Abstract
We designed, synthesized, and identified UIC-94017 (TMC114), a novel nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitor (PI) containing a 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane (bis-THF) and a sulfonamide isostere which is extremely potent against laboratory HIV-1 strains and primary clinical isolates (50% inhibitory concentration [IC50], ∼0.003 μM; IC90, ∼0.009 μM) with minimal cytotoxicity (50% cytotoxic concentration for CD4+ MT-2 cells, 74 μM). UIC-94017 blocked the infectivity and replication of each of HIV-1NL4-3 variants exposed to and selected for resistance to saquinavir, indinavir, nelfinavir, or ritonavir at concentrations up to 5 μM (IC50s, 0.003 to 0.029 μM), although it was less active against HIV-1NL4-3 variants selected for resistance to amprenavir (IC50, 0.22 μM). UIC-94017 was also potent against multi-PI-resistant clinical HIV-1 variants isolated from patients who had no response to existing antiviral regimens after having received a variety of antiviral agents. Structural analyses revealed that the close contact of UIC-94017 with the main chains of the protease active-site amino acids (Asp-29 and Asp-30) is important for its potency and wide spectrum of activity against multi-PI-resistant HIV-1 variants. Considering the favorable pharmacokinetics of UIC-94017 when administered with ritonavir, the present data warrant that UIC-94017 be further developed as a potential therapeutic agent for the treatment of primary and multi-PI-resistant HIV-1 infections.
Combination therapy or highly active antiretroviral therapy (HAART) with two or more reverse transcriptase inhibitors and protease inhibitors (PIs) has dramatically improved the quality of life and survival of patients infected with human immunodeficiency virus (HIV) type 1 (HIV-1) (15, 16, 23, 28). However, the ability to provide effective long-term antiretroviral therapy for HIV-1 infection has become a complex issue, since 40 to 50% of those who initially achieved favorable viral suppression to undetectable levels have experienced treatment failure (7, 21). Moreover, 10 to 40% of antiviral therapy-naive individuals infected with HIV-1 have persistent viral replication (plasma HIV RNA load, >500 copies/ml) while they are receiving HAART (5, 12, 26), possibly due to the transmission of drug-resistant HIV-1 variants (22, 27, 29). In addition, it is evident that with these anti-HIV drugs, only partial immunologic reconstitution is attained in patients with advanced HIV-1 infection.
In the development of new anti-HIV-1 therapeutics, we have faced a variety of challenges different from those faced in the design of the first-line drugs, invoking thoughts about selection pressure mechanisms, in addition to the conventional issues of potency, pharmacology, safety, and mechanism of drug action (3, 4, 17). The issue of the emergence of drug-resistant HIV-1 variants is one of the most formidable challenges in the era of HAART. Indeed, it is of note that the very features that contribute to the specificity and efficacy of reverse transcriptase inhibitors and PIs provide the virus with a strategy to develop resistance (3, 14), and it seems inevitable that this resistance issue will remain problematic for years to come.
A number of studies indicate that cross-resistance is a major obstacle inherent to antiviral therapy with PIs (9, 11). This observation should not be a surprise since all the inhibitors were designed to bind to the wild-type enzyme. Mutations have been found in every subsite of HIV-1 protease; however, not every subsite residue has been found to mutate for resistance to certain drugs. In this respect, novel PIs targeting the critical sites (or atoms) of the enzyme other than those sites (or atoms) with which the available PIs interact should be active against HIV-1 variants resistant to existing PIs.
The present paper presents the first results of antiviral analyses of a novel PI, UIC-94017 (TMC114), containing a 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane (bis-THF) and a sulfonamide isostere which is extremely potent against a wide spectrum of HIV strains in vitro, including a variety of multi-PI-resistant clinical strains. We also describe the results of crystallographic analysis of UIC-94017, which revealed that the close contact of UIC-94017 with the main chains of the protease active-site amino acids (Asp-29 and Asp-30), which differs from the binding profiles of the PIs available at present, is important for its potency and wide spectrum of activity against multi-PI-resistant HIV-1 variants.
MATERIALS AND METHODS
Cells and viruses.
MT-2 and MT-4 cells were grown in an RPMI 1640-based culture medium supplemented with 15% fetal calf serum (FCS; HyClone Laboratories, Logan, Utah), 50 U of penicillin per ml, and 50 μg of streptomycin per ml. The following HIV strains were used for the drug susceptibility assay (see below): HIV-1LAI, HIV-1Ba-L, HIV-1NL4-3, HIV-2EHO, HIV-2ROD, two clinical HIV-1 strains from drug-naive patients with AIDS (HIV-1ERS104pre and HIV-1MOKW) (13, 25), and seven HIV-1 clinical isolates that were originally isolated from patients with AIDS who had received 9 to 11 anti-HIV-1 drugs over the past 32 to 83 months and that were genotypically and phenotypically characterized as multi-PI-resistant HIV-1 variants (30, 31).
Antiviral agents.
