Abstract
High-level, acquired macrolide resistance in mycobacteria is conferred by mutation within the 23S rRNA gene. However, several mycobacteria are naturally resistant to macrolides, including the Mycobacterium smegmatis group and Mycobacterium tuberculosis complex. Thus, the aim of this study was to characterize this resistance. Intrinsic macrolide resistance in M. smegmatis was inducible and showed cross-resistance to lincosamides but not to streptogramin B (i.e., ML resistance). A similar phenotype was found with Mycobacterium microti and macrolide-resistant Mycobacterium fortuitum. A search of the DNA sequence data for M. smegmatis strain mc2155 identified a novel erm gene, erm(38), and expression analysis showed that erm(38) RNA levels increased >10-fold after a 2-h incubation with macrolide. Inducible ML resistance was not expressed by an erm(38) knockout mutant, and complementation of this mutant with intact erm(38) in trans resulted in high-level ML resistance (e.g., clarithromycin MIC of >512 μg/ml). Thus, the results indicate that erm(38) confers the intrinsic ML resistance of M. smegmatis. Southern blot analysis with an erm(38)-specific probe indicated that a similar gene may be present in macrolide-resistant M. fortuitum. This finding, with the presence of the erm(37) gene (Rv1988) in the M. tuberculosis complex, suggests that such genes are widespread in mycobacteria with intrinsic macrolide resistance.
Most nontuberculosis mycobacteria (NTM) are susceptible to macrolides, such as azithromycin, clarithromycin, and roxithromycin. Consequently, macrolides have become the cornerstones of therapy for NTM infections. Acquired, high-level resistance to macrolides in NTM is conferred by mutation in the 23S rRNA gene (12, 13, 20, 26), although there is some evidence that other mechanisms may be involved (8). However, intrinsic resistance to macrolides is expressed by a range of mycobacteria, the most notable example being Mycobacterium tuberculosis (3, 10). Inherent resistance to macrolides is also common with a range of rapidly growing pathogenic mycobacteria, including the Mycobacterium fortuitum third biovariant complex (sorbitol positive), the Mycobacterium smegmatis group, and Mycobacterium mageritense (5, 6, 24, 25).
The most common mechanism of resistance to macrolides in clinically important bacteria is the presence of a 23S rRNA methylase or erm gene (18). Resistance is conferred by the transfer of one or two methyl groups to an adenine residue in the peptidyltransferase region of the 23S rRNA. Methylation of this site leads to ribosomes with reduced binding of macrolides (18). Expression of erm genes confers cross-resistance to macrolide-lincosamide-streptogramin B (MLS) agents. Resistance to MLS agents is conferred by the expression of several other systems (18), including efflux pumps (e.g., mefA and msrA) and drug-inactivating enzymes (e.g., the ere and mph genes). However, these alternative systems tend to result in resistance restricted to one or two MLS agents.
Thus, the aim of this study was to characterize the intrinsic macrolide resistance of M. smegmatis. Although M. smegmatis is not considered an important human pathogen, it has been associated with disease (16, 17). However, studying this organism may provide an important insight into drug resistance in other mycobacteria of greater clinical importance but that are much less amenable to laboratory study, such as M. tuberculosis.
(This study was presented in part at the 102nd ASM General Meeting, Salt Lake City, Utah, 19 to 23 May 2002 [K. A. Nash, Abstr. 102nd Gen. Meet. Am. Soc. Micorbiol., abstr. A-141, 2002].)
MATERIALS AND METHODS
Bacteria.
M. smegmatis strain mc2155 (22) was provided by W. R. Jacobs, Jr. (Albert Einstein College of Medicine, Bronx, N.Y.). The mycobacterial host for transformation was a recA gene knockout variant (termed recA27) of M. smegmatis mc2155, created by allelic exchange with the suicide vector pKN10 (described below). All American Type Culture Collection designated organisms were obtained from the American Type Culture Collection (Manassas, Va.). Two clinical isolates of M. fortuitum were generously provided by A. E. Rosato (Virginia Commonwealth University, Richmond, Va.), and a clinical isolate of Mycobacterium abscessus (strain MAB30) was isolated at Children's Hospital Los Angeles. The broth medium used was 7HSF, which was comprised of Middlebrook 7H9 broth supplemented with 1 g of Trypticase casein digest (Difco)/liter, 0.05% Tween 80, and 10% oleic acid-albumin-dextrose-catalase (BD Diagnostic Systems, Sparks, Md.).
Susceptibility testing.
Susceptibility to antimicrobial agents was assessed by broth microdilution based on protocols described elsewhere (4). The results were scored after 2 to 4 days for rapidly growing mycobacteria, and after 7 to 14 days for slowly growing mycobacteria. To study inducible resistance, organisms were preincubated either in medium alone or in subinhibitory levels of antimicrobial agent (0.1 and 0.5 times the MIC) prior to assessing susceptibility to MLS and non-MLS agents. Preincubation times ranged between 2 and 20 h. To assess whether resistance was constitutive or inducible, organisms that grew out of the 16-μg/ml clarithromycin wells of a susceptibility assay were harvested for further study. The recovered organisms were washed with saline to remove residual clarithromycin and incubated for 18 h in medium alone or in medium containing 0.125 μg of clarithromycin/ml. After the preincubation, the drug susceptibility of the organisms was reassessed.
