Abstract
ABT-492 demonstrated potent antibacterial activity against most quinolone-susceptible pathogens. The rank order of potency was ABT-492 > trovafloxacin > levofloxacin > ciprofloxacin against quinolone-susceptible staphylococci, streptococci, and enterococci. ABT-492 had activity comparable to those of trovafloxacin, levofloxacin, and ciprofloxacin against seven species of quinolone-susceptible members of the family Enterobacteriaceae, although it was less active than the comparators against Citrobacter freundii and Serratia marcescens. The activity of ABT-492 was greater than those of the comparators against fastidious gram-negative species, including Haemophilus influenzae, Moraxella catarrhalis, Neisseria gonorrhoeae, and Legionella spp. and against Pseudomonas aeruginosa and Helicobacter pylori. ABT-492 was as active as trovafloxacin against Chlamydia trachomatis, indicating good intracellular penetration and antibacterial activity. In particular, ABT-492 was more active than trovafloxacin and levofloxacin against multidrug-resistant Streptococcus pneumoniae, including strains resistant to penicillin and macrolides, and H. influenzae, including β-lactam-resistant strains. It retained greater in vitro activity than the comparators against S. pneumoniae and H. influenzae strains resistant to other quinolones due to amino acid alterations in the quinolone resistance-determining regions of the target topoisomerases. ABT-492 was a potent inhibitor of bacterial topoisomerases, and unlike the comparators, DNA gyrase and topoisomerase IV from either Staphylococcus aureus or Escherichia coli were almost equally sensitive to ABT-492. The profile of ABT-492 suggested that it may be a useful agent for the treatment of community-acquired respiratory tract infections, as well as infections of the urinary tract, bloodstream, and skin and skin structure and nosocomial lung infections.
In the outpatient setting, β-lactams, cephalosporins, the combination of amoxicillin and clavulanate, and macrolides are commonly prescribed therapies for community-acquired respiratory tract infections (CA-RTIs). However, antimicrobial resistance in bacterial species involved in CA-RTIs is a growing clinical problem. From recent surveillance studies, the prevalence of penicillin resistance, macrolide resistance, and multidrug resistance in Streptococcus pneumoniae is increasing in isolates from patients with CA-RTIs (9, 17). β-Lactam resistance in Haemophilus influenzae and Moraxella catarrhalis is also a documented problem in the community (7, 8, 17). Moreover, multidrug-resistant, methicillin-resistant Staphylococcus aureus is now occasionally associated with CA-RTIs (1, 16).
The newer fluoroquinolones, such as levofloxacin, gatifloxacin, and moxifloxacin, have antibacterial potency, pharmacokinetics, and tolerability properties leading to regulatory approvals for their use in the treatment of CA-RTIs. They are being prescribed more frequently for these clinical indications, in part because the prevalence of fluoroquinolone resistance in isolates from CA-RTIs remains at <3% worldwide (2, 9, 17). However, increases in fluoroquinolone resistance in S. pneumoniae corresponding to increases in fluoroquinolone use have been reported (5). For example, 5.5% of S. pneumoniae strains in Hong Kong were levofloxacin resistant in 1998, but by 2000, 13% were levofloxacin resistant (18, 19). Fluoroquinolone-resistant H. influenzae strains have also been identified occasionally (2). Therefore, new fluoroquinolones that are active and effective against pathogens that cause CA-RTIs, including multidrug-resistant S. pneumoniae and fluoroquinolone-resistant strains, may maintain the clinical usefulness of this antibiotic class for these indications.
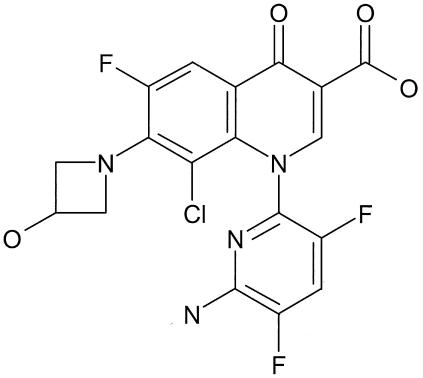
ABT-492 is a new fluoroquinolone that differs from other members of the class by two structural features: the 6-amino-3,5-difluoropyridine at the 1 position and the 3-hydroxyazetidine-1-yl substituent at the 7 position of the 6-fluoroquinolone core (Fig. 1). The antibacterial spectrum and potency of ABT-492 were compared with those of levofloxacin and ciprofloxacin as well as with those of trovafloxacin as an example of a potent and broad-spectrum fluoroquinolone. In addition, the ability of ABT-492 to form cleavable complexes in conjunction with DNA gyrase and topoisomerase IV isolated from E. coli and S. aureus was studied.
FIG. 1.
Structure of ABT-492.
(The contents of this work were presented at the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, 2002.)
MATERIALS AND METHODS
Antibiotics.
ABT-492 (formerly WQ-3034) was prepared by the Wakunaga Pharmaceutical Co., Hiroshima, Japan. Trovafloxacin, levofloxacin, and ciprofloxacin were prepared at Abbott Laboratories, Abbott Park, Ill. Erythromycin, clindamycin, penicillin, and vancomycin reference powders were purchased from the U.S. Pharmacopeial Convention, Inc., Rockville, Md. Ampicillin was purchased from the Sigma Chemical Company, St. Louis, Mo.
Determination of antibacterial activity.
The bacterial strains used for susceptibility studies were either clinical isolates from the Abbott Laboratories culture collection or reference strains obtained from the American Type Culture Collection (ATCC; Manassas, Va.).
The susceptibilities of aerobic, nonfastidious species, S. pneumoniae, and H. influenzae were determined visually by broth microdilution as described by the National Committee for Clinical Laboratory Standards (NCCLS) (26). The susceptibilities of Neisseria gonorrhoeae and Helicobacter pylori were determined by agar dilution, as described by NCCLS (26) Quality control, as specified by NCCLS, was performed for all tests that used NCCLS reference methods; our results met NCCLS standards (26). Susceptibilities were also determined in medium containing 50% (vol/vol) rat serum (Invitrogen Corp., Rockville, Md.) or human serum (Scantibodies Laboratory, Inc., Santee, Calif.) that had been heat inactivated for 30 min at 56°C. Minimum bactericidal concentrations (MBCs) were determined by broth microdilution in conjunction with MICs following NCCLS methods (6, 25, 26).
The susceptibilities of Legionella spp. were determined on buffered charcoal yeast extract agar with incubation at 35°C in an atmosphere containing 5% CO2 (6). The susceptibilities of Mycobacterium avium were determined on Middlebrook 7H10 agar supplemented with 0.5% (vol/vol) glycerol and 10% (vol/vol) oleic acid-albumin-dextrose-catalase enrichment (Difco Laboratories, Detroit, Mich.) with incubation at 35°C in an atmosphere containing 5% CO2 (37). The susceptibilities of anaerobic species were determined by agar dilution with Wilkins-Chalgren agar with incubation at 35°C in an oxygen-free atmosphere containing greater than 4% CO2 (39).