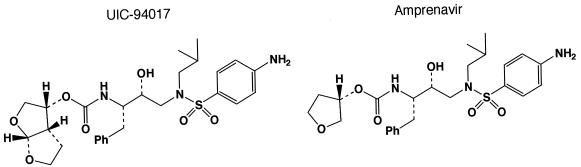
UIC-94017 (TMC114) (Fig. 1), a novel nonpeptidic PI containing a bis-THF and a sulfonamide isostere, was designed and synthesized in a convergent manner by coupling an optically active P2-bis-THF ligand and an (R)-hydroxyethylamino sulfonamide isostere. Detailed synthetic methods for UIC-94017 will be described elsewhere by A. K. Ghosh et al. 3′-Azido-2′,3′-dideoxythymidine (AZT; zidovudine) was purchased from Sigma (St. Louis, Mo.). Saquinavir (SQV) and ritonavir (RTV) were kindly provided by Roche Products Ltd. (Welwyn Garden City, United Kingdom) and Abbott Laboratories (Abbott Park, Ill.), respectively. Amprenavir (APV) was a kind gift from Glaxo-Wellcome, Research Triangle Park, N.C. Nelfinavir (NFV), indinavir (IDV), and lopinavir (LPV) were kindly provided by Japan Energy Inc., Tokyo, Japan.
FIG. 1.
Structures of UIC-94017 and amprenavir.
Drug susceptibility assay.
The susceptibilities of HIV-1LAI, HIV-1Ba-L, HIV-2EHO, HIV-2ROD, and the primary HIV-1 isolates to various drugs were determined as described previously (30), with minor modifications. Briefly, MT-2 cells (2 × 104/ml) were exposed to 100 50% tissue culture infectious doses (TCID50s) of HIV-1LAI, HIV-1Ba-L, HIV-2EHO, or HIV-2ROD in the presence or absence of various concentrations of drugs in 96-well microculture plates and were incubated at 37°C for 7 days. After 100 μl of the medium was removed from each well, 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (10 μl, 7.5 mg/ml in phosphate-buffered saline) was added to each well in the plate, followed by incubation at 37°C for 2 h. After incubation to dissolve the formazan crystals, 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 was added to each well and the optical density was measured in a kinetic microplate reader (Vmax; Molecular Devices, Sunnyvale, Calif.). All assays were performed in duplicate or triplicate.
To determine the sensitivities of the primary HIV-1 isolates to drugs, phytohemagglutinin-activated peripheral blood mononuclear cells (PHA-PBMCs; 106/ml) were exposed to 50 TCID50s of each primary HIV-1 isolate and cultured in the presence or absence of various concentrations of drugs in 10-fold serial dilutions in 96-well microculture plates. To determine the drug susceptibilities of certain laboratory HIV-1 strains, MT-4 cells were used as target cells, as described previously (30), with minor modifications. In brief, MT-4 cells (105/ml) were exposed to 100 TCID50s of drug-resistant HIV-1 strains in the presence or absence of various concentrations of drugs and were incubated at 37°C. On day 7 of culture, the supernatant was harvested and the amount of p24 Gag protein was determined by using a fully automated chemiluminescent enzyme immunoassay system (Lumipulse F; Fujirebio Inc., Tokyo, Japan) (13). The drug concentrations that suppressed the production of p24 Gag protein by 50% (50% inhibitory concentrations [IC50s]) were determined by comparison with the level of p24 production in drug-free control cell cultures. All assays were performed in triplicate.
Generation of PI-resistant HIV-1 in vitro.
MT-4 cells (105/ml) were exposed to HIV-1NL4-3 (500 TCID50s) and cultured in the presence of various PIs at an initial concentration of 0.01 to 0.03 μM. Viral replication was monitored by determination of the amount of p24 Gag produced by MT-4 cells. The culture supernatants were harvested on day 7 and were used to infect fresh MT-4 cells for the next round of culture in the presence of increasing concentrations of each drug. When the virus began to propagate in the presence of the drug, the drug concentration was generally increased two- to threefold. Proviral DNA samples obtained from the lysates of infected cells were subjected to nucleotide sequencing. This drug selection procedure was carried out until the drug concentration reached 5 μM.
Determination of nucleotide sequences.
Molecular cloning and determination of the nucleotide sequences of HIV-1 strains passaged in the presence of anti-HIV-1 agents were performed as described previously (30, 31). In brief, high-molecular-weight DNA was extracted from HIV-1-infected MT-4 cells by using the InstaGene Matrix (Bio-Rad Laboratories, Hercules, Calif.) and was subjected to molecular cloning, followed by sequence determination. The primers used for the first round of PCR with the entire Gag- and protease-encoding regions of the HIV-1 genome were LTR-F1 (5′-GAT GCT ACA TAT AAG CAG CTG C-3′) and PR12 (5′-CTC GTG ACA AAT TTC TAC TAA TGC-3′). The first-round PCR mixture consisted of 5 μl of proviral DNA solution, 2.0 U of Premix Taq (Ex Taq Version; Takara Bio Inc., Otsu, Japan), and 12.5 pmol of each of the first PCR primers in a total volume of 50 μl. The PCR conditions used were an initial 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 3 min at 72°C, with a final 8 min of extension at 72°C. The first-round PCR products (1 μl) were used directly in the second round of PCR with primers LTR F2 (5′-GAG ACT CTG GTA ACT AGA GAT C-3′) and Ksma2.1 (5′-CCA TCC CGG GCT TTA ATT TTA CTG GTA C-3′) under the same PCR conditions described above. The second-round PCR products were purified with spin columns (MicroSpin S-400 HR columns; Amersham Biosciences Corp., Piscataway, N.J.), cloned directly, and subjected to sequencing with a model 377 automated DNA sequencer (Applied Biosystems, Foster City, Calif.).
Crystallographic analyses.