Nucleic acid extraction, PCR, and RT-PCR.
Genomic DNA was isolated from mycobacteria by using the method described by Belisle and Sonnenberg (2). Total RNA was extracted from mycobacteria with the Qbiogene (Carlsbad, Calif.) FastRNA kit Blue coupled with the on-column DNase-treatment of the Qiagen (Valencia, Calif.) RNeasy system. Qiagen HotStarTaq DNA polymerase was used for PCR amplification of DNA targets, and the Qiagen OneStep reverse transcriptase (RT) PCR system was used for amplification of RNA targets. Table 1 shows the primers used in this study. Comparison of transcript levels was achieved by a template dilution analysis. Briefly, the RNA preparations were serially diluted (usually in 2- or 10-fold steps), and each series was amplified by RT-PCR. Following RT-PCR, the template dilutions that generated equivalent low yields of amplimer (∼5 ng/μl) were established. The difference in the template dilutions should be equivalent to the difference in target-specific RNA levels between the two RNA preparations. Comparisons were made at low amplimer yields to maximize accuracy by analyzing amplification reactions still in the exponential phase. To improve the confidence in the analysis, all RNA preparations were normalized to the amount of 23S rRNA as assessed by using a low cycle number RT-PCR (15 cycles). The RT-PCR approach was modified to be RNA strand specific by using only one primer during the RT step. The second primer was added during the denaturation step following the RT step.
TABLE 1.
Primers used in the PCR and RT-PCR analysis of M. smegmatis
| Primer | Sequence (5′→3′) | Target |
|---|---|---|
| MSX-1 | ACGAGCTCGGCCAGAACTTCCTGT | erm |
| MSX-2 | CGTCTTCGCGAGGTCCTGACACAA | folD |
| MSX-3 | GGTGAGCGGGGCAGTGGGTAG | erm |
| MSX-4 | CGGCGGATCTGCGTCAGACACT | erm |
| MSX-5 | GGTCACCGCGGACATGGTCAAG | folD-Rv3355 |
| MSX-6 | ACCGACAGCCGCGATGATGATG | folD |
| MS23-1 | CGAATGGCGTAACGACTTCTCA | 23S rRNA |
| MS23-2 | GCGACACCCCTTCCACTCAG | 23S rRNA |
| MSX-4acca | ACCTCCGGACGGCGGATCTGCGTCAGACACT | erm |
| MSX-5bspa | CGATCGATGGTCACCGCGGACATGGTCAAG | erm |
| MSpgaE-1 | CGGTCGGGGATGCTGACAT | pgaE |
| KN10_U2 | CGCAGCACCACCGAGAA | pYUB854 |
| KN10_L1 | AGGCAACTATGGATGAACGAAAT | pYUB854 |
| SACUP-2nota | AAGGCGGCCGCTTGCAATCCAAACGAGAGTCTA | pK19mobsacb |
| SACDN | AATAGGATATCGGCATTTTCTTTTG | pK19mobsacb |
Underlined bases indicate restriction sites (acc, AccIII; bsp, Bsp106I; not, NotI) used to clone the amplimers into the expression vector pMV261.
Cloning and expression of the erm gene.
The putative erm gene was isolated by PCR with the Herculase DNA polymerase mixture (Stratagene, La Jolla, Calif.) with the primers MSX-4acc and MSX-5bsp (Table 1). The resulting 1.4-kbp amplimer was restriction digested with AccIII and Bsp106I (Stratagene) and ligated to similarly restricted pMV261, an Escherichia coli mycobacterial shuttle expression vector (23). This arrangement placed the erm gene expression under the control of the vector's hsp60 promoter, which is constitutively expressed in mycobacteria. This construct was used to transform M. smegmatis.
Disruption of the erm gene by allelic exchange.
A 4.5-kbp suicide vector, pKN10, was created by replacing a nonessential 682-bp region of vector, pYUB854 (generously provided by S. Bardarov, Albert Einstein College of Medicine) with a 1,684-bp region containing the sacB gene of plasmid pK19mobsacb (21). This was achieved by generating amplimers of pYUB854 and pK19mobsacb with primer combinations of KN10_U2/KN10_L1 and SACUP-2not/SACDN (Table 1), respectively. The amplimers were restricted either with NotI (pYUB854) or with NotI-EcoRV (pK19mobsacb), and the resulting products were ligated together.
