Abstract
Antisense phosphorodiamidate morpholino oligomers (PMOs) were tested for the ability to inhibit gene expression in Escherichia coli. PMOs targeted to either a myc-luciferase reporter gene product or 16S rRNA did not inhibit luciferase expression or growth. However, in a strain with defective lipopolysaccharide (lpxA mutant), which has a leaky outer membrane, PMOs targeted to the myc-luciferase or acyl carrier protein (acpP) mRNA significantly inhibited their targets in a dose-dependent response. A significant improvement was made by covalently joining the peptide (KFF)3KC to the end of PMOs. In strains with an intact outer membrane, (KFF)3KC-myc PMO inhibited luciferase expression by 63%. A second (KFF)3KC-PMO conjugate targeted to lacI mRNA induced β-galactosidase in a dose-dependent response. The end of the PMO to which (KFF)3KC is attached affected the efficiency of target inhibition but in various ways depending on the PMO. Another peptide-lacI PMO conjugate was synthesized with the cationic peptide CRRRQRRKKR and was found not to induce β-galactosidase. We conclude that the outer membrane of E. coli inhibits entry of PMOs and that (KFF)3KC-PMO conjugates are transported across both membranes and specifically inhibit expression of their genetic targets.
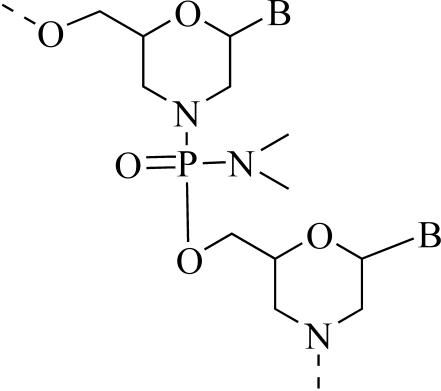
Phosphorodiamidate morpholino oligomers (PMOs) are DNA mimics that inhibit expression of specific mRNA in eukaryotic cells (1, 17, 22). They are synthesized by using the four natural bases, with a base sequence that is complementary (antisense) to a region of a specific mRNA. They are different than DNA in the chemical structure that links the bases together (Fig. 1). Ribose has been replaced with a morpholine group, and the phosphodiester is replaced with a phosphorodiamidate. These alterations make the antisense molecule resistant to nucleases (11) and free of charges at physiological pH, yet it retains the molecular architecture required for binding specifically to a complementary strand of nucleic acid (21, 22, 24). PMOs are currently in phase I and II human clinical trials as therapeutics for restenosis and cancers.
FIG. 1.
PMO structure. B is adenine, thymine, guanine, or cytosine.
Recently, another type of antisense DNA mimic, called peptide nucleic acid (PNA), was shown to inhibit the expression of bacterial genes in vitro and in pure culture (8, 9). However, entry of PNA into Escherichia coli was inefficient because of the outer membrane of this gram-negative bacterium (10). Entry was greatly improved (7) by coupling PNA to cationic peptides that were previously shown to permeabilize the gram-negative outer membrane (18, 25).
In this report, we show that PMOs inhibit gene expression in E. coli but are limited for cellular uptake by the outer membrane. In addition, uptake across the outer membrane was achieved by coupling PMOs to the peptide (KFF)3KC.
MATERIALS AND METHODS
Bacteria and growth conditions.
E. coli SM105 [thr-1 araC14 tsx-78 Δ(galK-attλ)99 hisG4 rfbD1 rpsL136 sylA5 mtl-1 thi-1] and its isogenic lpxA2(Ts) derivative SM101 were purchased from the E. coli Genetic Stock Center (New Haven, Conn.). E. coli BL21(DE3)pLysS was from Invitrogen, Inc. (Carlsbad, Calif.). Cultures were started from 1 to 4 colonies inoculated into 2 ml of broth and grown for 15 h with shaking. Fifteen-hour cultures were diluted 1/50 in broth and immediately mixed with PMO. Strains without recombinant plasmids were grown in Luria-Bertani (LB) broth (13), and strains transformed with pT7myc-luc or pSE380myc-luc were grown in LB plus 100 μg of ampicillin/ml. BL21(DE3)(pLysS/pT7myc-luc) was grown at 37°C in LB plus 50 μg of chloramphenicol/ml and 100 μg of ampicillin/ml. SM105 and SM101 were grown at 30°C.
PMOs.
PMOs were synthesized and purified at AVI BioPharma, Inc. (Corvallis, Oreg.) as previously described (23), dissolved in water, filtered through a 0.2-μm-pore-size membrane (HT Tuffryn; Gelman Sciences, Inc., Ann Arbor, Mich.), and stored at either 4 or −20°C. The purity of PMOs was analyzed by mass spectrometry and reverse-phase high-pressure liquid chromatography (HPLC). Sequences of PMOs and peptide-PMO conjugates used in this study are shown in Tables 1 and 2. The concentration of all PMOs and peptide-PMO conjugates was measured by UV spectrophotometric absorbance at 260 nm, with extinction coefficients calculated for nucleic acids as described in reference 3 and for peptides as described in reference 27. The contribution to absorbance at 260 nm of the 6 or 12 equivalents of phenylalanine per mol of peptide-PMO conjugate was less than 1 or 2%, respectively, of the total absorbance. The absorbance at 260 nm from the cross-linkers was negligible.
