI] [PubMed] [Google Scholar]
- 28.Junn, E., S. H. Han, J. Y. Im, Y. Yang, E. W. Cho, H. D. Um, D. K. Kim, K. W. Lee, P. L. Han, S. G. Rhee, and I. Choi. 2000. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 164:6287-6295. [DOI] [PubMed] [Google Scholar]
- 29.Kang, P. J., A. R. Hauser, G. Apodaca, S. M. Fleiszig, J. Wiener-Kronish, K. Mostov, and J. N. Engel. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249-1262. [DOI] [PubMed] [Google Scholar]
- 30.Kapas, S., A. Bansal, V. Bhargava, R. Maher, D. Malli, E. Hagi-Pavli, and R. P. Allaker. 2001. Adrenomedullin expression in pathogen-challenged oral epithelial cells. Peptides 22:1485-1489. [DOI] [PubMed] [Google Scholar]
- 31.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 32.Kazmierczak, B. I., and J. N. Engel. 2002. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase-activating protein for RhoA, Rac1, and Cdc42. Infect. Immun. 70:2198-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keyse, S. M. 2000. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi, H., M. Suzuki, N. Kanayama, T. Nishida, M. Takigawa, and T. Terao. 2002. Suppression of urokinase receptor expression by bikunin is associated with inhibition of upstream targets of extracellular signal-regulated kinase-dependent cascade. Eur. J. Biochem. 269:3945-3957. [DOI] [PubMed] [Google Scholar]
- 35.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, A., D. Chow, B. Haus, W. Tseng, D. Evans, S. Fleiszig, G. Chandy, and T. Machen. 1999. Airway epithelial tight junctions and binding and cytotoxicity of Pseudomonas aeruginosa. Am. J. Physiol. 277:L204-L217. [DOI] [PubMed] [Google Scholar]
- 37.Leonard, E. J., and T. Yoshimura. 1990. Human monocyte chemoattractant protein-1 (MCP-1). Immunol. Today 11:97-101. [DOI] [PubMed] [Google Scholar]
- 38.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. 3. Identity of the lethal toxins produced in vitro and in vivo. J. Infect. Dis. 116:481-489. [DOI] [PubMed] [Google Scholar]
- 39.Malangoni, M. A., R. Crafton, and F. C. Mocek. 1994. Pneumonia in the surgical intensive care unit: factors determining successful outcome. Am. J. Surg. 167:250-255. [DOI] [PubMed] [Google Scholar]
- 40.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 41.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, K. C., A. Cardoni, and Z. Z. Xiang. 2000. Vascular endothelial growth factor in bronchoalveolar lavage from normal subjects and patients with diffuse parenchymal lung disease. J. Lab. Clin. Med. 135:332-338. [DOI] [PubMed] [Google Scholar]
- 43.Mountz, J. D., H. G. Zhang, H. C. Hsu, M. Fleck, J. Wu, M. H. al-Maini, and T. Zhou. 1999. Apoptosis and cell death in the endocrine system. Rec. Prog. Horm. Res. 54:235-268. [PubMed] [Google Scholar]
- 44.Nakayama, K., A. Furusu, Q. Xu, T. Konta, and M. Kitamura. 2001. Unexpected transcriptional induction of monocyte chemoattractant protein 1 by proteasome inhibition: involvement of the c-Jun N-terminal kinase-activator protein 1 pathway. J. Immunol. 167:1145-1150. [DOI] [PubMed] [Google Scholar]
- 45.Philipp, W., L. Speicher, and C. Humpel. 2000. Expression of vascular endothelial growth factor and its receptors in inflamed and vascularized human corneas. Investig. Ophthalmol. Vis. Sci. 41:2514-2522. [PubMed] [Google Scholar]
- 46.Pritts, T., E. Hungness, Q. Wang, B. Robb, D. Hershko, and P. O. Hasselgren. 2002. Mucosal and enterocyte IL-6 production during sepsis and endotoxemia: role of transcription factors and regulation by the stress response. Am. J. Surg. 183:372-383. [DOI] [PubMed] [Google Scholar]
- 47.Rangnekar, V. V., S. Waheed, and V. M. Rangnekar. 1992. Interleukin-1-inducible tumor growth arrest is characterized by activation of cell type-specific “early” gene expression programs. J. Biol. Chem. 267:6240-6248. [PubMed] [Google Scholar]
- 48.Ratner, A. J., R. Bryan, A. Weber, S. Nguyen, D. Barnes, A. Pitt, S. Gelber, A. Cheung, and A. Prince. 2001. Cystic fibrosis pathogens activate Ca2+-dependent mitogen-activated protein kinase signaling pathways in airway epithelial cells. J. Biol. Chem. 276:19267-19275. [DOI] [PubMed] [Google Scholar]
- 49.Roncero, C., A. Darzins, and M. J. Casadaban. 1990. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J. Bacteriol. 172:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roux, P., J. M. Blanchard, A. Fernandez, N. Lamb, P. Jeanteur, and M. Piechaczyk. 1990. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell 63:341-351. [DOI] [PubMed] [Google Scholar]
- 51.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 52.Salyers, A. A., and D. D. Whitt. 1994. Bacterial pathogenesis: a molecular approach. ASM Press, Washington, D.C.