The wild type HIV-1 protease was expressed, purified, and crystallized as described previously (14). In brief, the inhibitor was dissolved in dimethyl sulfoxide (DMSO). Crystals were grown from a 5:1 ratio of inhibitor to protease. Crystals grew in hanging drops at room temperature with a well solution of 30 mM sodium acetate (pH 4.8), 10% glycerol, 10% DMSO, and 10% dioxane. X-ray diffraction data were collected with the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beam line at the Advanced Photon Source, Argonne National Laboratory. Data were processed in space group P21212 with unit cell dimensions of a equal to 58.26 Å, b equal to 85.91 Å, and c equal to 46.05 Å by using the HKL 2000 suite of programs (18). The structures were solved by molecular replacement with the AmoRe package (17a), refined with anisotropic B factors by using the high-resolution refinement program SHELX97 (24), and refitted by using the computer graphics program O 8.0 (10).
RESULTS
Anti-HIV-1 activity and cytotoxicity of UIC-94017.
We designed, synthesized, and examined ∼200 different bis-THF-containing nonpeptidic PIs for their anti-HIV activities and found that UIC-94017 had extremely potent activity against a laboratory HIV-1 strain, HIV-1LAI, compared to the activities of the clinically available Food and Drug Administration-approved PIs. As shown in Table 1, six of the available PIs suppressed the infectivity and replication of HIV-1LAI, with IC50s ranging from 0.017 to 0.047 μM with MT-2 cells as the target cells, while UIC-94017 had the most potent activity in terms of suppressing the infectivity and replication of the virus (IC50, 0.003 μM). The selectivity index of UIC-94017, which was 24,800, was much greater than those of SQV and IDV, although the 50% cytotoxic concentration (CC50) of APV was greater than 100 μM, and further testing was not conducted with APV.
TABLE 1.
Activity of UIC-94017 against HIV-1LAI and cytotoxicity of UIC-94017a
| Drug | IC50 (μM) | CC50 (μM) | Selectivity index |
|---|---|---|---|
| UIC-94017 | 0.003 ± 0.0001 | 74.4 ± 1.2 | 24,800 |
| SQV | 0.017 ± 0.003 | 11.3 ± 2.8 | 660 |
| APV | 0.036 ± 0.011 | >100 | >2,800 |
| IDV | 0.047 ± 0.008 | 70.3 ± 4.6 | 1,500 |
| NFV | 0.027 ± 0.004 | ND | ND |
| RTV | 0.045 ± 0.012 | ND | ND |
| LPV | 0.034 ± 0.006 | ND | ND |
MT-2 cells (2 × 103) were exposed to 100 TCID50s of HIV-1LAI and cultured in the presence of various concentrations of PIs, and the IC50s were determined by the MTT assay on day 7 of culture. All assays were conducted in duplicate, and the data shown represent mean ± 1 standard deviation derived from the results of three independent experiments. ND, not determined.
UIC-94017 was further tested against an R5 laboratory HIV-1 strain, HIV-1Ba-L, and two HIV-2 strains, HIV-2ROD and HIV-2EHO, in vitro (Table 2). UIC-94017 was also found to have potent activity against HIV-1Ba-L (IC50, 0.003 μM) when it was tested with PHA-PBMCs as the target cells and against the two HIV-2 strains (IC50s, 0.003 to 0.006 μM) when it was tested with MT-2 cells as the target cells. UIC-94017 had 6- to 13-fold greater activity against HIV-1Ba-L than the other PIs tested. In addition, UIC-94017 and SQV had more potent activities than the other four PIs against the two HIV-2 strains and suppressed the infectivity and replication of the two HIV-2 strains (Table 2). In particular, the activities of APV and RTV were much less potent than those of the other PIs against HIV-2.
TABLE 2.
Activities of selected anti-HIV agents against HIV-1Ba-L, HIV-2ROD, and HIV-2EHOa
| Virus | Cell | IC50 (μM)
|
||||||
|---|---|---|---|---|---|---|---|---|
| AZT | SQV | APV | IDV | NFV | RTV | UIC-94017 | ||
| HIV-1Ba-L | PBMC | 0.009 ± 0.001 | 0.018 ± 0.010 | 0.026 ± 0.005 | 0.025 ± 0.012 | 0.017 ± 0.004 | 0.039 ± 0.020 | 0.003 ± 0.0003 |
| HIV-2ROD | MT-2 | 0.018 ± 0.002 | 0.003 ± 0.0002 | 0.23 ± 0.01 | 0.014 ± 0.006 | 0.019 ± 0.003 | 0.13 ± 0.06 | 0.003 ± 0.0001 |
| HIV-2EHO | MT-2 | 0.011 ± 0.002 | 0.006 ± 0.002 | 0.17 ± 0.05 | 0.011 ± 0.002 | 0.029 ± 0.018 | 0.24 ± 0.006 | 0.006 ± 0.003 |
For HIV-1Ba-L, the IC50s were determined by using PHA-PBMCs and inhibition of p24 Gag protein production by the drug as an endpoint. For HIV-2ROD and HIV-2EHO, MT-2 cells were exposed to the virus and cultured, and the IC50s were determined by the MTT assay. All assays were conducted in duplicate or triplicate, and the data shown represent means ± 1 standard deviation derived from the results of three independent experiments.
UIC-94017 has potent activity against HIV-1 variants selected with PIs.
