The first report of Staphylococcus aureus with intermediate-level resistance to vancomycin (VISA) was from Japan in 1997 (20), raising the threat of incurable staphylococcal infections. Since then, a number of cases have been reported worldwide, with eight confirmed cases in the United States as of June 2002 (9, 13). The majority of these cases have occurred in patients who have had prolonged exposure to vancomycin (13). Furthermore, the majority of these strains appear to have evolved from methicillin-resistant S. aureus (MRSA) strains previously infecting the patient, a conclusion that may be drawn from the similarities observed between the pulsed-field gel electrophoresis patterns of the VISA strains and the preexisting MRSA strains (12, 34). Fortunately, since emerging 6 years ago, infection with VISA is still a rare event. However, the phenomenon of vancomycin heteroresistance in S. aureus (hVISA) has been described more frequently in the literature, although the best method to detect hVISA strains and their clinical significance are ill-defined.
DEFINING VISA AND HVISA
VISA organisms have been defined by the National Committee for Clinical Laboratory Standards (NCCLS) as those staphylococci requiring vancomycin concentrations of 8 to 16 μg/ml for inhibition. Vancomycin resistance is defined by an MIC of ≥32 μg/ml. The Centers for Disease Control and Prevention (CDC) has adopted three criteria to identify VISA strains: (i) broth microdilution vancomycin MICs of 8 to 16 μg/ml, (ii) Etest vancomycin MICs of ≥6 μg/ml, and (iii) growth within 24 h on commercial brain heart infusion agar (BHIA) screen plates containing 6 μg of vancomycin/ml (36). The first vancomycin-resistant S. aureus (VRSA) clinical isolate, defined as a strain for which the vancomycin MIC is ≥32 μg/ml, was reported in Michigan in June 2002 (7). For this highly resistant strain, which contained the vanA determinant which mediates resistance to vancomycin in enterococci. the vancomycin MIC was 1,024 μg/ml. Conjugative transfer of the vanA gene from a coinfecting vancomycin-resistant enterococcus (VRE) strain likely explains the origin of this VRSA strain and the second VRSA strain isolated in Pennsylvania in October 2002 (8, 9).
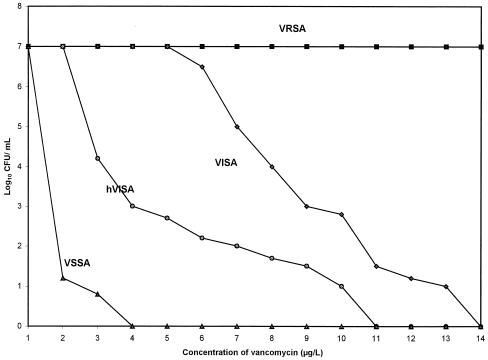
Heterogeneous VISA (hVISA) appears to be the stage that precedes the development of intermediate-level resistance in S. aureus or VISA. These are strains of S. aureus containing subpopulations of vancomycin-intermediate daughter cells; the MICs for the parent strains of these daughter cells fall within the susceptible range of 1 to 4 μg/ml (Fig. 1). Vancomycin creates a selective pressure that favors the outgrowth of rare, vancomycin-resistant clones leading to hVISA clones, and eventually, with continued exposure, to a uniform population of VISA clones. However, the criteria for identifying hVISA strains have not been standardized, complicating any determination of their clinical significance and role in treatment failures.
FIG. 1.
Population analysis of VRSA, VISA, hVISA, and VSSA. The population analysis shows how many cells in a fixed number of cells (usually about 107 CFU) of each strain are resistant to various concentrations of vancomycin. VRSA is a highly resistant and homogeneously resistant strain with 100% of the population growing at each of the vancomycin concentrations tested. VISA is intermediately resistant with 100% of the population growing at 4 μg of vancomycin per ml and also with significant subpopulations growing at 8 μg/ml. hVISA demonstrates heterogeneous resistance, having subpopulations of cells with various levels of resistance to vancomycin and including small populations of vancomycin-intermediate resistant cells with growth at 8 μg of vancomycin per ml.