The susceptibility of Mycoplasma pneumoniae was determined by broth microdilution in SP4 medium with incubation at 35°C in ambient air (15). The susceptibility of Borrelia burgdorferi was determined by broth macrodilution in Barbour-Stoenner-Kelly medium with incubation at 35°C in an atmosphere containing 5% CO2 (15, 21).
The susceptibility of Chlamydia trachomatis was determined by broth microdilution with infected McCoy cells incubated at 37°C in an atmosphere containing 5% CO2 for 48 h; intracellular C. trachomatis was visualized by fluorescent-antibody staining (Merifluor Chlamydia; Meridian Diagnostics, Inc., Cincinnati, Ohio) (32).
Determination of molecular mechanisms of resistance.
The molecular mechanisms of macrolide and vancomycin resistance were identified in selected gram-positive isolates by PCR amplification of the Mef efflux, Erm methylase, VanA, and VanB genes as described previously (11, 34, 35). β-Lactamase production was determined by nitrocefin hydrolysis (Cefinase; Becton Dickinson, Cockeysville, Md.).
Mutations in the quinolone resistance-determining regions of DNA gyrase and topoisomerase IV were determined by PCR amplification and DNA sequence analysis for S. pneumoniae and the staphylococci as described previously (1, 4). DNA sequences were compared with published sequences for parC (GenBank accession no. Z67739) (28) and gyrA (GenBank accession no. U49087) (38) for S. pneumoniae or with published sequences for grlA (GenBank accession no. L25288) (12) and gyrA (GenBank accession no. M86227) (24) for the staphylococci.
For isolates of S. pneumoniae, increased efflux pump activity was screened by using the reserpine antagonism screen, as described previously (3, 4).
Determination of plasma protein binding.
The plasma protein binding of the fluoroquinolones was determined by using ultracentrifugation to separate non-protein-bound drug from the protein-bound drug. Drugs were added to different solutions of rat or human serum to achieve final concentrations of 20 μg/ml. The solutions were allowed to equilibrate for 1 h at room temperature and were then centrifuged at 86,000 × g for 18 h. The resulting gradient was separated into fractions; the samples were extracted and analyzed by reverse-phase high-performance liquid chromatography.
Topoisomerase-mediated DNA cleavage assays.
E. coli DNA gyrase holoenzyme was purchased from Enzyco, Inc. (Denver, Colo.). ParC and ParE subunits of E. coli topoisomerase IV were obtained from N. Cozzarelli (University of California, Berkeley). The GyrA and GyrB subunits of DNA gyrase and the GrlA and GrlB subunits of topoisomerase IV from S. aureus were cloned and isolated at Abbott as polyhistidine-tagged fusion proteins (30). E. coli topoisomerase IV, S. aureus DNA gyrase, and S. aureus topoisomerase IV were reconstituted by incubating equimolar amounts of the respective subunits (GyrA-GyrB, ParC-ParE, or GrlA-GrlB) at room temperature for 10 min; the reconstituted enzymes were maintained on ice or stored at −20°C for less than 2 months. Human topoisomerase II (cloned HeLa cell topoisomerase II alpha) was obtained from N. Osheroff (Vanderbilt University, Nashville, Tenn.). ColE1 DNA substrate was isolated by using CsCl-gradient centrifugation.
Cleavage reactions catalyzed by S. aureus gyrase and topoisomerase IV were performed as described previously (30). Cleavage reactions catalyzed by E. coli enzymes were carried out in a 20-μl reaction mixture containing either 150 mM Tris-HCl (pH 7.5), 20 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 1.5 mM ATP, 5 mM spermidine, 50 μg of bovine serum albumin per ml, 13% sucrose, 0.15 μg of supercoiled ColE1 DNA, and 1 ng of gyrase or 50 mM Tris-HCl (pH 7.5), 70 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.5 mM ATP, 50 μg of bovine serum albumin per ml, 0.15 μg of supercoiled ColE1 DNA, and 2 ng of topoisomerase IV. The reaction mixtures were incubated at 37°C for 30 min. The reactions were then stopped by adding 2 μl of a mixture containing 3% sodium dodecyl sulfate and 4 mg of proteinase K per ml and reincubating the mixture at 37°C for 1 h. Human topoisomerase II cleavage reactions were carried out as described previously (33). Cleavage reactions contained various concentrations of drug. The conversion of supercoiled DNA to linear DNA was monitored by densitometry of agarose gels stained with ethidium bromide after electrophoresis, as described previously (30, 33). For the bacterial topoisomerase reactions, the drug concentrations (in micrograms per milliliter) at which half-maximal cleavage (DNA linearization) was obtained relative to the maximal cleavage obtained with 100 μg of ciprofloxacin per ml were determined. For human topoisomerase II reactions, the drug concentrations (in micrograms per milliliter) resulting in 7% more DNA cleavage than drug-free cleavage were determined; the latter concentrations were normalized to the value obtained with clinafloxacin, which has a predetermined standard value of 18.4 μg/ml.
RESULTS AND DISCUSSION
The in vitro antibacterial activity of ABT-492 was compared with those of three reference fluoroquinolones: trovafloxacin, levofloxacin, and ciprofloxacin. The MIC ranges and the MICs at which 50% (MIC50) and 90% (MIC90) of the tested strains are inhibited are presented in Table 1. NCCLS interpretive criteria were used to define categories of antibiotic susceptibility for penicillin, vancomycin, ampicillin, erythromycin, and the quinolones (27).
TABLE 1.