Next we tested the activities of UIC-94017 and five clinically available PIs against various HIV-1 variants selected for resistance in vitro with each of the five PIs. A laboratory HIV-1 strain, HIV-1NL4-3, was passaged in the presence of increasing concentrations of up to 5 μM each PI in MT-4 cells. The selection was carried out in a cell-free manner for a total of 27, 23, 22, 21, and 14 passages for SQV, APV, IDV, NFV, and RTV, respectively. When the concentration of each drug reached 5 μM, we found that the protease-encoding region of HIV-1 contained a variety of previously known PI resistance-associated amino acid substitutions (Table 3). When resistance was examined in MT-4 cells as the target cells, each PI-selected HIV-1 variant was, as expected, highly resistant to the PI with which the virus was selected, with IC50s exceeding 1 μM. UIC-94017 had potent activities against HIV-1NL4-3 selected with SQV at 5 μM (HIV-1SQV5μM) and HIV-1NFV5μM, although it was less potent against HIV-1IDV5μM and HIV-1RTV5μM. Considering that UIC-94017 shares a P2′ 4-aminobenzenesulfonamide with APV (Fig. 1), it was not surprising that UIC-94017 had the least potent activity against HIV-1APV5μM, with an IC50 73-fold greater than that for HIV-1NL4-3.
TABLE 3.
Activity of UIC-94017 against laboratory PI-resistant HIV-1a
| Virus | Amino acid substitutions in the protease | IC50 (μM)
|
|||||
|---|---|---|---|---|---|---|---|
| SQV | APV | IDV | NFV | RTV | UIC-94017 | ||
| HIV-1NL4-3 | Wild type | 0.009 ± 0.002 | 0.027 ± 0.008 | 0.011 ± 0.002 | 0.020 ± 0.005 | 0.018 ± 0.004 | 0.003 ± 0.0005 |
| HIV-1SQV5μM | L101, G48V, I54V, L90M | >1 (>111) | 0.17 ± 0.09 (6) | >1 (>91) | 0.30 ± 0.04 (15) | >1 (>56) | 0.005 ± 0.0009 (2) |
| HIV-1APV5μM | L10F, V32I, M46I, I54M, A71V, I84V | 0.02 ± 0.008 (2) | >1 (>37) | 0.31 ± 0.05 (28) | 0.21 ± 0.05 (11) | >1 (>56) | 0.22 ± 0.05 (73) |
| HIV-1IDV5μM | L10F, L24I, M46I, L63P, A71V, G73S, V82T | 0.015 ± 0.004 (2) | 0.33 ± 0.01 (12) | >1 (>91) | 0.74 ± 0.04 (37) | >1 (>56) | 0.029 ± 0.0007 (10) |
| HIV-1NFV5μM | L10F, D30N, K45I, A71V, T74S | 0.031 ± 0.009 (3) | 0.093 ± 0.003 (3) | 0.28 ± 0.08 (25) | >1 (>50) | 0.09 ± 0.06 (5) | 0.003 ± 0.0002 (1) |
| HIV-1RTV5μM | M46I, V82F, I84V | 0.013 ± 0.009 (1) | 0.61 ± 0.29 (23) | 0.31 ± 0.07 (28) | 0.24 ± 0.09 (12) | >1 (>56) | 0.025 ± 0.006 (8) |
MT-4 cells (104) were exposed to each HIV-1 (100 TCID50s), and the inhibition of p24 Gag protein production by the drug was used as an endpoint. The numbers in parentheses represent the fold changes of IC50s for each isolate compared to the IC50s for HIV-1NL4-3. The data shown are means ± 1 standard deviation derived from the results of three independent experiments conducted in triplicate.
UIC-94017 exerts potent activity against highly multi-PI-resistant clinical HIV-1 strains.
Highly multi-PI-resistant primary HIV-1 strains were isolated from seven patients with AIDS who had failed existing anti-HIV regimens after receiving 9 to 11 anti-HIV-1 drugs over the previous 32 to 83 months (30, 31) and served as a source of primary infectious HIV-1 strains. These primary strains contained 9 to 14 amino acid substitutions in the protease-encoding region which have reportedly been associated with HIV-1 resistance to various PIs. The mutations seen in these isolates included Leu-10→Ile (seven of seven isolates), Met-46→Ile or Leu (six of seven isolates), Ile-54→Val (five of seven isolates), Leu-63→Pro (seven of seven isolates), Ala-71→Val or Thr (six of seven isolates), Val-82→Ala or Thr (seven of seven isolates), and Leu-90→Met (five of seven isolates) (see footnote a of Table 4).
TABLE 4.