The first hVISA strain, Mu3, was isolated in Japan in 1996 from a 64-year-old man with MRSA pneumonia that did not respond to vancomycin (19). Although the vancomycin MIC for this isolate was 4 μg/ml, the isolate contained subpopulations that were able to grow in media containing 5 to 9 μg of vancomycin/ml, thus demonstrating heterogeneous resistance. Pulsed-field gel electrophoresis showed that Mu3 had a pattern indistinguishable from that of the VISA strain Mu50 that was recovered several months later in the same hospital, suggesting that the two strains were closely related. Furthermore, when Mu3 was serially passaged in increasing concentrations of vancomycin, it gave rise to subpopulations with levels of resistance comparable to that of Mu50. This in vitro phenomenon suggests that colonization or infection with VISA may be preceded by infection with hVISA, with repeated vancomycin exposure acting as a selection pressure favoring the development of a uniformly resistant population (18).
It should also be noted that heteroresistance to teicoplanin, a glycopeptide used widely outside of the United States, has been observed. Historically, S. aureus acquired teicoplanin resistance before it acquired vancomycin resistance, and there are MRSA strains that are resistant to teicoplanin but susceptible to vancomycin based on its MIC (5, 18). All VISA strains have been observed to have reduced susceptibility to teicoplanin.
MECHANISMS OF RESISTANCE TO VANCOMYCIN IN HVISA AND VISA
Currently, the mechanism of intermediate resistance in S. aureus is unknown. The transfer of the vanA gene resistance determinants from VRE to S. aureus by cell-to-cell mating has been demonstrated in vitro (29). As mentioned previously, conjugative transfer appears to be the mechanism of resistance in the two VRSA strains isolated thus far (7, 8). However, none of the VISA strains have been shown to have any of the van determinants (vanA, vanB, vanC1, vanC2, or vanC3) that are present in VRE; thus, interspecies transfer of resistant genes is not responsible for intermediate resistance to vancomycin in S. aureus. VISA strains have been observed to have lower growth rates and thicker cell walls than fully susceptible strains (34). Hanaki et al. (16) found that hVISA produced three- to fivefold-greater quantities of penicillin-binding proteins 2 and 2′ and three- to eightfold increased quantities of cell wall precursors than vancomycin-susceptible strains did (32). Cui et al. (10) noted that cell wall thickening correlated with increased vancomycin MICs and was a common phenotype observed in VISA strains. Increased cell wall thickness appears to play a role in resistance by sequestering vancomycin molecules in the cell wall peptidoglycan, thus reducing the susceptibility of S. aureus to vancomycin.
CLINICAL SIGNIFICANCE OF HVISA AND VISA
The clinical significance of hVISA and VISA has been difficult to assess. It is unknown whether these strains are fully virulent or perhaps even more virulent than vancomycin-susceptible strains of S. aureus and whether levels of resistance are responsible for treatment failures. A variety of complicating factors make it difficult to ascertain whether the reported deaths in patients with VISA infections are directly attributable to the organism. For example, a patient in Illinois with VISA mitral valve endocarditis died while bacteremic from VISA but had refused surgical intervention (6).
Treatment failures with vancomycin may occur fairly commonly even with vancomycin-susceptible S. aureus (VSSA) strains. Moise and Schentag (27) reviewed 23 cases of vancomycin treatment failures in lower respiratory tract S. aureus infections, representing a treatment failure rate of 40% in their institution over a 1-year period. In each case, the vancomycin MIC for the organism was within the susceptible range and the vancomycin concentration in serum was shown to be in the therapeutic range. Several of these patients received multiple courses of vancomycin as their infections relapsed upon discontinuation of the antibiotic. The isolates recovered in this study were not tested to determine if they might be hVISA.