TABLE 1.
PMOs
| Name | Target | AVI BioPharma identification no. | Base sequence (5′→3′)a |
|---|---|---|---|
| 16S | 16S rRNA | 1-23-17 | GCA AAG GTA TTA ACT TTA CTC |
| myc | Human c-myc | 1-22-126 | ACG TTG AGG GGC ATC GTC GC |
| myc-contr | Mismatch control for c-myc | 1-22-144 | ACT GTG AGG GCG ATC GCT GC |
| lacI | lacI repressor | 1-23-142 | TTC ACA TTC ACC ACC CTG AA |
| acp | Acyl carrier protein | 1-23-140 | TCT TCG ATA GTG CTC ATA CT |
| acp-scr | Scrambled control for acp | 1-23-141 | TTG TCC TGA ATA TCA CTT CG |
Mismatched bases are underlined. The complement of the start codon is shown in boldface type.
TABLE 2.
Peptide-PMO conjugates
| Name | Target | PMO end attached to peptide | Cross-linker | Base sequence (5′→3′)a |
|---|---|---|---|---|
| P150 | c-myc | 3′, 5′, and 3′ + 5′ KFFKFFKFFKC | SPDP | ACG TTG AGG GGC ATC GTC GC |
| P152 | c-myc | 3′, 5′, and 3′ + 5′ KFFKFFKFFKC | GMBS | ACG TTG AGG GGC ATC GTC GC |
| P173 | c-myc | 3′ KFFKFFKFFKC | SPDP | ACG TTG AGG GGC ATC GTC GC |
| P187 | c-myc control | 3′ KFFKFFKFFKC | GMBS | ACT GTG AGG GCG ATC GCT GC |
| 3 + 5′KFC-lacI | lacI | 3′, 5′, and 3′ + 5′ KFFKFFKFFKC | GMBS | TTC ACA TTC ACC ACC CTG AA |
| 3 + 5′KFC-lacI-scr | lacI control | 3′, 5′, and 3′ + 5′ KFFKFFKFFKC | GMBS | ATC CTC CCA ACT TCG ACA TA |
| 3′KFC-lacI | lacI | 3′ KFFKFFKFFKC | GMBS | TTC ACA T TC ACC ACC CTG AA |
| 3′KFC9-lacI-scr | lacI control | 3′ KFFKFFKFFKC | GMBS | ATC CTC CCA ACT TCG ACA TA |
| 5′KFC-lacI | lacI | 5′ KFFKFFKFFKC | GMBS | TTC ACA TTC ACC ACC CTG AA |
| 5′KFC-lacI-scr | lacI control | 5′ KFFKFFKFFKC | GMBS | ATC CTC CCA ACT TCG ACA TA |
| rTat-lacI | lacI | 5′ CRRRQRRKKR | GMBS | TTC ACA TTC ACC ACC CTG AA |
| rTat-lacI-scr | lacI control | 5′ CRRRQRRKKR | GMBS | ATC CTC CCA ACT TCG ACA TA |
Mismatched bases are underlined. The complement of the start codon is shown in boldface type.
Peptides KFFKFFKFFKC-CONH2 and rTat (CRRRQRRKKR-CONH2) were synthesized and purified (>90%) by Global Peptide Services (Ft. Collins, Colo.) and were attached to the PMO through a heterobifunctional cross-linker purchased from Pierce Chemical Co. (Rockford, Ill.) as follows.
3′-End conjugation.
β-Alanine was coupled to the 3′ end of PMO, which was still covalently attached to resin by its 5′ end. The attachment of β-alanine provided a primary amine to which peptides were cross-linked. The PMO-β-alanine was subsequently cleaved off the resin as described below. A bifunctional cross-linker containing maleimide and succinimide moieties was used to link the 3′ primary amine of the PMO-β-alanine with the sulfhydryl group of a peptide to form the PMO-peptide conjugate. The details of the modification at 3′ end of PMO are described below.
N-β-FMOC-β-alanine pentafluorophenyl ester.
N-β-FMOC (9-fluorenylmethoxy carbonyl)-β-alanine (Novabiochem, San Diego, Calif.) was converted to the pentafluorophenyl ester by using N,N′-dicyclohexylcarbodiimide chemistry.
3′ β-Alanine PMO.
N-β-FMOC-β-alanine pentafluorophenyl ester (0.1 M) was dissolved in 400 μl of N-methyl-2-pyrolidone (NMP). N-Ethyl morpholine was added to a final concentration of 1%, and the solution was mixed in a disposable chromatography column with PMO (approximately 5 to 10 μmol), which was still attached to the resin used for synthesizing the PMO. The mixture was incubated at 45°C for 20 min, and then another 400 μl of 0.1 M N-β-FMOC-β-alanine pentafluorophenyl ester solution was added to the column and followed by incubation at 45°C for 60 min, after which the column was washed with NMP. PMO was cleaved off the resin with 1 ml of cleavage solution (0.1 M dithiothreitol in NMP containing 10% triethylamine). The resin was washed with 300 μl of cleavage solution three times immediately followed by addition of 4 ml of concentrated ammonia hydroxide and incubation for 16 h at 45°C to remove base protection groups. PMO was precipitated out of the NMP-NH4OH solution with 8 volumes of acetone and precipitated by centrifugation (14,000 × g, 5 min). The pellet was washed with 15 ml of CH3CN. The washed pellet was redissolved in 4 ml of H2O and lyophilized. The product was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) (mass spectrometry) and HPLC. Seventy percent of the product corresponded to the full-length sequence that had β-alanine at its 3′ end and a triethylene glycol moiety at its 5′ end.