Activity of UIC-94017 against HIV-1 clinical isolates in PHA-PBMCsa
| Virus | IC50 (μM)
|
||||||
|---|---|---|---|---|---|---|---|
| AZT | SQV | APV | IDV | NFV | RTV | UIC-94017 | |
| HIV-1ERS104pre (wild-type X4) | 0.004 ± 0.002 | 0.010 ± 0.004 | 0.023 ± 0.005 | 0.018 ± 0.004 | 0.019 ± 0.002 | 0.027 ± 0.009 | 0.003 ± 0.0003 |
| HIV-1MOKW (wild-type R5) | 0.016 ± 0.009 | 0.004 ± 0.001 | 0.011 ± 0.002 | 0.018 ± 0.003 | 0.033 ± 0.008 | 0.032 ± 0.008 | 0.003 ± 0.0005 |
| HIV-1TM (MDR X4) | 0.73 ± 0.29 (183) | 0.23 ± 0.02 (23) | 0.39 ± 0.11 (17) | >1 (>56) | 0.54 ± 0.01 (28) | >1 (>37) | 0.004 ± 0.0008 (1) |
| HIV-1MM (MDR R5) | 0.37 ± 0.07 (93) | 0.30 ± 0.04 (30) | 0.34 ± 0.16 (15) | >1 (>56) | >1 (>53) | >1 (>37) | 0.02 ± 0.009 (7) |
| HIV-1JSL (MDR R5) | 0.08 ± 0.03 (20) | 0.35 ± 0.16 (35) | 0.75 ± 0.07 (33) | >1 (>56) | >1 (>53) | >1 (>37) | 0.029 ± 0.007 (10) |
| HIV-1A (MDR X4) | ND | 0.14 ± 0.06 (14) | 0.16 ± 0.05 (7) | >1 (>56) | 0.36 ± 0.10 (19) | >1 (>37) | 0.004 ± 0.0004 (1) |
| HIV-1B (MDR X4) | ND | 0.31 ± 0.01 (31) | 0.34 ± 0.01 (15) | >1 (>56) | >1 (>53) | >1 (>37) | 0.013 ± 0.006 (4) |
| HIV-1C (MDR X4) | ND | 0.037 ± 0.001 (4) | 0.28 ± 0.13 (12) | >1 (>56) | 0.44 ± 0.02 (23) | >1 (>37) | 0.003 ± 0.0006 (1) |
| HIV-1G (MDR X4) | ND | 0.029 ± 0.001 (3) | 0.25 ± 0.11 (11) | 0.39 ± 0.05 (22) | 0.32 ± 0.06 (17) | 0.44 ± 0.19 (16) | 0.004 ± 0.001 (1) |
The amino acid substitutions identified in the protease-encoding region of HIV-1ERS104pre, HIV-1TM, HIV-1MM, HIV-1JSL, HIV-1A, HIV-1B, HIV-1C, and HIV-1G compared to the consensus type B sequence cited from the Los Alamos database include L63P; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, and I93L; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, and Q92K; L10I, L24I, L33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, and V82A; L10I, I15V, E35D, N37E, K45R, I54V, L63P, A71V, V82T, L90M, I93L, and C95F, L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70O, V82A, and L89M; and L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M, respectively. HIV-1MOKW was confirmed to lack any known drug resistance-associated amino acid substitutions. The IC50s were determined by using PHA-PBMCs as target cells and the inhibition of p24 Gag protein production as an endpoint. All values were determined in triplicate, and those shown are derived from the results of three independent experiments. Numbers in parentheses represent the fold changes in IC50s for each isolate compared to the IC50s for HIV-1ERS104pre. MDR, multidrug resistant; ND, not determined.
Two wild-type primary strains, X4 HIV-1ERS104pre (25) and R5 HIV-1MOKW (13), were used as reference strains, against which all the drugs tested exerted potent antiviral activities, with IC50s ranging from 0.003 to 0.033 μM (Table 4). Three primary strains were highly resistant to AZT, with IC50s being 20- to 183-fold greater than the IC50 for HIV-1ERS104pre. SQV showed considerable activity against two multidrug-resistant primary strains, strains HIV-1C and HIV-1G, with the IC50s being three- to fourfold greater than those for HIV-1ERS104pre; however, its activity was less potent against the other five strains, with IC50s ranging from 0.14 to 0.35 μM. APV, IDV, NFV, and RTV were less potent in blocking the replication of all multi-PI-resistant strains, with IC50s ranging from 0.16 to more than 1 μM. These data confirm that virtually all the primary strains tested were highly resistant to all clinically available PIs except SQV, to which strains were susceptible in a few cases. It should be noted, however, that UIC-94017 potently blocked the replication of five of seven primary strains, with IC50s ranging from 0.003 to 0.013 μM, although the IC50s of UIC-94017 were 7- and 10-fold greater for HIV-1MM and HIV-1JSL, respectively.
Effects of human serum proteins on anti-HIV-1 activity of UIC-94017.
In an attempt to determine whether the anti-HIV-1 activity of UIC-94017 is affected by proteins in the bloodstream or body fluids, we examined the effects of α1-acid glycoprotein (AAG) or human serum (45%) on the activity of UIC-94017 against HIV-1ERS104pre in PHA-PBMCs (Table 5). In the presence of 10 μM AAG, the IC50s and IC90s of five clinically available PIs rose by factors of 3 to 16 and 2 to >12 compared to the IC50s and IC90s determined in the presence of 15% FCS, respectively. In the presence of 45% human serum, the IC50s and IC90s of the five PIs rose by factors of 4 to 49 and 8 to 15 (or >13) compared to the IC50s and IC90s obtained with 15% FCS, respectively. The IC50s and IC90s of UIC-94017 also rose by factors of 14 and 19, respectively, in the presence of 10 μM AAG and by factors of 11 and 12, respectively, in the presence of 45% human serum. It is worth noting that the IC50s and IC90s of UIC-94017 were the second lowest after those of SQV in the presence of AAG and were the lowest among those of the other PIs in the presence of 45% human serum.
TABLE 5.