Since the recognition of hVISA and VISA, it has been suggested that hVISA strains are responsible for clinical failures to vancomycin treatment of otherwise apparently susceptible S. aureus strains. Ariza et al. (1) reported that 86% (12 of 14) of orthopedic surgery patients with MRSA infections whose isolates tested positive for hVISA experienced treatment failure compared to 20% (1 of 5) of patients with MRSA-positive and hVISA-negative infections. For all of these MRSA strains, the vancomycin MICs were between 1 and 4 μg/ml. However, the interpretation of the results of this study was complicated by the presence of implanted orthopedic devices in 12 of the 13 failures and in 13 of the 14 hVISA-infected patients. The only hVISA-negative patient who experienced treatment failure had an implanted device; none of the four hVISA-negative patients who were cured had devices in place.
Moore et al. (28) found that hVISA was associated with treatment failure in a patient with endocarditis. Paired S. aureus isolates (the pretreatment and relapse clinical isolates) from this patient were tested. Both strains had similar genotypes, and the vancomycin MICs for both strains were ≤2 μg/ml; however, population analysis determined that the second isolate exhibited heterogeneous resistance to vancomycin. The strains were further tested with a rabbit model of endocarditis in which the pretreatment isolate was eradicated by vancomycin while the relapse hVISA isolate was not, suggesting that vancomycin treatment failure in this case was due to heterogeneous resistance.
The properties of the drug itself may be sufficient to account for these observed treatment failures. In vitro, its activity falls between that of classically bacteriostatic drugs, such as tetracyclines, and that of bactericidal drugs, such as penicillins. Vancomycin is less rapidly bactericidal than antistaphylococcal penicillins, such as nafcillin, and is therefore less efficacious for the treatment of methicillin-susceptible staphylococcal (MSSA) infections (33). Patients with MRSA endocarditis treated with vancomycin have a delayed clinical response to the drug, as evidenced by prolonged bacteremia and sustained fever, compared to that of patients with MSSA endocarditis treated with beta-lactams (25).
Whether vancomycin failure is due to an intrinsic property of the drug, the virulence of the organism itself, or perhaps some combination of both continues to be an area of much controversy. Further studies are needed to evaluate the relevance of hVISA in patients with clinical failure to vancomycin. In order to conduct such studies, a means of accurately identifying these strains is essential.
EPIDEMIOLOGY OF HVISA
Since the first report of hVISA in Japan, a number of groups have conducted epidemiologic studies to examine the prevalence of this organism in their region. Most of these studies were retrospective and screened isolates stored at hospitals or strain collection centers. The clinical background of the patient from which the strain was recovered was unknown in the majority of the studies. In addition, the definition of hVISA and the methods used to screen for it varied among studies, making it difficult to compare the prevalence statistics reported.
We reviewed 14 studies published between 1997 and 2001 and found 132 hVISA isolates out of 7,920 S. aureus strains tested, or a prevalence of 1.67% (Table 1). These isolates represent S. aureus strains from around the world, including Japan (19), Korea (24), Hong Kong (40), Thailand (37), France (3, 30), Spain (1), Greece (23), Germany (2, 14), Italy (26), and the United Kingdom (21, 31). Interestingly, some of these strains have been present in several countries since the early 1990s but were not identified by routine laboratory testing (1, 2). There was a wide array of prevalence statistics reported, ranging from 0% to as high as 74% in one study (1). Furthermore, prevalence appeared to vary with the setting from which the isolates were recovered. Hiramatsu (17) found a 9.3% prevalence of hVISA among 129 MRSA strains collected at eight university hospitals but a 1.3% prevalence among 970 strains collected at community hospitals and clinics. Higher antibiotic selection pressures at tertiary care academic centers may account for the higher prevalence of hVISA in these hospitals.
TABLE 1.