Addition of a cross-linker.
The noncleavable linker N-[γ-maleimidobutyryloxy]succinimide ester (GMBS) or the cleavable linker N-succinimidyl 3-[pyridyldithio]propionate (SPDP) (Pierce Chemical Co.) was predissolved in 50 μl of dimethyl sulfoxide and mixed with the PMO (2 to 5 mM) dissolved in sodium phosphate buffer (100 mM, pH 7.2) at a 1:2 PMO/GMBS molar ratio. The mixture was stirred at room temperature in the dark for 30 min, and the product was purified by passing through a P2 (Bio-Rad, Hercules, Calif.) gel filtration column to remove the excess cross-linker and by-products. PMO-GMBS was lyophilized and analyzed by MALDI-TOF and HPLC. The conversion from PMO to PMO-GMBS was >90%.
Conjugation.
PMO-GMBS (2 to 5 mM) was dissolved in phosphate buffer (50 mM phosphate [pH 6.5], 5 mM EDTA). The peptide was added to the PMO-GMBS solution at 2:1 peptide-to-PMO molar ratio. The reaction mixture was stirred at room temperature for 2 h. The conjugate was purified first through an Excellulose gel filtration column (Pierce Chemical Co.) to remove excess peptide and then through a CM-Sepharose (Sigma, St. Louis, Mo.) cation-exchange column to remove nonconjugated PMO and finally dialyzed against H2O to remove salt. The conjugate was lyophilized and analyzed by MALDI-TOF and HPLC. The final product contained about 60% material corresponding to a full-length PMO conjugated to a peptide, with the balance composed of shorter peptide-PMO conjugates and a small amount of nonconjugated PMOs of various lengths. The concentration calculation used for all experiments was based on the total absorbance at 260 nm.
5′-End conjugation.
To conjugate a peptide onto the 5′ end of PMO, the 3′ end was first acetylated by mixing in a disposable chromatography column excess acetic anhydride (0.1 M) in NMP and (approximately 10 μmol) PMO, which was still attached to the resin used for synthesis. The column was incubated at 45°C for 20 min and drained, and another similar portion of acetic anhydride was added to the column and incubated at 45°C for 60 min. The column was drained and washed with NMP. PMO was cleaved off the resin and worked up as described in “3′ β-Alanine PMO” above. The acetylated PMO had a piperazine group at the 5′ end, which has a secondary amine for reacting with the cross-linker. The reaction with cross-linker and the conjugation with a peptide were done as described in “3′-End conjugation” above.
3′-End and 5′-end conjugation.
To conjugate KFC peptide onto both ends of PMO, β-alanine was first attached to the 3′ end. After the PMO was cleaved off the resin and worked up as described in “3′ β-Alanine PMO,” the PMO-β-alanine had a primary amine at the 3′ end and a secondary amine (piperazine group) at the 5′ end. Both amines were reacted with either GMBS or SPDP cross-linker at a reaction ratio of 1:5 (PMO to cross-linker). The product was purified as described in “Addition of a cross-linker,” conjugated with KFC peptide at reaction ratio of 1:5 (PMO to peptide), and purified by the procedure described in “Conjugation” above.
Reporter gene construction.
Standard molecular biology procedures (2) were used for all constructions. A fusion sequence consisting of (from 5′ to 3′) 30 bp of human c-myc from −14 to +16 (where the first base of the translation start codon is +1), an 8-bp linker region of the sequence GTCGACTG, and bases +2 to +1653 of the gene (luc) for firefly luciferase was obtained by PCR of a preexisting plasmid (pCNmyclucΔA) with the forward primer 5′-TAGGCTAGCAGCCTCCCG and the reverse primer 5′-TTACAATAGCTAAGAATTTC and subcloned into pT7CTTOPO (Invitrogen, Inc.), creating pT7CTmyclucΔA. In vitro mutagenesis was subsequently performed on pT7CTmyclucΔA with QuikChange (Stratagene, Inc., La Jolla, Calif.) and oligonucleotides 5′-ATTTTGTTTAACTTTAAGAAGGAATCCCGCGACGATGCCCCTCAACG and 5′-CGTTGAGGGGCATCGTCGCGGGATTCCTTCTTAAAGTTAAACAAAAT in order to remove 22 bases that were between the ribosome-binding sequence (RBS) present in pT7CTmyclucΔA and the ATG of its myc leader, thus leaving 11 bases in that region, creating pT7myc-luc.