Effects of human serum proteins on anti-HIV-1 activity of UIC-94017a
| Drug | 15% FCS
|
10 μM AAG
|
45% human serum
|
|||
|---|---|---|---|---|---|---|
| IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | |
| UIC-94017 | 0.003 ± 0.0003 | 0.009 ± 0.0005 | 0.042 ± 0.011 (14) | 0.17 ± 0.10 (19) | 0.034 ± 0.0005 (11) | 0.11 ± 0.03 (12) |
| SQV | 0.010 ± 0.004 | 0.060 ± 0.008 | 0.033 ± 0.0006 (3) | 0.09 ± 0.002 (2) | 0.17 ± 0.12 (17) | 0.89 ± 0.13 (15) |
| APV | 0.023 ± 0.006 | 0.083 ± 0.004 | 0.35 ± 0.06 (15) | >1 (>12) | 0.35 ± 0.07 (15) | >1 (>12) |
| IDV | 0.018 ± 0.004 | 0.075 ± 0.002 | 0.051 ± 0.002 (3) | 0.35 ± 0.16 (5) | 0.069 ± 0.008 (4) | 0.61 ± 0.04 (8) |
| NFV | 0.019 ± 0.002 | 0.075 ± 0.0007 | 0.31 ± 0.01 (16) | 0.83 ± 0.01 (11) | 0.94 ± 0.06 (49) | >1 (>13) |
| RTV | 0.027 ± 0.009 | 0.082 ± 0.005 | 0.21 ± 0.16 (8) | 0.69 ± 0.19 (8) | 0.33 ± 0.05 (12) | >1 (>12) |
The IC50s and IC90s were determined by using PHA-PBMCs as target cells and the inhibition of p24 Gag protein production as an endpoint. All assays were conducted in triplicate. The results shown are means ± standard deviations from three independent assays. Numbers in parentheses represent fold increases in IC50 and IC90s in the presence of 10 μM AAG or 45% human serum compared to those determined in the presence of 15% FCS. The virus used was HIV-1ERS104pre.
Crystallographic analysis of UIC-94017 complexed with HIV-1 protease.
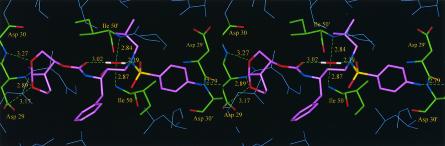
In order to investigate the mechanism of how UIC-94017 exerts its potent activity against a wide spectrum of multi-PI-resistant HIV-1 strains, an X-ray crystal structure of HIV-1 protease complexed with UIC-94017 was refined to an R factor of 0.15 and R free of 0.19 at a 1.30-Å resolution. (The crystallographic R factor is defined as folows: R = ∑(|Fobs − Fcalc|/|Fobs|), where Fobs represents the observed structure factors and Fcalc represents the calculated structure factors.) It was found that UIC-94017 was bound in two orientations, and the side chains of several amino acids showed disordered densities with two conformations, as observed previously for hydrophobic side chains near the peptide analogs (14). Equivalent interactions were also shown in both orientations of UIC-94017. The inhibitor's P1 and P1′ groups form van der Waals interactions with protease residues Leu-23, Gly-49, Ile-50, Pro-81, Val-82, and Ile-84. The P2 and P2′ groups interact with residues Ala-28, Asp-29, Asp-30, Val-32, Ile-47, and Ile-50. The inhibitor hydroxyl group interacts with all four carboxylate oxygens of the catalytic residues Asp-25 and Asp-25′. The carbonyl oxygen and one of the sulfonamide oxygen atoms form hydrogen bonds with the water molecule that interacts with the amides of Ile-50 and Ile-50′ on the flaps, as observed in most HIV-1 protease-inhibitor complexes (8). The two oxygen atoms of the bis-THF group of UIC-94017 are positioned to form hydrogen bond interactions with the main chain amides of Asp-29 and Asp-30 and the carboxylate oxygen of Asp-29. Furthermore, it was found that UIC-94017 formed new polar interactions with the amide of the main chain and the carboxylate oxygen of Asp-30.
DISCUSSION
In the present study, we demonstrated that a novel nonpeptidic HIV-1 protease inhibitor, UIC-94017 (TMC114), which contains a bis-THF and a sulfonamide isostere, exerts extremely potent activity against laboratory HIV-1 strains and primary clinical isolates with minimal cytotoxicity (Tables 1 to 3). In particular, UIC-94017 efficiently blocked the infectivity and replication of each of the HIV-1NL4-3 variants exposed to and selected for resistance to SQV, IDV, NFV, or RTV at concentrations up to 5 μM (Table 3). UIC-94017 was also potent against multi-PI-resistant clinical HIV-1 variants isolated from patients who had no response to existing antiviral regimens after having received a variety of antiviral agents (Table 4). It was noted that the activity of UIC-94017 was less potent against APV-resistant strain HIV-1APV5μM; however, it was thought that this was due to the structural relatedness of UIC-94017 to APV, in that they share the P2′ 4-aminobenzenesulfonamide. It is intriguing that the activity of SQV was fairly well maintained against various HIV-1 strains selected with PIs but not against the variant selected with SQV, a profile generally consistent with previous results (1, 6, 30, 31). Nevertheless, when the activity of SQV against multi-drug-resistant primary strains was examined, high concentrations of SQV were required to suppress the replication of five of seven primary strains tested (IC50 range, 0.14 to 0.35 μM) (Table 4). In contrast, UIC-94017 exerted highly potent activity against all seven primary strains examined. As shown in Table 4, even the replication of the most apparently resistant primary strain, HIV-1JSL, was successfully suppressed by a relatively low concentration of UIC-94017 (IC50, 0.029 μM). With regard to the pharmacokinetics of UIC-94017, Hoetelmans et al. (R. Hoetelmans, et al., 10th Conf. Retrovir. Opportunistic Infect., abstr. 549, 2003) have recently reported that when UIC-94017 is coadministered with RTV by use of dosing regimens of 200 mg of UIC-94017 plus 100 mg of RTV once a day and 1,200 mg of UIC-94017 plus 200 mg of RTV once a day, the levels of UIC-94017 in the plasma of patients with AIDS rose to 2.6 and 9.6 μM, respectively, and the trough levels were 0.65 and 1.8 μM, respectively. These pharmacokinetic data from a clinical trial show that the levels of UIC-94017 in plasma do not go below 15- and 43-fold above the AAG-adjusted IC50s (0.042 μM; Table 5) with the low and high dosing regimens, respectively. Indeed, it should be noted that Arasteh et al. (K. Arasteh, et al., 10th Conf. Retrovir. Opportunistic Infect., abstr. 8, 2003) most recently reported the early results of an open, randomized phase IIa clinical trial with 50 multiple PI-experienced patients with AIDS in which patients receiving various combination regimens of UIC-94017 and RTV had a significant reduction in the number of HIV-1 RNA copies in their plasma (range, −0.47 to −2.5 log10; median reduction, −1.35 log10). Treatment with UIC-94017 plus RTV was generally well tolerated.