Comparative antibacterial activities
| Species (resistance)a | No. of isolates | Antibiotic | MIC (μg/ml)
|
||
|---|---|---|---|---|---|
| Range | 50% | 90% | |||
| S. aureus (quinolone S) | 16 | ABT-492 | ≤0.002-0.008 | 0.004 | 0.008 |
| Trovafloxacin | 0.008-0.015 | 0.015 | 0.03 | ||
| Levofloxacin | 0.06-0.25 | 0.12 | 0.25 | ||
| Ciprofloxacin | 0.25-0.5 | 0.25 | 0.5 | ||
| S. aureus (quinolone R) | 19 | ABT-492 | 0.06-0.5 | 0.12 | 0.5 |
| Trovafloxacin | 0.5-2 | 1 | 2 | ||
| Levofloxacin | 4-32 | 8 | 16 | ||
| Ciprofloxacin | 8-128 | 32 | 128 | ||
| S. epidermidis (quinolone S) | 10 | ABT-492 | ≤0.008 | ≤0.008 | ≤0.008 |
| Trovafloxacin | ≤0.008-0.06 | 0.03 | 0.03 | ||
| Levofloxacin | 0.06-0.25 | 0.25 | 0.25 | ||
| Ciprofloxacin | 0.12-0.25 | 0.12 | 0.25 | ||
| CoNSb (quinolone R) | 10 | ABT-492 | 0.03-0.5 | 0.25 | 0.5 |
| Trovafloxacin | 1-16 | 2 | 8 | ||
| Levofloxacin | 4-128 | 8 | 64 | ||
| Ciprofloxacin | 8->32 | 32 | >128 | ||
| S. pneumoniae (quinolone S) | 29 | ABT-492 | 0.004-0.03 | 0.008 | 0.015 |
| Trovafloxacin | 0.015-0.06 | 0.03 | 0.03 | ||
| Levofloxacin | 0.25-1 | 0.5 | 0.5 | ||
| Ciprofloxacin | 1-8 | 1 | 2 | ||
| S. pneumoniae (quinolone R) | 30 | ABT-492 | 0.004-0.5 | 0.06 | 0.12 |
| Trovafloxacin | 0.12-16 | 4 | 8 | ||
| Levofloxacin | 0.5-16 | 8 | 16 | ||
| Ciprofloxacin | 2-64 | 32 | 64 | ||
| S. pyogenes | 16 | ABT-492 | 0.004-0.03 | 0.008 | 0.015 |
| Trovafloxacin | 0.03-0.12 | 0.06 | 0.12 | ||
| Levofloxacin | 0.25-1 | 0.5 | 0.5 | ||
| Ciprofloxacin | 0.25-1 | 0.5 | 0.5 | ||
| Viridans group streptococci | 10 | ABT-492 | 0.004-0.03 | 0.008 | 0.015 |
| Trovafloxacin | 0.06-0.5 | 0.12 | 0.25 | ||
| Levofloxacin | 1-4 | 1 | 2 | ||
| Ciprofloxacin | 2-16 | 2 | 4 | ||
| E. faecalis (quinolone S) | 13 | ABT-492 | 0.015-0.03 | 0.03 | 0.03 |
| Trovafloxacin | 0.06-0.25 | 0.12 | 0.12 | ||
| Levofloxacin | 0.5-1 | 0.5 | 1 | ||
| Ciprofloxacin | 0.25-1 | 0.5 | 1 | ||
| E. faecalis (quinolone R) | 12 | ABT-492 | 0.06-1 | 0.25 | 0.5 |
| Trovafloxacin | 0.25-16 | 4 | 16 | ||
| Levofloxacin | 1-32 | 16 | 32 | ||
| Ciprofloxacin | 2-128 | 64 | 128 | ||
| E. faecium (quinolone S) | 6 | ABT-492 | ≤0.008-0.25 | 0.25 | |
| Trovafloxacin | 0.03-2 | 1 | |||
| Levofloxacin | 0.5-2 | 1 | |||
| Ciprofloxacin | 0.5-1 | 1 | |||
| E. faecium (quinolone R) | 14 | ABT-492 | 0.25-64 | 4 | 32 |
| Trovafloxacin | 0.5-32 | 4 | 32 | ||
| Levofloxacin | 1-128 | 32 | 128 | ||
| Ciprofloxacin | 2->128 | 128 | >128 | ||
| L. monocytogenes | 15 | ABT-492 | 0.06-0.12 | 0.12 | 0.12 |
| Trovafloxacin | 0.12-0.25 | 0.25 | 0.25 | ||
| Levofloxacin | 1 | 1 | 1 | ||
| Ciprofloxacin | 1-2 | 1 | 1 | ||
| E. coli (quinolone S) | 20 | ABT-492 | 0.008-0.06 | 0.015 | 0.015 |
| Trovafloxacin | 0.004-0.03 | 0.008 | 0.03 | ||
| Levofloxacin | 0.008-0.03 | 0.008 | 0.015 | ||
| Ciprofloxacin | 0.004-0.008 | 0.004 | 0.008/PICK> | ||
| E. coli (quinolone R) | 10 | ABT-492 | 4-32 | 8 | 16 |
| Trovafloxacin | 32->128 | >128 | >128 | ||
| Levofloxacin | 16-128 | 32 | 64 | ||
| Ciprofloxacin | 32->128 | 128 | >128 | ||
| Salmonella spp. | 20 | ABT-492 | 0.008-0.5 | 0.03 | 0.06 |
| Trovafloxacin | 0.008-0.5 | 0.03 | 0.06 | ||
| Levofloxacin | 0.015-1 | 0.06 | 0.06 | ||
| Ciprofloxacin | 0.008-0.5 | 0.015 | 0.03 | ||
| Shigella spp. | 9 | ABT-492 | 0.002-0.008 | ||
| Trovafloxacin | 0.002-0.015 | ||||
| Levofloxacin | 0.004-0.015 | ||||
| Ciprofloxacin | 0.002-0.015 | ||||
| C. freundii | 20 | ABT-492 | 0.03->2 | 0.25 | 2 |
| Trovafloxacin | 0.015-0.5 | 0.06 | 0.5 | ||
| Levofloxacin | 0.015-0.25 | 0.12 | 0.25 | ||
| Ciprofloxacin | 0.008-0.12 | 0.03 | 0.12 | ||
| Klebsiella spp.c (quinolone S) | 20 | ABT-492 | ≤0.002-0.25 | 0.06 | 0.12 |
| Trovafloxacin | 0.004-1 | 0.03 | 0.06 | ||
| Levofloxacin | 0.015-1 | 0.03 | 0.06 | ||
| Ciprofloxacin | 0.004-0.25 | 0.015 | 0.03 | ||
| K. pneumoniae (quinolone R) | 10 | ABT-492 | 2-8 | 4 | 8 |
| Trovafloxacin | 4->128 | >128 | >128 | ||
| Levofloxacin | 8-64 | 32 | 64 | ||
| Ciprofloxacin | 64->128 | 128 | >128 | ||
| P. mirabilis | 19 | ABT-492 | 0.008-0.25 | 0.015 | 0.12 |
| Trovafloxacin | 0.12-1 | 0.5 | 1 | ||
| Levofloxacin | 0.015-0.25 | 0.06 | 0.12 | ||
| Ciprofloxacin | 0.015-0.12 | 0.03 | 0.12 | ||
| Enterobacter spp. | 20 | ABT-492 | ≤0.004-0.06 | 0.015 | 0.03 |
| Trovafloxacin | 0.008-0.25 | 0.03 | 0.06 | ||
| Levofloxacin | ≤0.004-0.06 | 0.03 | 0.06 | ||
| Ciprofloxacin | ≤0.004-0.03 | 0.008 | 0.015 | ||