The myc-luc fusion cassette (∼1.8 kb) was transferred from pT7myc-luc to the expression vector pSE380 (Invitrogen, Inc.) by copying 1.8 kb (from the RBS of pT7myc-luc through the last codon of luc) in a PCR programmed with forward primer 5′-TCGCTCCATGGAGAAGGAATCCCGCGACGATGCCCCT and reverse primer 5′-ATAGTCCCGGGTTACAATAGCTAAGAATTTCGTCAT. The PCR product was restricted with NcoI and XmaI and subcloned into pSE380, which had been restricted with the same two enzymes. In vitro mutagenesis was then carried out with the oligonucleotides 5′-AATTTCACACTGCAAACAGACCTTGGAGAAGGA and 5′-TCCTTCTCCAAGGTCTGTTTGCAGTGTGAAATT and QuikChange to mutagenize what had become an extra RBS within pSE380 (from AGGA to TGCA) as well as to alter a second ATG codon that occurred in the NcoI site upstream of the ATG codon in the c-myc region (from CCATGG to CCTTGG). The final construct, pSE380myc-luc, contains one RBS and one initiator Met ATG codon (located within the myc leader sequence).
Measurements of growth.
Immediately after diluting the 15-h cultures 1/50, PMO or peptide-PMO was added to a final concentration of 20 μM, except where indicated. Duplicate 100-μl aliquots were transferred to either 1.5-ml microcentrifuge tubes or ultra low attachment 96-well microtiter plates (Corning, Inc., Corning, N.Y.) and incubated with shaking (225 rpm) at either 30 or 37°C. After 8, 15, or 24 h, growth was measured by either the optical density at 600 nm (OD600) or viable cell count, which was done by diluting the cultures and plating them in triplicate on LB plates with or without the appropriate antibiotics. The plates were incubated for 15 to 24 h at 30 or 37°C, and the number of colonies was enumerated by visual inspection.
Measurements of luciferase.
Overnight cultures were diluted and grown for 8 h as described in the preceding paragraph. At various times of growth, the OD600 was measured and aliquots (10 to 50 μl) were removed, mixed with an equal volume of 2× cell culture lysis reagent (Promega, Inc., Madison, Wis.), and incubated at 20°C for 30 min. Luciferase light production was measured as relative light units (RLU) by mixing a small aliquot (1 to 10 μl) of the lysed culture with 25 to 50 μl of luciferase reagent (Promega, Inc.) in a 1.5-ml microcentrifuge tube, incubating the mixture at 20°C for 30 s, and then recording the relative light production in a Cardinal Associates, Inc. (Mt. View, Calif.) model TD-20e luminometer. Each sample was measured 2 to 3 times per experiment, and the mean of the measurements was calculated. Each experiment was done at least two times, and this number is indicated in the figure legends. The percent inhibition of luciferase was calculated by dividing the RLU of lysed culture that was treated with PMO or peptide-PMO by the RLU of the lysed culture that was treated with water, subtracting this value from 1, and multiplying it by 100%. The calculated percent inhibition was normalized by division by the OD of the culture, although differences in ODs were usually less than 5%.
β-Galactosidase.
Overnight cultures of SM101 or SM105 were diluted 1/50 and grown aerobically at 30°C for 24 h in LB broth with or without 20 μM peptide-PMO. β-Galactosidase was measured in duplicate or triplicate for each sample by the method of Miller (13), except that chloroform was substituted for toluene, and volumes were reduced proportionally for use with a 96-well microtiter plate (Ultra low cluster, Costar 3474; Corning, Inc.). After cells were permeabilized with chloroform, 100 μl of aqueous-phase mixture was transferred to the microtiter plate. Substrate (20 μl) and carbonate (100 μl) were added. The OD600 was measured on a Hitachi (Tokyo, Japan) U-2000 spectrophotometer with a path length of 1 cm. Measurements at 420 and 550 nm were made by using a Molecular Devices (Sunnyvale, Calif.) Versamax microtiter plate reader. Each experiment was repeated at least twice, and this number (n) is indicated in the figure legends.
Statistical analysis.
Means were statistically analyzed by using the unpaired, one-sided t test comparing the myc-treated cultures with the corresponding myc control-treated culture.
RESULTS
PMO targeted to reporter gene or 16S rRNA.
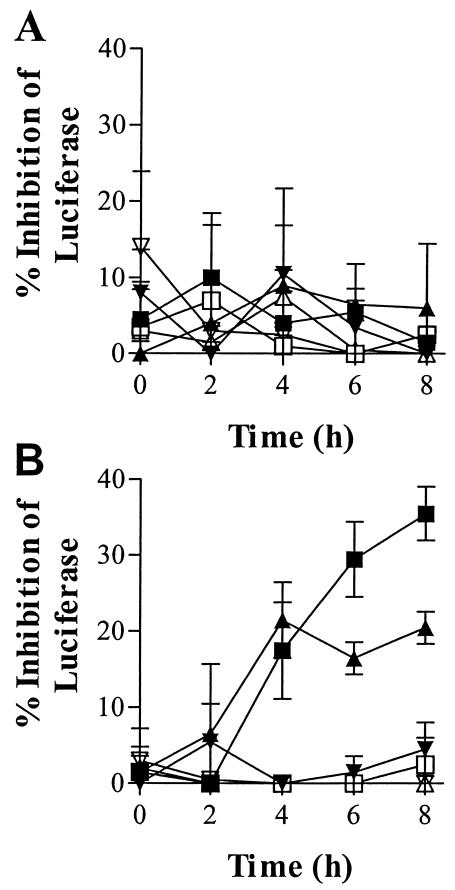
A reporter gene was constructed by fusing the 5′ end of human c-myc with luc, which codes for luciferase. E. coli SM105 was transformed with the reporter in an expression plasmid (pSE380myc-luc) and grown in the presence of various concentrations of myc PMO or a 6-base mismatched control (myc-contr). The sequence of myc PMO is complementary to bases −6 through +14 of the reporter construct. Samples were removed during 8 h of growth and analyzed for luciferase and OD (Fig. 2A). There was no significant difference in either growth rate (data not shown) or luciferase expression among the cultures.