The erm(38) gene was isolated by PCR with primers MSX-4 and MSX-2. This amplimer was restricted with NcoI and ligated to SspI-NcoI-restricted pKN10, forming plasmid pKNerm-1. A 536-bp deletion in the erm(38) gene was introduced by restricting pKNerm-1 with MluI and SmaI, blunt ending the MluI site, and then recircularizing the plasmid. This construct, pKNermKO-1, was electroporated into M. smegmatis mc2155, and the transformation reaction mixture was spread onto tryptic soy agar containing 50 μg of hygromycin B per milliliter. Organisms that had undergone allelic exchange between the chromosomal erm gene and the mutant erm gene (Δerm) of pKNermKO-1 were enriched by sucrose counterselection as described elsewhere (14, 15). One variant (ermKO4) was chosen, and the site of the deletion was confirmed by DNA sequencing. Loss of the suicide vector was assessed by Southern analysis.
Southern analysis.
Five micrograms of DNA was digested overnight with the restriction enzyme Bsp106I. The restricted DNA was subjected to Southern blot analysis with a probe specific for the M. smegmatis erm gene. The probe was a PCR product (primers MSX-1 and MSX-3) (Table 1) labeled with biotin with the BrightStar psoralen-biotin nonisotopic labeling kit (Ambion Inc., Austin, Tex.). Hybridization of the probe and blot occurred at 42°C with ULTRAhyb hybridization buffer (Ambion). Following one wash in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% sodium dodecyl sulfate and two washes in 0.1× SSC and 0.5% sodium dodecyl sulfate at 50°C, the bound probe was detected by use of the BrightStar BioDetect nonisotopic detection kit (Ambion) and exposure to Kodak BioMax Light film.
Sequence data.
Preliminary sequence data for M. smegmatis strain mc2155 was obtained from The Institute for Genomic Research website at http://www.tigr.org. The finalized sequence data for M. tuberculosis strain H37Rv (7) was obtained from the Sanger Centre website at http://www.sanger.ac.uk/Projects/M_tuberculosis/. Blast searching (1) of these genomes was through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/).
RESULTS
Intrinsic macrolide resistance.
M. smegmatis mc2155 grown in the absence of antimicrobial agents expressed a macrolide-susceptible phenotype, with MICs of clarithromycin and azithromycin of 2 and 32 μg/ml, respectively (Table 2). However, preincubating the organisms for 2 h with 0.125 μg of clarithromycin/ml resulted in a 16-fold increase in the MIC of clarithromycin and an 8-fold increase in the MIC of azithromycin (Table 2). In contrast, susceptibility to the other tested agents was unaffected by the clarithromycin treatment.
TABLE 2.
Effect of preincubation with clarithromycin (0.125 μg/ml) on susceptibility of M. smegmatis mc2155 to a range of antimicrobials
| Initial phenotypea | Preincubation medium (time [h]) | MIC (μg/ml)b
|
|||||
|---|---|---|---|---|---|---|---|
| AMK | AZM | CIP | CLR | EMB | RBT | ||
| Susceptible | Medium alone (2) | 1 | 32 | 0.25 | 2 | 1 | 1 |
| Clarithromycin (2) | 1 | 256 | 0.5 | 32 | 1 | 1 | |
| Resistant | Medium alone (18) | 1 | ND | 0.5 | 2 | 1 | 1 |
| Clarithromycin (18) | 1 | ND | 0.5 | >64 | 1 | 1 | |
Susceptible organisms were maintained in the absence of antimicrobial agent. Resistant organisms were harvested from the wells of a previous susceptibility assay containing 16 μg of clarithromycin per ml.
AMK, amikacin; AZM, azithromycin; CIP, ciprofloxacin; CLR, clarithromycin; EMB, ethambutol; RBT, rifabutin; ND, not determined.
The short preincubation time (i.e., less than the M. smegmatis generation time of 2.5 h) required to change the susceptibility to macrolides suggested that the resistant phenotype was inducible. Alternatively, the macrolide preincubation may have selected a constitutively resistant subpopulation of organisms. To distinguish these two possibilities, the resistant organisms that grew out of the 16-μg/ml clarithromycin wells of a susceptibility assay were harvested and used to determine whether the resistance was constitutive or transient (Table 2).
After an 18-h incubation in the absence of macrolide, the resistant organisms reverted to the susceptible state, i.e., a MIC of clarithromycin of 2 μg/ml (Table 2). In contrast, the MIC of clarithromycin increased to >64 μg/ml for the organisms maintained in clarithromycin. The incubation conditions had no effect on the susceptibility to the nonmacrolide agents. The relatively long incubation period in the absence of drug (i.e., ∼7 generations) should have both reduced the likelihood of complications caused by a postantibiotic effect and diluted out (by cell division) any induced resistance elements. However, this recovery period should have had little impact on constitutively expressed resistance. Thus, the results were consistent with the expression of inducible macrolide resistance.