| Providencia spp. | 20 | ABT-492 | 0.015-0.25 | 0.06 | 0.25 |
| Trovafloxacin | 0.12-1 | 0.25 | 0.5 | ||
| Levofloxacin | 0.12-1 | 0.25 | 0.25 | ||
| Ciprofloxacin | 0.015-0.5 | 0.06 | 0.25 | ||
| S. marcescens | 10 | ABT-492 | 0.5-2 | 0.5 | 2 |
| Trovafloxacin | 0.06-0.25 | 0.12 | 0.25 | ||
| Levofloxacin | 0.015-0.12 | 0.06 | 0.12 | ||
| Ciprofloxacin | 0.015-0.06 | 0.015 | 0.03 | ||
| P. aeruginosa (quinolone S) | 20 | ABT-492 | 0.03-0.5 | 0.06 | 0.25 |
| Trovafloxacin | 0.06-2 | 0.25 | 0.5 | ||
| Levofloxacin | 0.06-2 | 0.25 | 0.5 | ||
| Ciprofloxacin | 0.03-1 | 0.06 | 0.25 | ||
| P. aeruginosa (quinolone R) | 20 | ABT-492 | 8->128 | 32 | 128 |
| Trovafloxacin | 64->128 | >128 | >128 | ||
| Levofloxacin | 32->128 | 64 | >128 | ||
| Ciprofloxacin | 16->128 | 64 | 128 | ||
| B. cepacia | 3 | ABT-492 | >8 | ||
| Trovafloxacin | 8->8 | ||||
| Levofloxacin | 8->8 | ||||
| Ciprofloxacin | 8->8 | ||||
| S. maltophilia | 7 | ABT-492 | ≤0.008-2 | 1 | |
| Trovafloxacin | 0.03-0.5 | 0.5 | |||
| Levofloxacin | 0.25-2 | 1 | |||
| Ciprofloxacin | 0.25-4 | 2 | |||
| H. influenzae (quinolone S) | 25 | ABT-492 | ≤0.002-0.008 | ≤0.002 | 0.004 |
| Trovafloxacin | ≤0.002-0.015 | 0.004 | 0.015 | ||
| Levofloxacin | ≤0.002-0.015 | 0.008 | 0.008 | ||
| Ciprofloxacin | ≤0.002-0.008 | 0.004 | 0.008 | ||
| H. influenzae (quinolone R) | 6 | ABT-492 | ≤0.004-0.12 | ≤0.004 | |
| Trovafloxacin | 0.06-4 | 0.5 | |||
| Levofloxacin | 0.06-16 | 1 | |||
| Ciprofloxacin | 0.03-16 | 1 | |||
| M. catarrhalis | 17 | ABT-492 | ≤0.002-0.004 | ≤0.002 | ≤0.002 |
| Trovafloxacin | 0.004-0.008 | 0.008 | 0.008 | ||
| Levofloxacin | 0.008-0.03 | 0.015 | 0.03 | ||
| Ciprofloxacin | 0.008-0.015 | 0.015 | 0.015 | ||
| L. pneumophila | 14 | ABT-492 | 0.12 | 0.12 | 0.12 |
| Trovafloxacin | 0.25 | 0.25 | 0.25 | ||
| Levofloxacin | 0.5 | 0.5 | 0.5 | ||
| Ciprofloxacin | 0.5-1 | 1 | 1 | ||
| Legionella spp. | 5 | ABT-492 | 0.12 | 0.12 | |
| Trovafloxacin | 0.25 | 0.25 | |||
| Levofloxacin | 0.5 | 0.5 | |||
| Ciprofloxacin | 0.5-1 | 1 | |||
| N. gonorrhoeae | 10 | ABT-492 | ≤0.004 | ≤0.004 | ≤0.004 |
| Trovafloxacin | ≤0.004-0.03 | ≤0.004 | 0.03 | ||
| Levofloxacin | ≤0.004-0.06 | 0.015 | 0.06 | ||
| Ciprofloxacin | ≤0.004-0.06 | ≤0.004 | 0.008 | ||
| H. pylori | 45 | ABT-492 | 0.015-0.12 | 0.03 | 0.12 |
| Trovafloxacin | 0.03-0.5 | 0.12 | 0.25 | ||
| Levofloxacin | 0.12-1 | 0.5 | 0.5 | ||
| Ciprofloxacin | 0.06-1 | 0.25 | 0.5 | ||
| M. avium | 10 | ABT-492 | 2-16 | 8 | 16 |
| Trovafloxacin | 32-64 | 32 | 32 | ||
| Levofloxacin | 16-64 | 32 | 64 | ||
| Ciprofloxacin | 8-128 | 8 | 32 | ||
| M. pneumoniae | 18 | ABT-492 | 0.25-0.5 | 0.5 | 0.5 |
| Trovafloxacin | 0.12-0.25 | 0.12 | 0.25 | ||
| Levofloxacin | 1-2 | 1 | 2 | ||
| Ciprofloxacin | 1-2 | 2 | 2 | ||
| C. trachomatis | 2 | ABT-492 | 0.03-0.06 | ||
| Trovafloxacin | 0.06 | ||||
| Levofloxacin | 0.12-0.5 | ||||
| Ciprofloxacin | 0.5-1 | ||||
| B. fragilis | 16 | ABT-492 | 0.03-0.12 | 0.06 | 0.12 |
| Trovafloxacin | 0.12-0.5 | 0.25 | 0.5 | ||
| Levofloxacin | 1-2 | 2 | 2 | ||
| Ciprofloxacin | 2-16 | 4 | 8 | ||
| C. difficile | 12 | ABT-492 | ≤0.015 | ≤0.015 | ≤0.015 |
| Trovafloxacin | 1 | 1 | 1 | ||
| Levofloxacin | 2-4 | 2 | 4 | ||
| Ciprofloxacin | 16 | 16 | 16 | ||
| C. perfringens | 12 | ABT-492 | ≤0.015 | ≤0.015 | ≤0.015 |
| Trovafloxacin | 0.06-0.25 | 0.12 | 0.12 | ||
| Levofloxacin | 0.12-0.25 | 0.25 | 0.25 | ||
| Ciprofloxacin | 0.5-1 | 0.5 | 1 | ||
S, susceptible; R, resistant.
CoNS, coagulase negative staphylococci; S. epidermidis (n = 8) and S. haemolyticus (n = 2).
K. pneumoniae (n = 15), K. oxytoca (n = 3), and K. rhinosceromatis (n = 2).
Quinolone-susceptible gram-positive pathogens.
ABT-492 was consistently more potent than trovafloxacin, levofloxacin, and ciprofloxacin against quinolone-susceptible gram-positive pathogens. For all species, the rank order of potency was ABT-492 > trovafloxacin > levofloxacin > ciprofloxacin. The MIC90s of ABT-492 for quinolone-susceptible strains of staphylococci were 0.008 μg/ml or less; this potency was fourfold greater than that of trovafloxacin. Similarly, ABT-492 was fourfold more active than trovafloxacin against quinolone-susceptible enterococci, with an MIC90 for Enterococcus faecalis of 0.03 μg/ml and an MIC50 for Enterococcus faecium of 0.25 μg/ml. ABT-492 was two- to eight-fold more active than trovafloxacin against viridans group streptococci, Streptococcus pyogenes, and Listeria monocytogenes.