Prevalence of hVISA in 14 epidemiologic studies
| Study authors (reference) | MRSA strains
|
MSSA strains
|
All strains
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | No. of hVISA | % hVISA | No. of isolates | No. of hVISA | % hVISA | Total | No. of hVISA | % hVISA | |
| Hiramatsu et al. (19) | 1,149 | 35 | 3.05 | 0 | 0 | 0 | 1,149 | 35 | 3.05 |
| Franchi et al. (11) | 30 | 0 | 0 | 0 | 0 | 0 | 30 | 0 | 0 |
| Ariza et al. (1) | 19 | 14 | 73.68 | 0 | 0 | 0 | 19 | 14 | 73.68 |
| Kantzanou et al. (23) | 72 | 1 | 1.39 | 25 | 0 | 0 | 97 | 1 | 1.03 |
| Geisel et al. (14) | 85 | 7 | 8.24 | 0 | 0 | 0 | 85 | 7 | 8.24 |
| Wong et al. (39) | 52 | 3 | 5.77 | 112 | 0 | 0 | 164 | 3 | 1.83 |
| Bierbaum et al. (2) | 367 | 2 | 0.54 | 90 | 0 | 0 | 457 | 2 | 0.44 |
| Schmitz et al. (31) | 302 | 0 | 0 | 0 | 0 | 0 | 302 | 0 | 0 |
| Marchese et al. (26) | 179 | 2 | 1.12 | 0 | 0 | 0 | 179 | 2 | 1.12 |
| Kim et al. (24) | 3,371 | 59 | 1.75 | 1,172 | 0 | 0 | 4,543 | 59 | 1.30 |
| Bobin-Dubreux et al. (3) | 0 | 0 | 0 | 469 | 1 | 0.21 | 469 | 1 | 0.21 |
| Reverdy et al. (30) | 171 | 3 | 1.75 | 0 | 0 | 0 | 171 | 3 | 1.75 |
| Trakulsomboon et al. (37) | 155 | 5 | 3.23 | 0 | 0 | 0 | 155 | 5 | 3.23 |
| Wootton et al. (41) | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 |
| Total | 6,052 | 131 | 2.16 | 1,868 | 1 | 0.05 | 7,920 | 132 | 1.67 |
The majority of hVISA isolates reported are methicillin resistant, with only one reported case of methicillin-susceptible hVISA described in the literature. However, this finding may be biased because 9 of the 14 studies screened MRSA isolates exclusively, and of the 7,920 strains screened, 6,052 were methicillin resistant. MRSA isolates are speculated to be more likely to harbor the hVISA phenotype as they represent strains that emerged from heavy antibiotic selection pressures and then were subjected to additional selection pressures through treatment with vancomycin. The one reported case of methicillin-susceptible hVISA occurred in a 35-year-old woman evaluated in an outpatient clinic for conjunctivitis (3). She was an otherwise healthy woman who had not received any glycopeptides or other antimicrobial therapy in the preceding 3 months. She had not been in contact with anyone with risk factors for vancomycin resistance, for example, dialysis patients. The isolation of methicillin-susceptible hVISA isolates suggests that any S. aureus strain can potentially have reduced susceptibility to vancomycin. Heteroresistance to vancomycin may be an intrinsic property of the organism that occurs at a low frequency in the population and that is detected during chance sampling. An alternative explanation is that the strain was previously methicillin resistant but underwent deletion of the mecA gene, a process which has been described for VISA (15).
SCREENING METHODS FOR IDENTIFYING HVISA
PAP.
Currently, no standardized method for identifying hVISA exists. Population analysis profiling (PAP) has been proposed as the most precise method of determining heteroresistance. Tenfold serial diluents of a starting cell suspension of 108 CFU/ml are plated onto BHIA plates containing increasing concentrations of vancomycin. The number of viable colonies at 48 h for each antibiotic concentration is counted and plotted against the vancomycin concentration on a semilogarithmic graph. Figure 1 illustrates typical population analysis curves for VRSA, VISA, hVISA, and VSSA. One group took this method a step further and calculated the area under the concentration-time curve (AUC) for each strain in order to distinguish among VISA, hVISA, and VSSA (41). The ratios of the AUC of the test strains to the AUC of the Mu3 control were then determined; the resulting ratios were ≤0.90 for VSSA, 0.90 to 1.3 for hVISA, and ≥1.3 for VISA. However, the labor-intensive nature of this assay makes PAP an impractical means of screening a large number of isolates. In addition, it requires a $30,000 spiral plating apparatus which very few laboratories have. Moreover, this method has not been validated as being superior to others.