FIG. 2.
Effect of PMO on reporter gene expression. Myc PMO (solid symbols) or a mismatched, negative control (open symbols) at final concentrations of 100 μM (squares), 20 μM (triangles), or 4 μM (inverted triangles) was added to a growing culture of E. coli SM105(pSE380myc-luc) (A) or SM101(pSE380myc-luc) (B). Samples were removed after 0, 2, 4, 6, and 8 h of growth and analyzed for luciferase inhibition. The percent inhibition of luciferase was calculated relative to an identical culture that was not treated with PMO. Error bars show standard deviations for results of two experiments.
A similar kinetic, dose-response analysis was done with a PMO complementary to bases 446 to 466 of 16S rRNA. The results indicate that there was no significant difference in growth rate at any concentration tested (up to 50 μM) between cultures grown in the presence or absence of the 16S PMO (data not shown).
These results show that the myc and 16S PMOs did not significantly inhibit their complementary targets in E. coli.
Permeable strain SM101.
E. coli SM101 was transformed with the myc-luc reporter construct and grown in the presence of various concentrations of myc or control PMOs. SM101 is an isogenic mutant of SM105, with a defect in lipid A biosynthesis that causes the outer membrane to be permeable to high-molecular-weight substances (6, 26). Samples were removed during 8 h of growth and analyzed for luciferase and OD. Myc inhibited luciferase at 20 and 100 μM (Fig. 2B). Inhibition was first evident at 4 h. At the same concentration, myc-contr did not inhibit at any time. A statistical analysis indicates a significant difference (P < 0.05, n = 2) in luciferase values between samples from cultures that contained 20 or 100 μM myc and their corresponding control PMOs at 4 h and a highly significant (P < 0.01) difference at 6 and 8 h.
Myc inhibited in a dose-dependent manner. After 8 h of incubation in the presence of 100, 20, and 4 μM myc PMO, luciferase was inhibited by 36, 20, and 5%, respectively. Luciferase expression in cultures with control PMO was not inhibited at any concentration.
The results suggest that the intact outer membrane of E. coli SM105 restricts entry of PMO, but transport of PMO across the plasma membrane is possible.
Target essential gene.
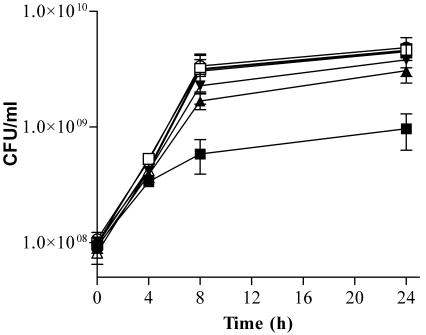
SM101 was grown in the presence of 3 concentrations of a PMO (acp) targeted to the essential gene acpP, which encodes acyl carrier protein. Control cultures of SM101 were grown in the presence of the same concentrations of a PMO with a scrambled base sequence (acp-scr). The cultures were analyzed for viable cells at 0, 4, 8, and 24 h. The acp PMO inhibited growth in a time- and concentration-dependent manner (Fig. 3). Maximum inhibition occurred as the cultures finished the exponential phase and entered the stationary phase (8 h). Inhibition of growth by the acp PMO was highly significant (P < 0.01) at 4, 8, and 24 h for the 40 μM concentration and significant (P < 0.05) at 24 h for the 20 μM concentration.
FIG. 3.
Effect of acp PMO on growth of permeable E. coli SM101. Exponential-phase cultures of E. coli SM101 were mixed with 10 μM (inverted triangles), 20 μM (triangles), or 40 μM (squares) acp PMO (solid symbols) or a scrambled sequence control (open symbols) or with an equivalent volume of water (○) and grown for 24 h. Aliquots of each culture were removed at 0, 4, 8, and 24 h, diluted, and plated for the determination of viable cell counts. Error bars indicate standard deviations of the results of two experiments.
Peptide-PMO conjugates.
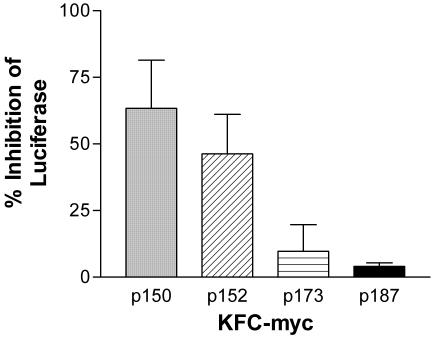
To facilitate the movement of PMO through the outer membrane, the peptide KFFKFFKFFKC(which we abbreviate as KFC) was covalently coupled to myc PMO. A similar peptide is known to permeabilize the outer membrane of E. coli to high-molecular-weight substances (25) and was used to facilitate entry of peptide nucleic acids into E. coli (7). KFC was coupled to both ends of the PMO with a cross-linker that is cleavable (p150) or not cleavable (p152) with reducing agents or to only the 3′ end of myc PMO (p173) with a cleavable cross-linker. A control conjugate (p187) was synthesized by coupling KFC to only the 3′ end of a myc-contr by using the noncleavable cross-linker.