The MIC90 for quinolone-susceptible strains of S. pneumoniae was 0.015 μg/ml, demonstrating that ABT-492 was 2-, 32-, and 64-fold more potent than trovafloxacin, levofloxacin, and ciprofloxacin, respectively. However, multidrug resistance in S. pneumoniae is well described, including patterns in which penicillin resistance and macrolide resistance are associated (9). Neither penicillin nonsusceptibility nor macrolide resistance had an effect on the susceptibilities of the strains to ABT-492. The MIC90 of 0.015 μg/ml for the 19 penicillin-nonsusceptible isolates was nearly identical to the MIC90 of 0.008 μg/ml for the 10 penicillin-susceptible isolates. Similarly, the MIC90 of 0.015 μg/ml for the 19 erythromycin-resistant isolates, comprised of 10 strains with the Mef(A) macrolide-specific efflux pump and 9 strains with the Erm(B) ribosome methylase, was identical to the MIC90 of 0.015 μg/ml for the 10 erythromycin-susceptible isolates.
Quinolone-resistant gram-positive pathogens.
ABT-492 was more active than the three comparators against quinolone-resistant strains of staphylococci, streptococci, and enterococci, although all four drugs showed reduced activities in comparison with their activities against quinolone-susceptible isolates of the same species. In particular, ABT-492 was highly active in vitro against quinolone-resistant strains of S. pneumoniae, with an MIC90 of 0.12 μg/ml, in comparison with MIC90s of 8, 16, and 64 μg/ml for trovafloxacin, levofloxacin, and ciprofloxacin, respectively. Mutations were detected in parC and gyrA of all resistant pneumococci. These mutations altered amino acids in the quinolone resistance-determining regions of the topoisomerases: two contained only a ParC change at Ser79, one contained only a ParC change at Asn91, 26 contained a ParC change at Ser79 or Asp83 plus a GyrA change at Ser81 or Glu85, and one contained three changes comprised of ParC at Ser79 and Asp83 plus a GyrA change at Ser81. None of the strains demonstrated detectable changes in quinolone efflux (3, 4). As was found with the quinolone-susceptible isolates, concurrent penicillin or macrolide resistance [Mef(A) and Erm(B)] did not alter the susceptibilities of the quinolone-resistant strains (data not shown).
Similarly, ABT-492 was more potent than the comparators against quinolone-resistant isolates of the staphylococci; all isolates were inhibited by 0.5 μg or less of ABT-492 per ml. The quinolone-resistant isolates of S. aureus carried mutations resulting in amino acid changes in the quinolone resistance-determining regions of the topoisomerases. Changes were identified at both Ser80 of GrlA and Ser84 of GyrA in 16 isolates, at both Ser84 of GrlA and Glu88 of GyrA in 1 isolate, and at Ser80 of GrlA and Ser84 plus Ser85 in 1 isolate. One resistant S. aureus isolate had a change in Ala116 plus upregulation of the NorA efflux pump (22). All eight quinolone-resistant isolates of Staphylococcus epidermidis contained ParC changes at Ser80 and Glu84 plus a GyrA change at Ser84. The quinolone resistance-determining region of parC could not be amplified with our PCR primers from the two isolates of Staphylococcus haemolyticus; however, they both had a GyrA change at Ser84, with one isolate having a second change at Asp88. Quinolone resistance is highly correlated with oxacillin resistance in staphylococci (31). In this study, 17 of 19 quinolone-resistant isolates of S. aureus were oxacillin resistant and all 10 of the quinolone-resistant isolates of coagulase-negative staphylococci were oxacillin resistant.
ABT-492 was also the most active compound tested against quinolone-resistant enterococci (mutations in the quinolone resistance-determining regions of the enterococcal topoisomerases were not determined). The MIC90 for quinolone-resistant E. faecalis was 0.5 μg/ml, which made it greater than 16-fold more potent than trovafloxacin; the 12 isolates included 3 VanA and 6 VanB strains. All compounds were poorly active against quinolone-resistant strains of E. faecium, with MIC90s of 32 μg/ml for ABT-492 and trovafloxacin; the 14 isolates included 4 VanA and 5 VanB strains.
Quinolone-susceptible gram-negative pathogens.
The potency of ABT-492 was assessed against quinolone-susceptible isolates of nine species of the family Enterobacteriaceae. The MIC90s of ABT-492 for E. coli, Salmonella, Shigella, Klebsiella, Proteus mirabilis, Enterobacter, and Providencia were 0.25 μg/ml or less and were comparable to the MIC90s of trovafloxacin, levofloxacin, and ciprofloxacin. Isolates of Citrobacter freundii and Serratia marcescens were somewhat less susceptible to ABT-492 than to the comparators. Although the ABT-492 MIC50s for these species were 0.25 and 0.5 μg/ml, respectively, the MIC90s were 2 μg/ml.
ABT-492 was as active as ciprofloxacin against ciprofloxacin-susceptible isolates of Pseudomonas aeruginosa; the MIC90s of both compounds were 0.25 μg/ml. All four quinolones were less active against ciprofloxacin-susceptible isolates of Stenotrophomonas maltophilia, with MIC50s ranging from 0.5 to 2 μg/ml.
ABT-492 was more potent than trovafloxacin, levofloxacin, and ciprofloxacin against fastidious gram-negative species. The MIC90 of ABT-492 for H. influenzae was 0.004 μg/ml; this group contained three β-lactamase producers and four β-lactamase-negative, ampicillin-resistant strains. The MIC90s of ABT-492 were ≤0.002, ≤0.004, and 0.12 μg/ml for M. catarrhalis, N. gonorrhoeae, and Legionella pneumophila, respectively.
Quinolone-resistant gram-negative pathogens.
In general, ciprofloxacin-resistant gram-negative strains demonstrated reduced susceptibilities to ABT-492, trovafloxacin, and levofloxacin in comparison to those of ciprofloxacin-susceptible strains of the same species. However, the MIC90s of ABT-492 for ciprofloxacin-resistant strains of E. coli and Klebsiella pneumoniae were 16 and 8 μg/ml, respectively, making it more than fourfold more potent than trovafloxacin, levofloxacin, and ciprofloxacin. All four quinolones had little to no in vitro activity against quinolone-resistant isolates of P. aeruginosa and three ciprofloxacin-resistant isolates of Burkholderia cepacia. Two isolates of S. maltophilia were low-level ciprofloxacin resistant, with ciprofloxacin MICs of 4 μg/ml. The MICs of ABT-492 for these isolates were 1 and 2 μg/ml, respectively.