Simplified population analysis.
The most common screening method in the literature was first described by Hiramatsu et al. (19) in the characterization of the prototype hVISA and VISA strains, Mu3 and Mu50. Known as simplified population analysis, this method involves inoculating 10 μl of a 108-CFU/ml bacterial suspension onto BHIA containing 4 μg of vancomycin per ml (BHIA-V4). Growth at 24 h was considered “potential VISA,” while growth at 48 h was considered “potential hVISA.” Strains were “confirmed VISA” if the vancomycin MICs for them were 8 μg/ml; they were “confirmed hVISA” if the strain produced subclones for which the vancomycin MICs were 8 μg/ml after selection with vancomycin and remained stably resistant for >9 days on drug-free medium.
A variety of screening methods have been described in the literature (Table 2). Many studies used variations of Hiramatsu's simplified population analysis method: some studies screened with Mueller-Hinton agar (MHA) (1, 3, 11, 22, 30) instead of BHIA-V4,; others applied a different inoculum size (100 versus 10 μl) to the screening plate (26, 37), while others used higher concentrations of bacterial suspension (e.g., McFarland standard of 1 versus 0.5) (23, 26). Several studies screened with media containing 2 instead of 4 μg of vancomycin per ml (3, 11, 30). One study chose the Etest as their initial screening method, followed by PAP on MHA containing increasing concentrations of vancomycin (11). Some studies distinguished between potential and confirmed hVISA and VISA, while others did not (2, 14, 19, 26, 40). Whether this distinction is useful remains controversial. Some studies have argued that Hiramatsu's confirmatory method involving passage of hVISA subclones may, in fact, be selecting for resistance in vitro rather than screening for it (35, 38). A few studies screened for vancomycin resistance by culturing isolates in vancomycin-salt agar containing an aztreonam disk, again raising the question of whether such a test induces resistance instead of detecting it (30, 40).
TABLE 2.
Laboratory detection of hVISA
| Laboratory (reference) | Detection methoda
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Simplified PAP on BHIA-V4 | Simplified PAP on MHA | Broth microdilution (MIC) | Agar dilution (MIC) | Etest | PAP | Disk-agar | MicroScan | Resistant mutant emergence | |
| Hiramatsu et al. (19) | I | CΨ | |||||||
| Franchi et al. (11) | I | C* | |||||||
| Ariza et al. (1) | X | X | |||||||
| Kantzanou et al. (23) | I♣ | CΨ | C | C | |||||
| Geisel et al. (14) | I | CΨ | |||||||
| Wong et al. (39) | I | CΨ | CΨ | I | |||||
| Bierbaum et al. (2) | I | CΨ | C | ||||||
| Schmitz et al. (31) | X | X | X | X | |||||
| Marchese et al. (26) | Iδ♣ | CΨ | C | ||||||
| Kim et al. (24) | I | CΨ | C | ||||||
| Bobin-Dubreux et al. (3) | Iφ | C | C | C | |||||
| Reverdy et al. (30) | Iφ | C | C | C | I | ||||
| Trakulsomboon et al. (37) | Iδ | C | C | C | |||||
| Wootton et al. (41) | X | X | X | X | |||||
Disk-agar, aztreonam disk on vancomycin salt; MicroScan, Microscan conventional gram-positive panels; I, initial screening test; C, confirmatory test; X, for the tests performed, did not distinguish between initial and confirmatory tests; *, PAP performed on MHA; Ψ, MIC determined on subclone recovered from initial screen; δ, 100 μl of starting cell suspension (instead of 10 μl) plated; φ, MHA with 2 μg of vancomycin per ml; ♣, McFarland standard of 1.0 (instead of 0.5) used.