- 53.Sauvonnet, N., B. Pradet-Balade, J. A. Garcia-Sanz, and G. R. Cornelis. 2002. Regulation of mRNA expression in macrophages after Yersinia enterocolitica infection: role of different Yop effectors. J. Biol. Chem. 277:25133-25142. [DOI] [PubMed] [Google Scholar]
- 54.Simpson, L., J. Li, D. Liaw, I. Hennessy, J. Oliner, F. Christians, and R. Parsons. 2001. PTEN expression causes feedback upregulation of insulin receptor substrate 2. Mol. Cell. Biol. 21:3947-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 56.Stacey, K. J., L. F. Fowles, M. S. Colman, M. C. Ostrowski, and D. A. Hume. 1995. Regulation of urokinase-type plasminogen activator gene transcription by macrophage colony-stimulating factor. Mol. Cell. Biol. 15:3430-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stocco, C. O., L. F. Lau, and G. Gibori. 2002. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20α-hsd genes by prostaglandin F2α in ovarian cells. J. Biol. Chem. 277:3293-3302. [DOI] [PubMed] [Google Scholar]
- 58.Tannenbaum, C. S., J. Major, E. Poptic, P. E. DiCorleto, and T. A. Hamilton. 1989. Lipopolysaccharide-inducible macrophage early genes are induced in BALB/c 3T3 cells by platelet-derived growth factor. J. Biol. Chem. 264:4052-4057. [PubMed] [Google Scholar]
- 59.Thomson, S., L. C. Mahadevan, and A. L. Clayton. 1999. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin. Cell Dev. Biol. 10:205-214. [DOI] [PubMed] [Google Scholar]
- 60.Ueda, S., K. Nishio, N. Minamino, A. Kubo, Y. Akai, K. Kangawa, H. Matsuo, Y. Fujimura, A. Yoshioka, K. Masui, N. Doi, Y. Murao, and S. Miyamoto. 1999. Increased plasma levels of adrenomedullin in patients with systemic inflammatory response syndrome. Am. J. Respir. Crit. Care Med. 160:132-136. [DOI] [PubMed] [Google Scholar]
- 61.Vassen, L., W. Wegrzyn, and L. Klein-Hitpass. 1999. Human insulin receptor substrate-2 (IRS-2) is a primary progesterone response gene. Mol. Endocrinol. 13:485-494. [DOI] [PubMed] [Google Scholar]
- 62.Vinals, F., and J. Pouyssegur. 1999. Confluence of vascular endothelial cells induces cell cycle exit by inhibiting p42/p44 mitogen-activated protein kinase activity. Mol. Cell. Biol. 19:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 64.Waterhouse, C. C., R. R. Joseph, G. L. Winsor, T. A. Lacombe, and A. W. Stadnyk. 2001. Monocyte chemoattractant protein-1 production by intestinal epithelial cells in vitro: a role for p38 in epithelial chemokine expression. J. Interferon Cytokine Res. 21:223-230. [DOI] [PubMed] [Google Scholar]
- 65.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589-607. [DOI] [PubMed] [Google Scholar]
- 66.Zhao, Y., and Z. Y. Zhang. 2001. The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J. Biol. Chem. 276:32382-32391. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, J. H., S. R. Broussard, K. Strle, G. G. Freund, R. W. Johnson, R. Dantzer, and K. W. Kelley. 2001. IL-10 inhibits apoptosis of promyeloid cells by activating insulin receptor substrate-2 and phosphatidylinositol 3′-kinase. J. Immunol. 167:4436-4442. [DOI] [PubMed] [Google Scholar]