Quinolone resistance in H. influenzae is found very rarely (2). ABT-492 retained the greatest potency of the four compounds, with the MIC for the least-susceptible strain being 0.12 μg/ml. In contrast, the trovafloxacin MIC for this strain was 1 μg/ml and the levofloxacin and ciprofloxacin MICs were 16 μg/ml. The six strains carried amino acid changes in GyrA at Ser84 and Asp88 with or without concomitant changes in ParC at Gly82, Ser84, or Glu88 (2).
Other pathogens.
The susceptibilities of additional pathogenic species to ABT-492 were also evaluated. ABT-492 was two- to fourfold more potent than trovafloxacin, levofloxacin, and ciprofloxacin against H. pylori. Moreover, the potency of ABT-492 against macrolide-resistant H. pylori strains was equivalent to that against the macrolide-susceptible strains, with MIC90s of 0.12 and 0.06 μg/ml, respectively; the macrolide-resistant strains have point mutations in domain V of the 23S rRNA gene at the residue equivalent to positions 2058 and 2059 in E. coli which reduce the level of binding of macrolides to the ribosome (36). Although ABT-492 was two- to fourfold more potent than trovafloxacin, levofloxacin, and ciprofloxacin against M. avium, all four compounds were poorly active in vitro.
ABT-492 was fourfold more potent than levofloxacin and ciprofloxacin against M. pneumoniae, with an MIC90 of 0.5 μg/ml; however, trovafloxacin was twofold more potent than ABT-492.
Chlamydia spp. are obligate intracellular bacteria, and the susceptibility test methods require the use of infected mammalian cells (32). The MICs of ABT-492 for two strains of C. trachomatis in this study were 0.03 and 0.06 μg/ml, respectively, which were comparable to the MICs of trovafloxacin, while levofloxacin and ciprofloxacin were less active. These results indicate that ABT-492 accumulates and retains antibacterial activity within mammalian cells.
Anaerobes.
Early quinolones such as ciprofloxacin typically had poor in vitro activities against anaerobic species; however, trovafloxacin demonstrates improved potency and is indicated for the treatment of infections caused by anaerobes (20). The in vitro activity of ABT-492 was at least 10-fold greater than that of trovafloxacin against trovafloxacin-susceptible isolates of three species of anaerobes: the MIC90 of ABT-492 was 0.12 μg/ml for Bacteroides fragilis and was less than 0.03 μg/ml for Clostridium perfringens and Clostridium difficile. Levofloxacin and ciprofloxacin were less active than ABT-492 and trovafloxacin.
Bactericidal activity.
The MICs of ABT-492 were compared with the MBCs for examples of gram-positive and gram-negative pathogens by the broth microdilution method (Table 2). ABT-492 demonstrated potent bactericidal activity, since the MBCs were less than 0.12 μg/ml and were no more than fourfold higher than the corresponding MICs for all 10 isolates. In particular, ABT-492 was bactericidal for all five strains of S. pneumoniae, including isolates which were resistant to quinolones, penicillin, and macrolides. Ciprofloxacin was also bactericidal for all isolates tested.
TABLE 2.
Comparison of MICs and MBCs
| Species | Resistancea | Strain | MIC or MBC (μg/ml)
|
|||
|---|---|---|---|---|---|---|
| ABT-492
|
Ciprofloxacin
|
|||||
| MIC | MBC | MIC | MBC | |||
| S. pneumoniae | Susceptible | 2486 | ≤0.002 | ≤0.002 | 4 | 8 |
| Pen R | 6502 | ≤0.002 | ≤0.002 | 4 | 8 | |
| Mef and Pen I | 5649 | 0.004 | 0.004 | 16 | 32 | |
| ErmB | 5979 | ≤0.002 | ≤0.002 | 8 | 8 | |
| Quinolone R | 7240 | 0.03 | 0.06 | 128 | 128 | |
| S. aureus | Susceptible | 1662 | ≤0.002 | ≤0.002 | 0.06 | 0.06 |
| E. faecalis | Susceptible | 1967 | 0.03 | 0.03 | 0.5 | 0.5 |
| E. coli | Susceptible | 1298 | 0.008 | 0.03 | 0.004 | 0.008 |
| H. influenzae | Susceptible | 1435 | ≤0.002 | ≤0.002 | 0.008 | 0.015 |
| M. catarrhalis | 2604 | ≤0.002 | ≤0.002 | 0.008 | 0.015 | |
Pen, penicillin; R, resistant; I, intermediate.
Effect of serum on antibacterial activity.
Plasma protein binding was determined as the percentage of bound drug and was assessed by ultracentrifugation (Table 3). The results for protein binding in human plasma for trovafloxacin and ciprofloxacin are in agreement with those reported in the U.S. package inserts for those drugs, 69 and 20 to 40%, respectively. The protein binding profile of ABT-492 is similar to that of trovafloxacin: significantly greater binding than ciprofloxacin in all plasma samples and greater protein binding in rat plasma than in human plasma.
TABLE 3.
Plasma protein binding
| Antibiotic | % Plasma protein binding
|
|
|---|---|---|
| Rat serum | Human serum | |
| ABT-492 | 89.0 | 77.0 |
| Trovafloxacin | 96.5 | 68.7 |
| Ciprofloxacin | 27.9 | 32.6 |
The effect of serum protein binding was assessed by determination of changes in the in vitro antibacterial activities against indicator strains of bacteria by broth microdilution (Table 4). The in vitro activities of ABT-492 and trovafloxacin were reduced when they were tested in 50% (vol/vol) heat-inactivated rat or human serum. The effect of rat serum on the antibacterial activities was greater than that of human serum, which paralleled the protein binding results. Serum had little to no effect on the in vitro activity of ciprofloxacin, which correlated with the significantly lower levels of protein binding.
TABLE 4.
Effect of serum on antibacterial activity
| Species | Resistance | Serum (50% [vol/vol]) | MIC (μg/ml)
|
||
|---|---|---|---|---|---|
| ABT-492 | Trovafloxacin | Ciprofloxacin | |||
| S. pneumoniae ATCC 6303 | Susceptible | None | 0.004 | 0.03 | 1 |
| Rat | 0.03 | 0.25 | 0.5 | ||
| Human | 0.008 | 0.06 | 1 | ||
| S. pneumoniae 7215 | Quinolone Ra | None | 0.008 | 0.25 | 8 |
| Rat | 0.25 | 2 | 4 | ||
| Human | 0.03 | 0.5 | 8 | ||
| S. pneumoniae 7257 | Quinolone R | None | 0.03 | 2 | 16 |
| Rat | 0.5 | >2 | 32 | ||
| Human | 0.06 | >2 | 16 | ||
| H. influenzae 1435 | Susceptible | None | ≤0.001 | 0.004 | ≤0.008 |
| Rat | ≤0.001 | 0.008 | ≤0.008 | ||
| Human | 0.06 | 0.12 | 0.03 | ||
| S. aureus NCTC 10649 | Susceptible | None | ≤0.001 | 0.004 | 0.12 |
| Rat | 0.015 | 0.12 | 0.25 | ||
| Human | 0.008 | 0.25 | 2 | ||
| E. coli Juhl | Susceptible | None | 0.008 | 0.008 | ≤0.008 |
| Rat | 0.12 | 0.25 | ≤0.008 | ||
| Human | 0.03 | 0.015 | ≤0.008 | ||
| E. faecalis PIU 1967 | Susceptible | None | 0.015 | 0.06 | 0.25 |
| Rat | 0.5 | 0.5 | 0.5 | ||
| Human | 0.06 | 0.12 | 0.5 | ||
R, resistant.