It was recently reported that certain PIs can exert potent activities against clinical HIV-1 strains which acquired high levels of resistance to multiple PIs (2, 30, 31). One such agent, JE-2147, represents a dipeptide PI which, like UIC-94017, is potent in vitro against a wide spectrum of HIV-1 strains, including HIV-1 variants highly resistant to multiple PIs (31). JE-2147, unlike UIC-94017, possesses a P2′ benzylamide group that has two rotatable bonds between the amide and the phenyl ring. This flexible P2′ substituent, which can adapt to structurally altered active sites of mutant HIV-1 proteases and maintain its tight binding to protease, is believed to be responsible for the potent activity of JE-2147 against multi-PI-resistant HIV-1 variants (31). In contrast, the observed extreme potency of UIC-94017 seen in the present in vitro study appears to stem from the formation of strong hydrogen bonds between the two oxygens of UIC-94017's bis-THF and the main chains of Asp-29 and Asp-30 in the S2 subsite (Fig. 2). More importantly, the reason that UIC-94017 exerts potent activity against multi-PI-resistant variants is also presumably due to its interaction with the main chains (not the side chains, with which other PIs interact) of these two aspartic acids. Indeed, the Asp-30→Asn substitution emerges as a primary substitution in HIV-1 following long-term exposure to NFV, which is known to interact with the side chain of Asp-30, and presumably alters the NFV-protease interaction and confers on the virus resistance to NFV (19, 20). Although HIV-1 selection experiments with UIC-94017 are under way in H.M.’s laboratory, it is worth noting that HIV-1 strains exposed to UIC-94003 (30), an analog of UIC-94017, did not undergo a substitution at codon 30, probably because side chain changes do not confer on the virus resistance to UIC-94003, as described previously (30). Instead, HIV-1 strains exposed to UIC-94003 acquired a unique Ala→Ser substitution at residue 28 at the active site of the enzyme, which can produce a steric and electrostatic expulsion and which may alter the conformation of the enzymatic active site composed of Asp-29 and Asp-30 (30). The unique characteristics exhibited by the bis-THF group of UIC-94017—namely, its close position and tight binding to the main chains of the active-site amino acids, Asp-29 and Asp-30, which differ from the profiles of other Food and Drug Administration-approved PIs—should be important for the potency of UIC-94017 and its activity against multi-PI-resistant HIV-1 variants. These structural properties of UIC-94017 may also provide a new framework for the development of PIs which hardly permit or delay the emergence of PI-resistant HIV-1 variants.
FIG. 2.
Stereoview of X-ray crystal structure of HIV-1 protease complexed with UIC-94017. UIC-94017 is depicted in purple, and significant HIV-1 protease residues are shown in green with the corresponding three-letter amino acid codes and sequence numbers. Nitrogen and oxygen atoms are shown in blue and red, respectively. Hydrogen bonds between UIC-94017 and each amino acid and those between UIC-94017 and the water molecule are indicated by green dashed lines.
Taken together, the present data strongly suggest that UIC-94017 might have potent efficacy in patients harboring multidrug-resistant HIV-1 variants; however, it is likely that HIV-1 will eventually become resistant to UIC-94017, and the potential usefulness of UIC-94017 should be interpreted in the context of placebo-controlled, double-blind clinical trials.
Acknowledgments
We thank Toshikazu Miyakawa, Shigeyoshi Harada, and Manabu Aoki for helpful discussions.
This work was supported in part by grants from the National Institutes of Health (grant GM 53386 to A.K.G. and grant GM62920 to I.T.W.); Hungarian grant OTKA F35191 to P.B.; a grant from a Research for the Future Program of the Japan Society for the Promotion of Science (grant JSPS-RFTF 97L00705 to H.M.); a Grant-in-Aid for Scientific Research (Priority Areas; to H.M.) from the Ministry of Education, Culture, Sports, Science, and Technology (Monbu-Kagakusho) of Japan; and a Grant for Promotion of AIDS Research from the Ministry of Health, Labor and Welfare (Kosei-Rodosho) of Japan (to H.M.). Use of the Advanced Photon Source was supported by the Basic Energy Sciences, Office of Science, U.S. Department of Energy, under contract no. W-31-109-Eng-38.