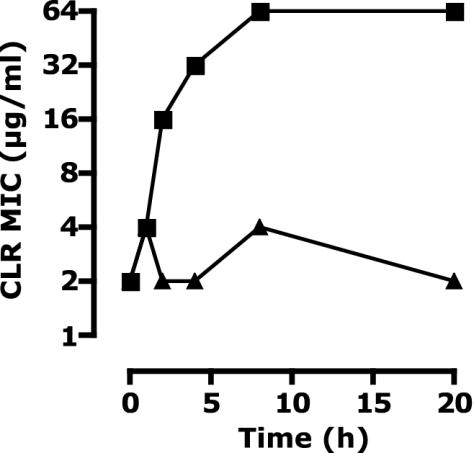
To characterize the induction kinetics of the macrolide resistance, clarithromycin susceptibility was monitored over time for organisms cultured in either medium alone or medium containing 0.125 μg of clarithromycin/ml (Fig. 1). Extending the induction resulted in an increase in the MIC of clarithromycin, reaching a steady state after 8 h. This prolonged induction of the resistance was consistent with a relatively slow accumulation of a resistance determinant (e.g., modified ribosomes) rather than the expression of a drug-degrading enzyme or a drug export system.
FIG. 1.
The change in the MIC of clarithromycin (CLR) over time for M. smegmatis mc2155 cultures incubated either in medium alone (triangles) or in medium containing 0.125 μg of clarithromycin/ml (squares).
Inducers and the scope of resistance.
To explore the characteristics of the macrolide resistance further, the susceptibility of clarithromycin-induced organisms to a wider range of MLS agents was assessed (Table 3). The clarithromycin-induced organisms expressed cross-resistance to clarithromycin, erythromycin, and spiramycin (macrolides) and clindamycin (a lincosamide). However, increased resistance was not expressed to quinupristin (streptogramin B), dalfopristin (streptogramin A), or rifabutin (a rifamycin).
TABLE 3.
MICs of MLS and non-MLS agentsa for M. smegmatis following overnight incubation with and without clarithromycin
| Preincubation medium | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
| MLS agent
|
Non-MLS agent
|
||||||
| CLN | CLR | ERY | Q | SPM | D | RBT | |
| Medium alone | 32 | 1 | 64 | 64 | 8 | 128 | 0.5 |
| Clarithromycin (0.125 μg/ml) | 1,024 | 256 | 1,024 | 64 | 512 | 128 | 0.5 |
CLN, clindamycin; CLR, clarithromycin; D, dalfopristin; Q, quinupristin; SPM, spiramycin; RBT, rifabutin; ERY, erythromycin.
Clarithromycin resistance (MIC > 64 μg/ml) was induced by preincubation with clindamycin, erythromycin, and spiramycin as well as with clarithromycin. In contrast, M. smegmatis remained susceptible to clarithromycin (MIC < 4 μg/ml) following overnight preincubation in amikacin, ciprofloxacin, dalfopristin, ethambutol, quinupristin, and rifabutin. Thus, induction of macrolide resistance was not a general antimicrobial stress response. Rather, it was associated with the nature of the resistance.
Inducible macrolide resistance in other mycobacteria.
Inducible resistance was assessed in a range of mycobacteria and defined as a >2-fold-higher MIC of clarithromycin in organisms preincubated overnight in clarithromycin (at ≤0.5 times the MIC) compared to organisms preincubated in medium alone. No evidence of inducible resistance was found for M. abscessus (MAB30), Mycobacterium africanum ATCC 25420, Mycobacterium avium 101, Mycobacterium bovis ATCC 35734 (BCG Pasteur), Mycobacterium chelonae ATCC 35752, Mycobacterium flavescens ATCC 14474, Mycobacterium haemophilum ATCC 29548, Mycobacterium kansasii ATCC 12478, Mycobacterium marinum ATCC 927, Mycobacterium peregrinum ATCC 14467, Mycobacterium phlei ATCC 354, Mycobacterium terrae ATCC 15755, and Mycobacterium xenopi ATCC 19250. However, M. africanum did constitutively express an intermediate level of resistance to clarithromycin (MIC, 8 to 16 μg/ml). This is typical of members of the M. tuberculosis complex.
In contrast to the other mycobacteria, M. fortuitum (strain ATCC 6841 and 2 clinical isolates), M. microti ATCC 19422, and M. smegmatis (strains mc2155 and ATCC 14468) cultures preincubated with macrolide all presented with a MIC of clarithromycin which was >32-fold higher than that of controls.
Further analysis of the resistance demonstrated that M. smegmatis ATCC 14468 expressed an inducible macrolide-lincosamide (ML) phenotype, similar to strain mc2155 (data not shown). Although clarithromycin resistance in M. fortuitum and M. microti crossed to spiramycin and clindamycin, the high noninduced MIC of quinupristin (256 μg/ml) precluded a satisfactory interpretation of whether these organisms expressed MLS resistance.
Sequence analysis of the M. smegmatis genome.