Formation of cleavable complexes with bacterial and human topoisomerases.
Fluoroquinolones interact with DNA gyrase and topoisomerase IV, the two bacterial type II topoisomerases. DNA gyrase is involved in maintaining appropriate DNA supercoiling during DNA synthesis and transcription, while topoisomerase IV is involved in decatenation of chromosomal DNA and chromosome segregation (10, 14). Besides inhibiting the catalytic activities of the enzymes, quinolones form a stable enzyme-DNA-quinolone cleavable complex. The formation of enzyme-quinolone-DNA complexes reversibly inhibits DNA synthesis and initially causes bacteriostatic growth inhibition. Irreversible, lethal single- and double-stranded breaks are released when the complex dissociates. The interaction of fluoroquinolones with the enzyme-DNA complex also interferes with chromosome replication and chromosome segregation (10, 13, 14, 40). ABT-492 was a potent inhibitor of the topoisomerases of both E. coli and S. aureus, as measured by the formation of cleavable complexes (Table 5).
TABLE 5.
Formation of cleavable complexes with bacterial and human topoisomerases
| Species | DNA topo- isomerase | Concn (μg/ml) for inhibition of cleavable complex formationa
|
||
|---|---|---|---|---|
| ABT-492 | Trovafloxacin | Ciprofloxacin | ||
| E. coli | DNA gyrase | 0.8 | 0.43 | 0.24 |
| Topoisomerase IV | 1.1 | 4.5 | 1.8 | |
| S. aureus | DNA gyrase | 0.57 | 1.6 | 3.5 |
| Topoisomerase IV | 1.7 | 0.19 | 0.18 | |
| Human | Topoisomerase II | >100 | >100 | >100 |
For bacterial topoisomerases, the results are reported as the drug concentration causing half-maximal DNA cleavage. For human topoisomerase II, the results are reported as the drug concentration causing 7% more DNA cleavage than that seen without drug treatment.
Topoisomerase IV is typically more sensitive to quinolone interactions than DNA gyrase in gram-positive species (10, 30). Trovafloxacin and ciprofloxacin showed 8- and 19-fold greater potencies, respectively, against S. aureus topoisomerase IV than against S. aureus DNA gyrase. In contrast, the interaction of ABT-492 with both S. aureus topoisomerases was nearly equivalent, with threefold greater activity against DNA gyrase than against topoisomerase IV. By direct comparison, ABT-492 was nearly threefold more active than trovafloxacin and was about sixfold more active than ciprofloxacin against S. aureus DNA gyrase. Although fluoroquinolones interact with both enzymes, DNA gyrase may be a more effective target than topoisomerase IV because the interaction between fluoroquinolones and DNA gyrase, which acts ahead of the replication fork to remove positive supercoils, inhibits DNA replication more rapidly than the interaction with topoisomerase IV, which acts behind it (13, 40). Thus, the more potent interaction with DNA gyrase demonstrated with the S. aureus enzyme is likely to be responsible for the greater antibacterial activity of ABT-492 against gram-positive organisms compared with those of ciprofloxacin and trovafloxacin.
For gram-negative species, DNA gyrase is typically more sensitive than topoisomerase IV to the fluoroquinolone interaction. In this study, ciprofloxacin and trovafloxacin interacted nearly 10-fold more potently with DNA gyrase than with topoisomerase IV. However, the interactions of ABT-492 with both E. coli topoisomerases were nearly equivalent. All three quinolones were highly active against E. coli DNA gyrase and demonstrated half-maximal DNA cleavage at concentrations less than 1 μg/ml, with ciprofloxacin having the greatest activity. However, in addition to activity equivalent to those of the other quinolones against DNA gyrase, ABT-492 was twofold more potent than ciprofloxacin and fourfold more potent than trovafloxacin against E. coli topoisomerase IV.
ABT-492 demonstrated an atypical pattern of enzyme interactions, with nearly equivalent interactions with the DNA gyrase and topoisomerase IV enzymes of both E. coli and S. aureus. Susceptibility to fluoroquinolones is driven by the interaction of the antibiotic with the most sensitive topoisomerase. Moreover, stepwise accumulation of mutations in the quinolone resistance-determining regions progressively decreases the antibacterial activities of the quinolones (40). The near equivalence of DNA gyrase and topoisomerase IV as targets has been postulated to reduce the likelihood that resistance will develop, since a mutation in both targets, a statistically rare event, is required to effectively express reduced susceptibility (29, 40). For example, S. pneumoniae mutants for which MICs were reduced twofold or less were selected only very rarely (frequency, 5 × 10−10) in vitro with clinafloxacin, a fluoroquinolone which targets both DNA gyrase and topoisomerase IV (29, 30). Thus, the near equivalency of DNA gyrase and topoisomerase IV as intracellular targets for ABT-492 may help prevent the selection of topoisomerase mutations during therapy. Confirmation of this hypothesis will require studies with ABT-492-resistant mutants generated in vitro.
Like the bacterial topoisomerases, human topoisomerase II catalyzes relaxation and chromosome segregation reactions via double-stranded DNA breaks in human cells. It also has partial homology to the bacterial topoisomerases (23). In contrast to its potent interactions with the bacterial topoisomerases, ABT-492 did not demonstrate an interaction with human topoisomerase II at the highest concentration tested (Table 5) and, therefore, has a greater than 50-fold greater selectivity for bacterial topoisomerases. Etoposide, a known poison of human topoisomerase II, demonstrated 7% DNA cleavage at 1.7 μg/ml in this assay.
Conclusions.
The in vitro antibacterial activity of ABT-492 was significantly greater than that of levofloxacin against quinolone-susceptible pathogens involved in CA-RTIs. In addition, ABT-492 had improved in vitro activity against antibiotic-resistant respiratory tract pathogens, including multidrug-resistant S. pneumoniae strains and S. pneumoniae and H. influenzae strains with mutations in DNA gyrase and topoisomerase IV that render them resistant to levofloxacin. ABT-492 was also more potent than trovafloxacin and ciprofloxacin against most quinolone-susceptible pathogens responsible for nosocomial respiratory tract, urinary tract, bloodstream, and skin and skin structure infections and against anaerobic pathogens responsible for infections and was significantly more active than the comparators against quinolone-resistant gram-positive strains. ABT-492 was active against C. trachomatis, indicating good intracellular penetration and antibacterial activity. The enhanced antibacterial activity of ABT-492 relative to those of ciprofloxacin, levofloxacin, and trovafloxacin is likely to be explained, in part, by its potent interactions with bacterial topoisomerases, in particular, DNA gyrase. Moreover, the equivalence of DNA gyrase and topoisomerase IV as drug targets for ABT-492 may help prevent the selection of resistant mutants during therapy, as has been postulated for other quinolones. Thus, ABT-492 may be a useful bactericidal agent for the treatment of CA-RTIs as well as other infections for which broad-spectrum antibiotics are indicated.