Equal concentrations (20 μM) of peptide-PMO conjugates p150, p152, p173, or p187, the free KFC peptide, or an equivalent volume of water were mixed separately with growing cultures of E. coli BL21(DE3)(pLysS/pT7myc-luc). This strain has a normal, intact outer membrane. After 4 h, viable cells and luciferase were measured. Small differences (within 5%) in OD were used to normalize the luciferase readings. p150, p152, p173, and p187 inhibited luciferase by 63, 46, 10, and 4%, respectively (Fig. 4), relative to the water-treated control. Statistical analysis indicates a highly significant difference (P < 0.01) between the effects of p150 and either p173 or p187, and a significant difference (P < 0.05) between p152 and either p173 or p187. There was no statistically significant difference (P > 0.05) between the effects of p150 and p152.
FIG. 4.
Effects of KFC-myc on intact (nonleaky) E. coli. Three variations of KFC-myc (p150, p152, and p173) and a scrambled sequence KFC-control (p187) PMO were mixed separately (20 μM) with exponentially growing cultures of E. coli BL21(DE3)(pLysS/pT7myc-luc), which expresses the myc-luc reporter construct. After 4 h, the OD of each culture was read, samples of each culture were lysed, and luciferase activity was measured. Inhibition of luciferase activity was calculated relative to an untreated culture. Error bars indicate standard deviations for the results of five experiments for p150 and p152 and two experiments for p173 and p187.
Control cultures that contained 20 μM free peptide failed to grow significantly in 8 h. This shows that the free peptide was toxic and that conjugation of the peptide to the PMO apparently eliminated the toxicity associated with the free peptide.
KFC-lacI PMO.
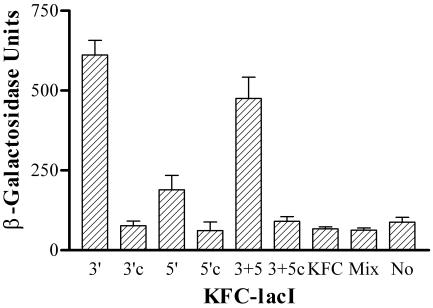
The effects of KFC-myc suggested that attachment of KFC to the 5′ end or to both ends of the PMO produced a more effective gene-specific inhibitor than the 3′ equivalent. To test this further, KFC was coupled with the same, noncleavable cross-linker to the 5′, 3′, or both ends of another PMO that is targeted to lacI and to a scrambled-base control. LacI was targeted because inhibition would produce a positive signal against a low background of β-galactosidase in the absence of inducer, and the target is not expressed from a multicopy plasmid. Each KFC-PMO was added (20 μM total PMO) to a growing culture of SM105. After 24 h of aerobic growth, the OD of the culture and β-galactosidase activity were measured. Each KFC-lacI PMO induced a significantly higher β-galactosidase activity than the corresponding control or mock treatment (Fig. 5). Attachment of KFC to the 3′ or both ends of lacI PMO produced a more effective inducer of β-galactosidase activity than attachment to the 5′ end. Qualitatively similar results were found when the cultures were analyzed at 4 and 8 h of growth (data not shown).
FIG. 5.
Comparison of 3′, 5′, and 3′ plus 5′ site of attachment of KFC to lacI PMO and a mixture of KFC and lacI. Cultures of E. coli SM105 were mixed with various KFC-lacI peptide-PMO conjugates (20 μM), KFC (20 μM), a mixture (Mix) of KFC plus lacI PMO (20 μM each), or neither PMO nor peptide (No) and grown in LB medium at 30°C for 24 h. The end of the PMO that is attached to the peptide is indicated. 3+5 indicates a mixture of peptide-PMO conjugates that included peptide linked to the 3′ and 5′ ends of the peptide and both ends of the PMO, although the total concentration of all forms was 20 μM. Scrambled controls are indicated (c). Error bars indicate standard deviations of the results of three experiments, except for 3+5, which shows the results of two experiments.
Is covalent attachment of KFC necessary, or can β-galactosidase be induced with a mixture of PMO and KFC? Although 20 μM KFC was toxic and slowed growth, cultures treated with 20 μM KFC or a mixture of 20 μM (each) KFC and lacI PMO grew to the same final culture density as an untreated culture after 24 h of growth. β-Galactosidase was not induced in the culture treated with the mixture of free KFC and lacI PMO (Fig. 5).
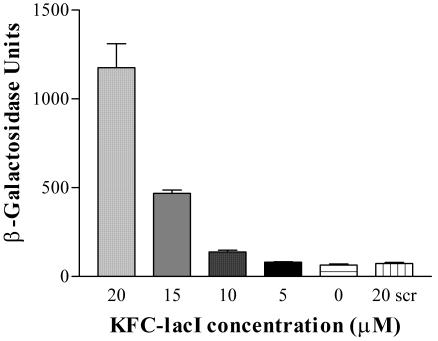
A dose-response analysis was measured with the 3′ KFC-lacI and strain SM105. The results show that induction of β-galactosidase was proportional to the concentration of peptide-PMO (Fig. 6).
FIG. 6.
Dose response. Exponential-phase cultures of E. coli SM105 were mixed with various concentrations of 3′KFC-lacI as indicated and grown in LB medium for 24 h at 30°C. The amount of β-galactosidase was measured. Error bars indicate standard deviations of the results of three experiments.