The DNA sequence data for M. smegmatis was searched for the presence of a possible rRNA methylase by using two consensus amino acid sequences, rADc and RrnaAD (defined in the conserved domain database at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
This analysis identified a theoretical protein in M. smegmatis with >30% identity to 21 rRNA methylases associated with macrolide resistance. Figure 2 shows the two best-fit alignments: with Erm(31) (PikR2) of Streptomyces venezuelae (57% identity; GenBank accession no. T17407) and with Erm(X) of Corynebacterium jeikeium strain CJ21 (53% identity; GenBank accession no. AAK28907). Since the M. smegmatis gene is <80% identical to any other known erm gene, it represents a new erm gene class following the guidelines of Roberts et al. (18). The sequence for this gene, designated erm(38), has been deposited with the erm registry at http://faculty.washington.edu/marilynr/, which is maintained by Marilyn C. Roberts.
FIG. 2.
ClustalW alignment of the RNA methylases of M. smegmatis mc2155 (MS_Erm), C. jeikeium CJ21 [Erm(X), GenBank accession no. AAK28907], and S. venezuelae [Erm(31), GenBank accession no. T17407]. The bar above the alignment indicates a possible fusion (insertion) site for the erm gene within the M. smegmatis genome.
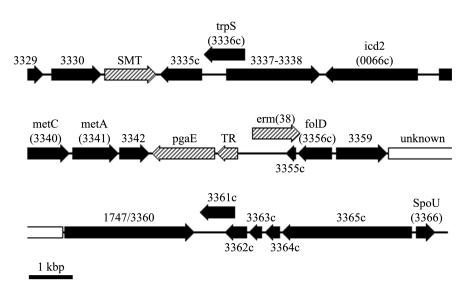
A search of 30 kbp of the DNA sequence data surrounding the erm(38) gene failed to identify any known or putative mobile elements (Fig. 3). In addition, this analysis indicated that there were no other drug resistance genes in the locale of the erm gene.
FIG. 3.
Genetic organization in the region of erm(38) for M. smegmatis mc2155 (contig 3311, complement of bases 1,221,668 to 1,251,667). Homologues of M. tuberculosis genes are indicated by block shading (black) and by numbering equivalent to the Rv designation of strain H37Rv. The crosshatched open reading frames are similar to genes in other bacteria but not to known mycobacterial sequences. SMT, putative small-molecule (cation) transporter (C terminus of protein shows 49% identity with a probable transporter of Pseudomonas aeruginosa [accession no. C83410]); pgaE, putative polyketide oxygenase (34% amino acid identity to PgaE of a Streptomyces sp. strain [accession no. AAK57522]); TR, putative transcriptional regulator (45% amino acid similarity to pfam00440, the TetR transcriptional regulator conserved domain; 34% identical to CalR1, a calicheamicin synthesis regulator [accession no. AAM94766]). The open box indicates a region with no convincing similarity to any other known amino acid sequence (i.e., <20% identity).
The genome surrounding erm(38) shows many open reading frames with a convincing degree of identity (>50%) to M. tuberculosis H37Rv. Alignment of the amino acid sequences of the M. smegmatis and M. tuberculosis homologues showed a median identity of 72% (range, 54 to 87%) with 95% (range, 78 to 98%) of residues being related (i.e., identical and similar). Furthermore, the gene order is largely conserved. However, there are several places within the erm(38) locale that are divergent from other known mycobacterial sequences. The most notable are the pgaE, the putative transcriptional regulator, and erm(38) genes. The three encoded amino acid sequences show convincing similarity (>30% identity) to genes of Streptomyces species, although the three genes are not adjacent in any other known DNA sequence.
Perhaps the most intriguing detail about the folD region in M. smegmatis was that erm(38) appears to span the Rv3355 gene and partially overlap the folD gene, albeit in the opposite orientation. This suggested that the erm gene represents a fusion between the source erm gene and part of the M. smegmatis chromosome. Based on an alignment of the Rv3355 gene homologues and the surrounding DNA of M. tuberculosis and M. smegmatis, a possible fusion site appeared to be between codons 258 and 261 of the erm(38) gene (Fig. 2).
Expression of erm(38).
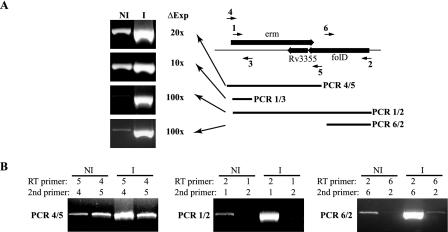
Fig. 4A shows RT-PCR analysis of RNA preparations isolated from M. smegmatis incubated for 2 h in either medium alone or medium containing 1 μg of erythromycin/ml. The level of RNA associated with the erm, Rv3355, and folD genes was assessed by comparative RT-PCR with four amplification reactions (PCRs 1/2, 1/3, 4/5, and 6/2; numbers refer to the MSX primers listed in Table 1). All four RT-PCRs indicated that exposure to a macrolide resulted in a 10- to 100-fold increase in the RNA levels of the erm-folD region. However, these results may not accurately reflect the change in erm gene expression following macrolide exposure, since the amplification reactions may detect Rv3355 and folD transcripts.