The variety of the screening methods used to detect hVISA makes the interpretation of any prevalence statistics difficult. Indeed, some groups have used several methods to test for heteroresistance, with variable results. Wong et al. (39) found that 21 (40%) of 52 MRSA strains tested positive for hVISA by the initial BHIA-V4 screen. Only three (5.7%) strains were confirmed positive according to Hiramatsu's method, and five (9.6%) strains tested positive by the aztreonam disk-agar method.
The reliability of the BHIA-V4 screen has also been questioned. Wootton et al. (41) found that this method had poor reproducibility. They tested Mu50 and Mu3 in 10 different batches and noted that while Mu50 demonstrated consistent growth at 24 h, Mu3 grew at 48 h only 80% of the time. They also tested 100 MRSA isolates in quintuplicate, identifying 16 as hVISA on one of five occasions and 1 additional strain as positive on two occasions (21). The same group tested this method against PAP in the evaluation of VSSA, hVISA, and VISA strains to determine the sensitivity and specificity of the BHIA-V4 screen, which were found to be 71 and 88%, respectively (38). In our experience, out of 239 isolates tested by simplified population analysis, 50 (21%) isolates were identified as potential hVISA as defined by growth on BHIA-V4 at 48 h on a single assay. However, only 16 (6.7%) isolates demonstrated growth in duplicate assays (unpublished data). None of the studies that we reviewed reported testing isolates more than once.
Tenover et al. (36) also noted that vancomycin-containing media prepared in-house showed occasional growth of susceptible strains. Four of five lots of BHIA containing 6 μg of vancomycin per ml prepared in-house grew vancomycin-susceptible American Type Culture Collection control strains. In contrast, prequalified commercially prepared media consistently inhibited growth of susceptible control strains, suggesting that strict quality control of test media is necessary for accurate results. None of the studies specified whether or not the BHIA media was prequalified prior to use in screening.
One additional difficulty in screening for hVISA is that the vancomycin resistance phenotype has been observed to be an unstable one. Although some VISA strains remain stable after multiple serial passages in drug-free culture medium (32), other strains revert after passage in vancomycin-free media. Boyle-Vavra et al. (4) serially passaged VISA isolates on nutrient agar with and without vancomycin supplementation. After 15 days of passage on nonselective media, vancomycin-susceptible revertant mutants were isolated from each VISA strain. If the strains were passaged on vancomycin-containing media, the VISA phenotype was preserved with no decreases in the MIC noted. In the latter experiment, subinhibitory concentrations of vancomycin were used, i.e., 2 μg/ml for Mu3 and 4 μg/ml for Mu50. This reversion phenomenon suggests that the VISA phenotype is unstable and perhaps impairs the fitness of the organism and that it is maintained only under continued selective pressure with vancomycin. Clinical laboratories may not be able to identify the organism since isolates are routinely incubated on nonselective media before susceptibility testing, thus providing an opportunity for reversion to occur. This observation may explain the difficulty in establishing an association between the presence of hVISA and observed clinical failures to vancomycin therapy.
The issue of reversion has led some researchers to propose that potential hVISA and VISA isolates be cultured on media containing low levels of vancomycin prior to susceptibility testing to prevent the phenomenon from occurring. Howe et al. (21) demonstrated that preincubation of hVISA and VISA strains with vancomycin significantly increased expression of vancomycin resistance, allowing some strains to grow in the presence of 16 μg of vancomycin per ml. However, whether this method truly detects or instead selects for vancomycin resistance once again remains unclear.
Alternative screening methods.
Alternatives to Hiramatsu's method that can be applied practically in a clinical laboratory setting have been proposed. Walsh et al. (38) tested 284 vancomycin-susceptible MRSA strains and 45 vancomycin-intermediate staphylococcal species (VISS) and heterogeneous vancomycin-intermediate staphylococcal species (hVISS) with a variety of assays, including Hiramatsu's single-plate agar method, BHIA containing 4 μg of vancomycin per ml, BHIA containing 6 μg of vancomycin per ml, MHA containing 5 μg of vancomycin per ml, broth microdilution, agar dilution, and the Etest. All methods were performed in duplicate and compared to population analysis as the gold standard. The study found that the Etest performed with an inoculum concentration corresponding to a McFarland standard of 2.0 on BHIA gave the most precise results, with high sensitivity (93%) and specificity (97%). Tenover et al. (36) tested 12 VISS and hVISS isolates for susceptibility to vancomycin by five commercial methods: Vitek, MicroScan conventional panels, MicroScan rapid panels, Sensitre panels, and the Etest. They noted that the most effective methods for detecting VISS and hVISS isolates were the Etest and MicroScan conventional panels. The Etest consistently produced MIC readings within 0.5 to 1 dilution of the results by the NCCLS broth dilution referenced method.