Effect of peptide sequence.
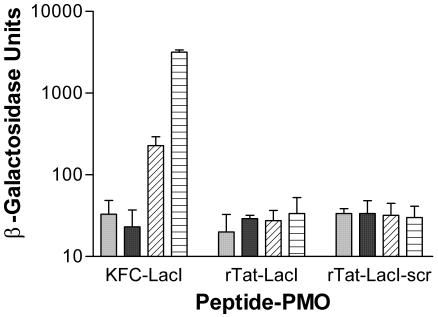
The specificity of KFC to enable covalently attached PMOs to translocate across the outer membrane was tested. Another cationic peptide, but without the hydrophobic residues, rTat (CRRRQRRKKR), was attached to lacI PMO or a scrambled PMO control. The rTat-PMOs were added separately to growing cultures of E. coli SM105. As a positive control, 3′-KFC-lacI was added separately to an identical culture. Neither rTat-PMO induced β-galactosidase, although the positive control induced significant amounts of activity (Fig. 7). Increasing the concentration of rTat-lacI from 20 to 100 μM did not increase induction of β-galactosidase activity (data not shown).
FIG. 7.
Effect of rTat-LacI on β-galactosidase induction. Peptide-PMO conjugates rTat-LacI, rTat-LacI-scr, or 3′KFC-LacI were added at 0 μM (light solid bar), 10 μM (dark solid bar), 20 μM (diagonal striped bar), or 40 μM (horizontal striped bar) to growing cultures of E. coli SM105. After 24 h, the OD and β-galactosidase activity of each culture were measured. Error bars indicate standard deviations of the results of two experiments.
DISCUSSION
We have found that PMOs enter E. coli with a permeable outer membrane (Fig. 2B). The outer membrane of gram-negative bacteria acts as a sieve and excludes polar solutes above about 1,000 Da (15, 16). The size of all PMOs used in this study was about 7,000 Da. The lack of target inhibition in strains with an intact outer membrane (Fig. 2A) suggests that an insufficient amount of PMO gets into the cytoplasm, at least for targets expressed from multicopy plasmids. In other experiments (data not shown), the lacI PMO failed to induce β-galactosidase in intact strains. Because lacI PMO targets the single-copy lacI and small increases in β-galactosidase are easy to detect against a low background signal in uninduced cultures, the results suggest that PMO does not enter the cytoplasm of intact E. coli. We infer that a normal outer membrane prevents high-molecular-weight PMOs from entering the cell.
The results show that the outer membrane barrier was overcome by attaching the peptide KFFKFFKFFKC to PMOs (Fig. 4 to 6). A mixture of KFC and PMO could not substitute for the conjugate of the same two molecules (Fig. 5). This result is similar to that shown for peptide nucleic acids (7). It is inferred from the results that covalently attached KFC peptide enables the KFC-PMO conjugate to traverse the outer membrane. Indeed, the KFC peptide conjugated to peptide nucleic acids has been shown to increase antisense efficacy by increasing the kinetics of outer membrane permeability (4). Regardless of the mechanism, the conjugated KFC-PMO was necessary for inhibition of the target in the cytoplasm of E. coli with an intact outer membrane.
Our results show a response that is directly proportional to the dose of KFC-PMO (Fig. 6). Together with the lack of any response to control conjugates with the same base composition but a different base sequence (Fig. 4 to 6), this shows that the response is a result of the conjugate and that the PMO portion of the conjugate confers specificity to its target.
The sequence of the KFC peptide is important for its role in facilitating delivery of the PMO conjugate into E. coli. Peptides that disrupt the gram-negative outer membrane often share an amphipathic motif of positively charged residues spaced between hydrophobic regions (12, 20, 25, 28). Other cationic peptides that lack amphipathic character, such as the human immunodeficiency virus Tat peptide, facilitate delivery of biologically active molecules into eukaryotic cells (5, 19), but their effects on bacteria are uncharacterized. We showed (Fig. 7) that a positively charged peptide that lacks amphipathic character (CRRRQRRKKR) is insufficient for enabling PMO conjugates to enter E. coli. In comparison to our results with KFC-PMO conjugates (Fig. 4 to 7), this demonstrates that the amino acid sequence of KFC is important for its ability to enable uptake of peptide-PMOs and suggests that the amphipathic character of KFC is important for enabling entry of peptide-PMO conjugates into E. coli. It was shown recently that a KFC-PMO conjugate is nearly unable to access the cytosol of eukaryotic cells (14). These characteristics of KFC make it attractive for the development of conjugated peptide-PMO antibiotics, which ideally would be deliverable into bacteria but not host cells. We speculate that the KFC peptide to which the PMO is attached can be optimized further for improved performance.
In addition to the peptide sequence of the conjugates, we have found that the end of the PMO to which the peptide is attached significantly affects its performance (Fig. 4 and 5). We do not yet understand the conceptual basis for this, but we speculate that this difference may be the result of different rates of transport across the membrane(s), or differences in the interactions with the target mRNA. Because the results (Fig. 4 and 5) differed between KFC-myc and KFC-lacI, it suggests that the base sequence of the PMO may contribute significantly to this effect. However, other factors may account for this result, such as differences in secondary structure of mRNA targets.