FIG. 4.
(A) RT-PCR analysis of erm(38) expression in M. smegmatis mc2155 either noninduced (NI) or induced (I) for 2 h with 1 μg of erythromycin/ml. The difference in expression (ΔExp) was assessed by RNA dilution analysis. (B) Analysis of the different transcript orientations by RT-PCR with only a single primer during the RT step; the second primer was added during the post-RT denaturation step.
Distinction of the erm(38) transcript from the Rv3355 and folD transcripts was accomplished by using an RNA strand-specific RT-PCR. To distinguish the two possible RNA orientations, selected amplification reactions (1/2, 4/5, and 6/2) were set up with only one primer in the RT step. The second primer was added during the denaturation step following the RT reaction. Figure 4B presents the results of these analyses. The negative (no template) control reactions failed to generate any detectable amplimers (data not shown).
The erm gene transcript-specific amplification reactions (RT primer 2 or 5) confirmed that the level of this RNA strand was >10-fold higher in the organisms incubated with macrolide. Furthermore, the results for the RT-PCR with primers 1 and 2 (gel image in center) indicated that the erm gene transcript was at least 1.9 kb long and completely spanned the folD gene. This suggests that expression of erm(38) may affect expression of Rv3355 and folD by antisense interference.
Analysis of the Rv3355 RNA strand (RT primer 4 and second primer 5) (Fig. 4B, left) demonstrated that the RNA spanning this gene was expressed at equivalent levels in the noninduced and macrolide-induced organisms. Furthermore, assuming the amplification efficiency was independent of which primer was used in the RT step, the result suggested that the Rv3355 transcript was at a slightly higher level than the erm(38) transcript in the noninduced organisms. Evidence of a folD-spanning transcript came from the strand-specific RT-PCR with RT primer 6 and second primer 2 (Fig. 4B, right), evidenced by a faint band in the macrolide-induced material.
Cloning of the M. smegmatis erm gene.
To confirm that erm(38) can confer ML resistance, this gene was expressed in trans in M. smegmatis recA27 [i.e., with a functional chromosomal copy of erm(38)]. The susceptibilities of organisms carrying the extrachromosomal copies of erm(38), and organisms carrying the vector alone, are shown in Table 4. The organisms were not preincubated in erm gene-inducing agent (i.e., ML agent) prior to assessing drug susceptibilities. Clearly, constitutive expression of the erm gene from a multicopy plasmid increased the MICs of the ML agents but not of quinupristin, dalfopristin, or rifabutin (included to control for nonspecific effects). Thus, expression of erm(38) in trans conferred a similar phenotype as clarithromycin-induced M. smegmatis (Table 3). In support of this conclusion, the erm(38) knockout (Δerm) variant was more susceptible to the ML agents (except spiramycin), and the MICs did not increase following overnight incubation with subinhibitory concentrations of clarithromycin or erythromycin. Reconstituting the Δerm mutant with intact erm(38) in trans conferred high MICs of the ML agents.
TABLE 4.
MICs of MLS and non-MLS agentsb for M. smegmatis expressing erm(38) in trans, an erm(38) knockout mutant, and a 23S rRNA gene mutant
| Variant | Genotypea | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| MLS agent
|
Non-MLS agent
|
||||||
| CLNc | CLR | Q | SPM | D | RBT | ||
| MV261 (WT) | pMV261 | 32 | 1 | 64 | 8 | 128 | 0.5 |
| MVACCerm1.4 | pMV261::erm | >1,024 | >512 | 64 | >1,024 | 128 | 0.5 |
| ermKO_4 | Δermd | 4 | 0.125 | 64 | 8 | 128 | 0.5 |
| MVOTERM-1 | Δerm, pMV261::erm | >1,024 | >512 | 64 | >1,024 | 128 | 0.5 |
| CLR-1 | Δerm, rrn mutant | >1,024 | >512 | 64 | >1,024 | 128 | 0.5 |
pMV261::erm, erm(38) expressed in trans from pMV261 expression vector; Δerm, erm(38) knockout variant; rrn mutant, A2058→C 23S rRNA gene mutation.
CLN, clindamycin; CLR, clarithromycin; D, dalfopristin; Q, quinupristin; SPM, spiramycin; RBT, rifabutin.
MICs of clindamycin and erythromycin were identical.
MICs did not change for the erm mutant following 18 h of incubation either in 0.06 μg of clarithromycin/ml or in 1 μg of erythromycin/ml.
The MLS resistance profile of an M. smegmatis variant (CLR-1) carrying an A2058→C mutation in the 23S rRNA gene was similar to that conferred by expression of erm(38). This result suggests that the binding site of quinupristin does not overlap the A2058 residue of the 23S rRNA and thus has a significantly different target site from that of ML agents. This provides a explanation for why erm(38) confers ML resistance without streptogramin B resistance.