CONCLUSIONS
While infection with VISA remains a rare event, data suggest that heteroresistance may be more common. However, a critical issue that has emerged is the reliability of the screening methods used to detect hVISA. Important questions regarding the prevalence and clinical significance of hVISA cannot be accurately addressed without an improved, standardized, practical, and validated means of testing for the organism. There are many areas of controversy surrounding the most appropriate way of identifying hVISA. Etest seems promising as an alternative to agar plating methods but will need further study before it can be adopted for standard use. What, if any, additional tests are needed to confirm a strain as hVISA after an initial positive screen? What quality control measures are needed to ensure precise results? What resistance breakpoint should be used to identify a strain as hVISA? Do strains need to be preincubated on vancomycin-containing media prior to susceptibility testing to prevent reversion? If so, what concentration of vancomycin should be used to limit the possibility of selecting for resistance?
The lack of a reliable method of detecting hVISA currently limits our ability to understand the role of the heterogeneous resistance phenotype in clinical treatment failures, to predict them, and to prevent them. Elucidation of the molecular and genetic bases of the hVISA phenotype may enable genotype testing, providing for a more definitive means of identification. The correlation of in vitro test results indicating the presence of the hVISA phenotype with efficacy studies in experimental animal models or with clinical outcomes and a propensity for increasing levels of resistance with vancomycin exposure are also important. In the absence of any of these data, the methods used to detect hVISA isolates and estimates of their prevalence must be interpreted cautiously.
Acknowledgments
This research was supported by grant AI43959 from the National Institutes of Health.
REFERENCES
- 1.Ariza, J., M. Pujol, J. Cabo, C. Pena, N. Fernandez, J. Linares, J. Avats, and F. Gudiol. 1999. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587-1588. [DOI] [PubMed] [Google Scholar]
- 2.Bierbaum, G., K. Fuchs, W. Lenz, C. Szekat, and H. G. Sahl. 1999. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 18:691-696. [DOI] [PubMed] [Google Scholar]
- 3.Bobin-Dubreux, S., M.-E. Reverdy, C. Nervi, M. Rougier, A. Bolmström, F. Vandenesch, and J. Etienne. 2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet, F., G. Vedal, F. Dreyfus, J. F. Vaxelaire, T. Giraud, B. Schremmer, and J. F. Monsallier. 1990. Failure of teicoplanin therapy in two neutropenic patients with staphylococcal septicemia who recovered after administration of vancomycin. Eur. J. Clin. Microbiol. Infect. Dis. 9:145-147. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2000. Staphylococcus aureus with reduced susceptibility to vancomycin—Illinois, 1999. Morb. Mortal. Wkly. Rep. 48:1165-1167. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 9.Chang, S., D. M. Sievert, J. Hageman, M. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 10.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M.-N. Kim, M.-C. Ploy, N. El Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchi, D., M. Climo, A. Wong, M. Edmond, and R. Wenzel. 1999. Seeking vancomycin resistant Staphylococcus aureus among patients with vancomycin-resistant enterococci. Clin. Infect. Dis. 29:1556-1558. [DOI] [PubMed] [Google Scholar]
- 12.Fridkin, S. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 13.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, and F. C. Tenover. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 14.Geisel, R., F. J. Schmitz, L. Thomas, G. Berns, O. Zetsche, B. Ulrich, A. C. Fluit, H. Labischinsky, and W. White. 1999. Emergence of heterogeneous intermediate vancomycin resistance in Staphylococcus aureus isolates in the Dusseldorf area. J. Antimicrob. Chemother. 43:846-848. [DOI] [PubMed] [Google Scholar]