REFERENCES
- 1.Carrillo, A., K. Stewart, H. Sham, D. Norbeck, W. Kohlbrenner, J. Leonard, D. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colonno, R. J., A. Thiry, K. Limoli, and N. Parkin. 2003. Activities of atazanavir (BMS-232632) against a large panel of human immunodeficiency virus type 1 clinical isolates resistant to one or more approved protease inhibitors. Antimicrob. Agents Chemother. 47:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Clercq, E. 2002. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discovery 1:13-25. [DOI] [PubMed] [Google Scholar]
- 4.Erickson, J. W., and S. K. Burt. 1996. Structural mechanisms of HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 36:545-571. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer, E., D. Podzamczer, M. Arnedo, E. Fumero, P. McKenna, A. Rinehart, J. L. Perez, M. J. Barbera, T. Pumarola, J. M. Gatell, and F. Gudiol. 2003. Genotype and phenotype at baseline and at failure in human immunodeficiency virus-infected antiretroviral-naive patients in a randomized trial comparing zidovudine and lamivudine plus nelfinavir or nevirapine. J. Infect. Dis. 187:687-690. [DOI] [PubMed] [Google Scholar]
- 6.Gong, Y. F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P. F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabar, S., C. Pradier, E. Le Corfec, R. Lancar, C. Allavena, M. Bentata, P. Berlureau, C. Dupont, P. Fabbro-Peray, I. Poizot-Martin, and D. Costagliola. 2000. Factors associated with clinical and virological failure in patients receiving a triple therapy including a protease inhibitor. AIDS 14:141-149. [DOI] [PubMed] [Google Scholar]
- 8.Gustchina, A., C. Sansom, M. Prevost, J. Richelle, S. Y. Wodak, A. Wlodawer, and I. T. Weber. 1994. Energy calculations and analysis of HIV-1 protease-inhibitor crystal structures. Protein Eng. 7:309-317. [DOI] [PubMed] [Google Scholar]
- 9.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 10.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt 2):110-119. [DOI] [PubMed] [Google Scholar]
- 11.Kemper, C. A., M. D. Witt, P. H. Keiser, M. P. Dube, D. N. Forthal, M. Leibowitz, D. S. Smith, A. Rigby, N. S. Hellmann, Y. S. Lie, J. Leedom, D. Richman, J. A. McCutchan, and R. Haubrich. 2001. Sequencing of protease inhibitor therapy: insights from an analysis of HIV phenotypic resistance in patients failing protease inhibitors. AIDS 15:609-615. [DOI] [PubMed] [Google Scholar]
- 12.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 13.Maeda, K., K. Yoshimura, S. Shibayama, H. Habashita, H. Tada, K. Sagawa, T. Miyakawa, M. Aoki, D. Fukushima, and H. Mitsuya. 2001. Novel low molecular weight spirodiketopiperazine derivatives potently inhibit R5 HIV-1 infection through their antagonistic effects on CCR5. J. Biol. Chem. 276:35194-35200. [DOI] [PubMed] [Google Scholar]
- 14.Mahalingam, B., J. M. Louis, J. Hung, R. W. Harrison, and I. T. Weber. 2001. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 43:455-464. [DOI] [PubMed] [Google Scholar]
- 15.Mitsuya, H., and J. Erickson. 1999. Discovery and development of antiretroviral therapeutics for HIV infection, p. 751-780. In T. C. Merigan, J. G. Bartlet, and D. Bolognesi (ed.), Textbook of AIDS medicine. The Williams & Wilkins Co., Baltimore, Md.
- 16.Murphy, E. L., A. C. Collier, L. A. Kalish, S. F. Assmann, M. F. Para, T. P. Flanigan, P. N. Kumar, L. Mintz, F. R. Wallach, and G. J. Nemo. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 135:17-26. [DOI] [PubMed] [Google Scholar]
- 17.Murphy, R. L. 2000. New antiretroviral drugs in development. AIDS 14(Suppl. 3):S227-S234. [PubMed] [Google Scholar]
- 17a.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. D50:157-163. [Google Scholar]
- 18.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 19.Patick, A., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 22.Salomon, H., M. A. Wainberg, B. Brenner, Y. Quan, D. Rouleau, P. Cote, R. LeBlanc, E. Lefebvre, B. Spira, C. Tsoukas, R. P. Sekaly, B. Conway, D. Mayers, J. P. Routy, et al. 2000. Prevalence of HIV-1 resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or injecting drug use. AIDS 14:F17-F23. [DOI] [PubMed] [Google Scholar]
- 23.Sepkowitz, K. A. 2001. AIDS—the first 20 years. N. Engl. J. Med. 344:1764-1772. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick, G. M., and T. R. Schneider. 1997. High resolution refinement. Methods Enzymol. 277:319-343. [PubMed] [Google Scholar]
- 25.Shirasaka, T., M. F. Kavlick, T. Ueno, W. Y. Gao, E. Kojima, M. L. Alcaide, S. Chokekijchai, B. M. Roy, E. Arnold, R. Yarchoan, and H. Mitsuya. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. USA 92:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, N. M. Ruiz, et al. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N. Engl. J. Med. 341:1865-1873. [DOI] [PubMed] [Google Scholar]
- 27.Wainberg, M. A., and G. Friedland. 1998. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 279:1977-1983. [DOI] [PubMed] [Google Scholar]
- 28.Yeni, P. G., S. M. Hammer, C. C. Carpenter, D. A. Cooper, M. A. Fischl, J. M. Gatell, B. G. Gazzard, M. S. Hirsch, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, M. S. Saag, M. Schechter, R. T. Schooley, M. A. Thompson, S. Vella, and P. A. Volberding. 2002. Antiretroviral treatment for adult HIV infection in 2002: updated recommendations of the International AIDS Society—USA Panel. JAMA 288:222-235. [DOI] [PubMed] [Google Scholar]
- 29.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura, K., R. Kato, M. F. Kavlick, A. Nguyen, V. Maroun, K. Maeda, K. A. Hussain, A. K. Ghosh, S. V. Gulnik, J. W. Erickson, and H. Mitsuya. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 76:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. USA 96:8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]