Species distribution of the erm(38) gene.
An erm(38)-specific PCR (primers MSX-1 and MSX-3) was used to screen DNA preparations isolated from a selection of mycobacteria. An amplification product was generated with the M. smegmatis ATCC 14468 DNA but not with DNA isolated from M. fortuitum ATCC 697, M. abscessus MAB30, M. chelonae ATCC 35753, M. microti ATCC 19422, M. avium 101, or M. bovis BCG (data not shown). This was particularly interesting since M. fortuitum and M. microti were found to have an inducible ML-resistant phenotype. Sequencing of the first 100 codons of the M. smegmatis ATCC 14468 putative erm gene showed that it was 100% identical to the erm gene of strain mc2155 (GenBank accession no. AY154656 and AY154657).
Using PCR to assess for the presence of a gene or homologue can be misleading because this technology can be affected by minor sequence differences, especially at the primer binding sites. Therefore, Southern blot analysis was applied by using a probe derived from an erm(38)-specific amplification product.
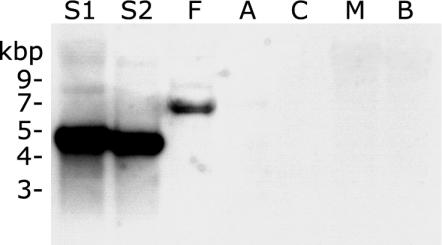
As expected, an erm-specific band (slightly smaller than 5 kbp) was detected in the DNA of the two M. smegmatis strains (Fig. 5); the expected size of the Bsp106I fragment containing the erm(38) gene was 4.7 kbp. Interestingly, an ∼7-kbp band was detected in the M. fortuitum DNA. This suggests that DNA with a significant degree of identity to erm(38) gene is present in M. fortuitum. Clearly, this was consistent with the inducible phenotype of M. fortuitum.
FIG. 5.
Southern analysis with an erm(38)-specific probe. Lanes: S1, M. smegmatis mc2155; S2, M. smegmatis ATCC 14468; F, M. fortuitum ATCC 6841; A, M. abscessus MAB30; C, M. chelonae ATCC 35752; M, M. microti ATCC 19422; B, M. bovis BCG.
DISCUSSION
Intrinsic macrolide resistance is common in several pathogenic mycobacterial groups, e.g., M. tuberculosis complex, M. smegmatis group, and M. fortuitum third biovariant complex (5, 6, 24). This report describes the role of a novel erm gene, erm(38), in the intrinsic resistance of M. smegmatis and shows that the phenotype is inducible. This M. smegmatis erm gene is distinct from the erm(37) gene (Rv1988) identified in the M. tuberculosis genome (K. Buriankova, J.-L. Pernodet, O. Dorson, J. Weiser, J.-C. Ghnassia, and F. Doucet-Populaire, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1816, 2001).
The resistance conferred by erm genes tends to cross to members of the MLS group of agents; thus, it is intriguing that erm(38) did not confer resistance to quinupristin (streptogramin B). One possibility is that Erm(38) is a monomethylase, as monomethylation of 23S rRNA confers increasing levels of resistance in the order of streptogramin B < macrolide < lincosamide (9). However, the high-level resistance to both macrolides and lincosamides conferred by Erm(38) (MICs of clindamycin and erythromycin were similar) is consistent with this protein being a dimethylase (as are most clinically important Erm alleles) (18). Furthermore, mutation at position A2058 of the 23S rRNA in M. smegmatis did not confer increased resistance to quinupristin. This suggests that the binding site of streptogramin B in mycobacteria does not overlap the A2058 residue of 23S rRNA, which is the methylation site of Erm enzymes (18).
In evolutionary terms, an important question is why the erm gene is present in M. smegmatis? Although M. smegmatis can cause disease in humans (16, 17), infections are uncommon and thus it seems unlikely that the erm gene was acquired as a response to antimicrobial therapy. This suggests that evolutionary pressures in the normal environment of this organism are responsible for the selection and maintenance of the erm gene. Perhaps it is not surprising, therefore, that M. smegmatis and some antimicrobial agent-producing bacteria (e.g., Streptomyces) share an ecological niche, i.e., soil. Thus, it is possible that in this shared environment, the erm gene provides a fitness benefit to M. smegmatis.
Many erm genes, including erm(X) of C. jeikeium (19), are associated with mobile elements (e.g., plasmids and transposons). Thus, an important finding about the M. smegmatis erm gene is that it is not in the proximity of any known or putative mobile elements. However, the site of the erm gene on the chromosome (i.e., spanning the Rv3355 and folD genes) suggests that it was inserted from an exogenous source, possibly by recombination. Evidence of insertion without an adjacent transposon or integrase was found for erm(X) in C. jeikeium strain CJ12 (19).