- 15.Hageman, J., D. A. Pegues, C. Jepson, R. L. Bell, M. Guinan, K. W. Ward, M. D. Cohen, J. A. Hindler, F. C. Tenover, S. K. McAllister, M. E. Kellum, and S. K. Fridkin. 2001. Vancomycin-intermediate Staphylococcus aureus in a home health-care patient. Emerg. Infect. Dis. 7:1023-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu, K. 1998. The emergence of Staphylococcus aureus with reduced susceptibility to vancomycin in Japan. Am. J. Med. 104:7S-10S. [DOI] [PubMed] [Google Scholar]
- 18.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 21.Howe, R. A., M. Wootton, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 1999. Expression and detection of hetero-vancomycin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 44:675-678. [DOI] [PubMed] [Google Scholar]
- 22.Hubert, S. K., J. M. Mohammed, S. K. Fridkin, R. P. Gaynes, J. E. McGowan, Jr., and F. C. Tenover. 1999. Glycopeptide-intermediate Staphylococcus aureus: evaluation of a novel screening method and results of a survey of selected U.S. hospitals. J. Clin. Microbiol. 37:3590-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantzanou, M., P. Tassios, A. Tseleni-Kotsovili, N. Legakis, and A. Vatopoulos. 1999. Reduced susceptibility to vancomycin of nosocomial isolates of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 43:729-731. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M.-N., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine, D. P., B. S. Fromm, and B. R. Reddy. 1991. Slow response to vancomycin or vancomycin plus rifampin in methicillin-resistant Staphylococcus aureus endocarditis. Ann. Intern. Med. 115:674-680. [DOI] [PubMed] [Google Scholar]
- 26.Marchese, A., G. Balistreri, E. Tonoli, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moise, P., and J. Schentag. 2000. Vancomycin treatment failures in Staphylococcus aureus lower respiratory tract infections. Int. J. Antimicrob. Agents 16(Suppl. 1):S31-S34. [DOI] [PubMed] [Google Scholar]
- 28.Moore, M. R., F. Perdreau-Remington, and H. F. Chambers. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylocococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble, W., Z. Virani, and R. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 30.Reverdy, M. E., S. Jarraud, S. Bobin-Dubreux, E. Burel, P. Girardo, G. Lina, F. Vandenesch, and J. Etienne. 2001. Incidence of Staphylococcus aureus with reduced susceptibility to glycopeptides in two French hospitals. Clin. Microbiol. Infect. 7:267-272. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz, F.-J., A. Krey, R. Geisel, J. Verhoef, H. P. Heinz, and A. Fluit. 1999. Susceptibility of 302 methicillin-resistant Staphylococcus aureus isolates from 20 European university hospitals to vancomycin and alternative antistaphylococcal compounds. Eur. J. Clin. Microbiol. Infect. Dis. 18:528-530. [DOI] [PubMed] [Google Scholar]
- 32.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 33.Small, P. M., and H. F. Chambers. 1990. Vancomycin for Staphylocococcus aureus endocarditis in intravenous drug users. Antimicrob. Agents Chemother. 34:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, W. R. Jarvis, et al. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 35.Tenover, F., J. Biddle, and M. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, S., P. L. Ho, P. C. Woo, and K. Y. Yuen. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 29:760-767. [DOI] [PubMed] [Google Scholar]
- 40.Wong, S. S., T. K. Ng, W. C. Yam, D. N. Tsang, P. C. Woo, S. K. Fung, and K. Y. Yuen. 2000. Bacteremia due to Staphylococcus aureus with reduced susceptibility to vancomycin. Diagn. Microbiol. Infect. Dis. 36:261-268. [DOI] [PubMed] [Google Scholar]
- 41.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-404. [DOI] [PubMed] [Google Scholar]