Another factor that may have affected the performance of the peptide-PMO was the cross-linker. Peptide-PMO mixtures with a cleavable cross-linker (p150) produced more inhibition than those with a noncleavable cross-linker (p152). One speculative explanation is that upon entry into the cytoplasm, KFC would be cleaved from p150 by the reductive environment. Removal of KFC upon entry into the cytoplasm might affect membrane transport or binding to the target mRNA.
In conclusion, PMOs are attractive for development as an antibiotic because of their genetic specificity and the ease with which they can be customized to any genetic target. This study showed that the strategy of conjugating PMO to a peptide with membrane-permeabilizing properties enabled entry into E. coli. Currently the characteristics of the peptide and its linkage to the PMO are being optimized. Peptide-PMOs may be useful as therapeutic agents and as reagents for functional genomics.
Acknowledgments
We thank Doreen Weller, Murali Reddy, and Susie Hatlevig (AVI BioPharma) for excellent analytical support.
This work was funded by AVI BioPharma, Inc.
REFERENCES
- 1.Arora, V., D. C. Knapp, B. L. Smith, M. L. Statdfield, D. A. Stein, M. T. Reddy, D. D. Weller, and P. L. Iversen. 2000. c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J. Pharmacol. Exp. Ther. 292:921-928. [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Borer, P. N. 1975. Optical properties of nucleic acids, adsorption, and circular dichroism spectra, p. 589. In G. D. Fasman (ed.), Handbook of biochemistry and molecular biology, 3rd ed., vol. 1. Nucleic acids. CRC Press, Cleveland, Ohio.
- 4.Eriksson, M., P. E. Nielsen, and L. Good. 2002. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277:7144-7147. [DOI] [PubMed] [Google Scholar]
- 5.Fawell, S., J. Seery, Y. Daikh, C. Moore, L. L. Chen, B. Pepinsky, and J. Barsoum. 1994. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl. Acad. Sci. USA 91:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galloway, S., and C. R. H. Raetz. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394-6402. [PubMed] [Google Scholar]
- 7.Good, L., S. K. Awasthi, R. Dryselius, O. Larsson, and P. E. Nielsen. 2001. Bacterial antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360-364. [DOI] [PubMed] [Google Scholar]
- 8.Good, L., and P. E. Nielsen. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355-358. [DOI] [PubMed] [Google Scholar]
- 9.Good, L., and P. E. Nielsen. 1998. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. USA 95:2073-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good, L., R. Sandberg, O. Larsson, P. E. Nielsen, and C. Wahlestedt. 2000. Antisense PNA effects in Escherichia coli are limited by the outer membrane LPS layer. Microbiology 146:2665-2670. [DOI] [PubMed] [Google Scholar]
- 11.Hudziak, R., L. Barofsky, D. Barofsky, D. L. Weller, S.-B. Huang, and D. D. Weller. 1996. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 6:267-272. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, P. M., and H. J. Vogel. 1998. Structure-function relationships of antimicrobial peptides. Biochem. Cell Biol. 76:235-246. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 14.Moulton, H. M., M. C. Hase, C. M. Smith, and P. L. Iversen. 2003. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 13:31-43. [DOI] [PubMed] [Google Scholar]
- 15.Nikaido, H. 1993. Transport across the bacterial outer membrane. J. Bioenerg. Biomembr. 25:581-589. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido, H., and M. Vaara. 1997. Outer membrane, p. 7-22. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology. Washington, D.C.
- 17.Qin, G., M. Taylor, Y. Y. Ning, P. Iversen, and L. Kobzik. 2000. In vivo evaluation of a morpholino antisense oligomer directed against tumor necrosis factor-α. Antisense Nucleic Acid Drug Dev. 10:11-16. [DOI] [PubMed] [Google Scholar]
- 18.Rustici, A., M. Velucchi, R. Faggioni, M. Sironi, P. Ghezzi, S. Quataert, B. Green, and M. Porro. 1993. Molecular mapping and detoxification of the lipid A binding site by synthetic peptides. Science 259:361-365. [DOI] [PubMed] [Google Scholar]
- 19.Schwarze, S. R., A. Ho, A. Vocero-Akbani, and S. F. Dowdy. 1999. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285:1569-1572. [DOI] [PubMed] [Google Scholar]
- 20.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 21.Stein, D., E. Foster, S.-B. Huang, D. Weller, and J. Summerton. 1997. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 22.Summerton, J., D. Stein, S. B. Huang, P. Mathews, D. Weller, and M. Partridge. 1997. Morpholino and phosphorothioate antisense oligomers compared in cell-free and in-cell systems. Antisense Nucleic Acid Drug Dev. 7:63-70. [DOI] [PubMed] [Google Scholar]
- 23.Summerton, J., and D. Weller. February 1993. Uncharged morpholino-based polymers having phosphorus containing chiral inter-subunit linkages. U.S. patent 5,185,444.
- 24.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187-195. [DOI] [PubMed] [Google Scholar]
- 25.Vaara, M., and M. Porro. 1996. Groups of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vuorio, R., and M. Vaara. 1992. The lipid A biosynthesis mutant lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 36:826-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wetlaufer, D. B. 1962. Ultraviolet spectra of proteins and amino acids. Adv. Protein Chem. 17:303-390. [DOI] [PubMed] [Google Scholar]
- 28.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]