Perhaps a more important medical issue is whether erm genes are widespread in mycobacteria. Certainly, the Southern analysis presented here suggests that at least some M. fortuitum strains possess DNA with a significant level of identity to the erm(38) gene. Clinically, M. tuberculosis is the most important Mycobacterium species, and isolates tend to present with an intermediate level of macrolide resistance (MIC of clarithromycin, 16 to 32 μg/ml) that is not associated with a 23S rRNA mutation (13). Although there is no evidence that the erm(38) gene is present in members of the M. tuberculosis complex, within the M. tuberculosis H37Rv genome is another putative erm gene, erm(37), or Rv1988. This gene is present in other members of the M. tuberculosis complex, including M. bovis, M. africanum, and M. microti (K. A. Nash, unpublished data), but is absent from the M. bovis BCG genome, as it is within the RD2 deletion region (11). The finding of putative erm genes in divergent mycobacteria suggests that these entities may be widespread in this genus.
Clearly, erm genes are relevant to drug resistance in mycobacteria, which usually acquire resistance by mutation within endogenous genes or regulatory regions (27). Further study, including the putative erm gene of the M. tuberculosis complex, will lead to a better understanding of the factors that affect the antimycobacterial activity of macrolides and may lead to the development of new macrolide antimycobacterial agents.
Acknowledgments
I thank Adriana Rosato for assistance in this study and Clark B. Inderlied and Marilyn C. Roberts for suggestions in preparing the manuscript.
Funding for this study was provided by the Department of Pathology and Laboratory Medicine, Children's Hospital Los Angeles, and by the NIH/NIAID grant RO1 AI052291-01. Sequencing of M. smegmatis strain mc2155 was accomplished with support from the U.S. National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 101:31-44. [DOI] [PubMed] [Google Scholar]
- 3.Bosne-David, S., V. Barros, S. C. Verde, C. Portugal, and H. L. David. 2000. Intrinsic resistance of Mycobacterium tuberculosis to clarithromycin is effectively reversed by subinhibitory concentrations of cell wall inhibitors. J. Antimicrob. Chemother. 46:391-395. [DOI] [PubMed] [Google Scholar]
- 4.Brown, B. A., J. M. Swenson, and R. J. Wallace, Jr. 1993. Broth microdilution MIC test for rapidly growing mycobacteria, p. 5.11.1. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, D.C.
- 5.Brown-Elliott, B. A., D. E. Griffith, and R. J. Wallace, Jr. 2002. Newly described or emerging human species of nontuberculous mycobacteria. Infect. Dis. Clin. North Am. 16:187-220. [DOI] [PubMed] [Google Scholar]
- 6.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Doucet-Populaire, F., C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1995. Acquired resistance in Mycobacterium avium complex strains isolated from AIDS patients and beige mice during treatment with clarithromycin. J. Antimicrob. Chemother. 36:129-136. [DOI] [PubMed] [Google Scholar]
- 9.Liu, M., and S. Douthwaite. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luna-Herrera, J., V. Reddy, D. Daneluzzi, and P. Gangadharam. 1995. Antituberculosis activity of clarithromycin. Antimicrob. Agents Chemother. 39:2692-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier, A., P. Kirschner, B. Springer, V. A. Steingrube, B. A. Brown, R. J. Wallace, Jr., and E. C. Böttger. 1994. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob. Agents Chemother. 38:381-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash, K. A., and C. B. Inderlied. 1995. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob. Agents Chemother. 39:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavelka, M. S., Jr., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 16.Pennekamp, A., G. E. Pfyffer, J. Wuest, C. A. George, and C. Ruef. 1997. Mycobacterium smegmatis infection in a healthy woman following a facelift: case report and review of the literature. Ann. Plast. Surg. 39:80-83. [DOI] [PubMed] [Google Scholar]
- 17.Pierre-Audigier, C., E. Jouanguy, S. Lamhamedi, F. Altare, J. Rauzier, V. Vincent, D. Canioni, J. F. Emile, A. Fischer, S. Blanche, J. L. Gaillard, and J. L. Casanova. 1997. Fatal disseminated Mycobacterium smegmatis infection in a child with inherited interferon gamma receptor deficiency. Clin. Infect. Dis. 24:982-984. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosato, A. E., B. S. Lee, and K. A. Nash. 2001. Inducible macrolide resistance in Corynebacterium jeikeium. Antimicrob. Agents Chemother. 45:1982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander, P., T. Prammananan, and E. C. Bottger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 21.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 22.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 23.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 24.Wallace, R. J., Jr., B. A. Brown, V. A. Silcox, M. Tsukamura, D. R. Nash, L. C. Steele, V. A. Steingrube, J. Smith, G. Sumter, Y. S. Zhang, et al. 1991. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 163:598-603. [DOI] [PubMed] [Google Scholar]
- 25.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace, R. J., Jr., A. Meier, B. A. Brown, Y. Zhang, P. Sander, G. O. Onyi, and E. C. Bottger. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-254. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. American Society for Microbiology, Washington, D.C.