Acknowledgments
We thank the members of Core Sequencing and Drug Metabolism groups at Abbott Laboratories for providing us the DNA sequence and protein binding data, respectively.
REFERENCES
- 1.Almer, L. S., V. D. Shortridge, A. M. Nilius, N. B. Soni, J. M. Beyer, G. G. Stone, and R. K. Flamm. 2002. Antimicrobial susceptibility and molecular characterization of community-acquired methicillin resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 43:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Biedenbach, D. J., and R. N. Jones. 2000. Fluoroquinolone-resistant Haemophilus influenzae: frequency of occurrence and analysis of confirmed strains in the SENTRY Antimicrobial Surveillance Program (North and Latin America). Diagn. Microbiol. Infect. Dis. 36:255-259. [DOI] [PubMed] [Google Scholar]
- 3.Brenwald, N. P., M. J. Gill, and R. Wise. 1998. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brueggemann, A. B., S. L. Coffman, P. Rhomberg, H. Huynh, L. Almer, A. Nilius, R. Flamm, and G. V. Doern. 2002. Fluoroquinolone resistance in Streptococcus pneumoniae in United States since 1994-1995. Antimicrob. Agents Chemother. 46:680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D. K., A. McGreer, J. C. de Azavedo, and D. E. Low. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 6.Clement, J. J., S. K. Tanaka, J. Alder, C. Vojtko, J. Beyer, D. Hensey, N. Ramer, D. McDaniel, and D. T. W. Chu. 1994. In vitro and in vivo evaluations of A-80556, a new fluoroquinolone. Antimicrob. Agents Chemother. 38:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doern, G. V., A. B. Brueggemann, G. Pierce, T. Hogan, H. P. Holley, Jr., and A. M. Rauch. 1996. Prevalence of antimicrobial resistance among 723 outpatient clinical isolates of Moraxella catarrhalis in the United States in 1994 and 1995: results of a 30-center national surveillance study. Antimicrob. Agents Chemother. 40:2884-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doern, G. V., A. B. Brueggemann, G. Pierce, G. H. P. Holley, Jr., and A. M. Rauch. 1996. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 to 1995 and detection of β-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a 30-center national surveillance study. Antimicrob. Agents Chemother. 41:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Breuggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 13.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootz, T. D., and K. E. Brighty. 1996. Fluoroquinolone antibacterials: SAR, mechanism of action, resistance, and clinical aspects. Med. Res. Rev. 16:433-486. [DOI] [PubMed] [Google Scholar]
- 15.Gump, D. W. 1996. Antimicrobial susceptibility testing for some atypical microorganisms: chlamydiae, mycoplasmas, Rickettsia, and spirochetes, p. 212-229. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 16.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 17.Hoban, D. J., G. V. Doern, A. C. Fluit, M. Roussel-Delvallez, and R. N. Jones. 2001. Worldwide prevalence of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32:S81-S93. [DOI] [PubMed] [Google Scholar]
- 18.Ho, P.-L., T.-L. Que, D.-C. Tsang, T.-K. Ng, K.-H. Chow, and W.-H. Seto. 1999. Emergence of fluoroquinolone resistance among multiply resistant strains of Streptococcus pneumoniae in Hong Kong. Antimicrob. Agents Chemother. 43:1310-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, P.-L., R. W. H. Yung, D.-C. Tsang, T.-L. Que, M. Ho, W.-H. Seto, T.-K. Ng, W. C. Yam, and W. W. S. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, D. C. 2000. New uses for new and old quinolones and the challenge of resistance. Clin. Infect. Dis. 30:243-254. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, S. E., G. C. Klein, G. P. Schmid, and J. C. Feeley. 1984. Susceptibility of the Lyme disease spirochete to seven antimicrobial agents. Yale J. Biol. Med. 57:549-553. [PMC free article] [PubMed] [Google Scholar]
- 22.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madhusudan, K., and V. Nagaraja. 1996. Alignment and phylogenetic analysis of type II DNA topoisomerases. J. Biosci. 21:613-629. [Google Scholar]
- 24.Margerrison, E. E., R. Hopewell, and L. M. Fischer. 1992. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J. Bacteriol. 174:1596-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 1992. Methods for determining bactericidal activity of antimicrobial agents. Tentative guideline M26-T. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 26.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing; ninth informational supplement. Approved standard M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Pan, X., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan, X.-S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saiki, A. Y. C., L. L. Shen, C.-M. Chen, J. Baranowski, and C. G. Lerner. 1999. DNA cleavage activities of Staphylococcus aureus gyrase and topoisomerase IV stimulated by quinolones and 2-pyridones. Antimicrob. Agents Chemother. 43:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz, F.-J., A. C. Fuit, S. Brisse, J. Verhoef, K. Köhrer, and D. Milatovic. 1999. Molecular epidemiology of quinolone resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol. Med. Microbiol. 26:281-287. [DOI] [PubMed] [Google Scholar]
- 32.Segreti, J., H. A. Kessler, K. S. Kapell, and G. M. Trenholme. 1989. In vitro activity of A-56268 (TE-031) and four other antimicrobial agents against Chlamydia trachomatis. Antimicrob. Agents Chemother. 31:100-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, L. L., J. Baranowski, J. Fostel, D. A. Montgomery, and P. A. Lartey. 1992. DNA topoisomerases from pathogenic fungi: targets for the discovery of antifungal drugs. Antimicrob. Agents Chemother. 36:2778-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29:1186-1188. [DOI] [PubMed] [Google Scholar]
- 35.Shortridge, V. D., R. K. Flamm, N. Ramer, J. Beyer, and S. K. Tanaka. 1996. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 26:73-78. [DOI] [PubMed] [Google Scholar]
- 36.Stone, G. G., D. Shortridge, R. K. Flamm, J. Versalovic, J. Beyer, K. Idler, L. Zulawinski, and S. K. Tanaka. 1996. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter 1:227-228. [DOI] [PubMed] [Google Scholar]
- 37.Swenson, J. M., C. Thornsberry, and V. A. Silcox. 1982. Rapidly growing mycobacteria: testing of susceptibility to 34 antimicrobial agents by broth microdilution. Antimicrob. Agents Chemother. 22:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tankovic, J., B. Perichon, J. Duval, and P. Courvalin. 1996. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40:2505-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wexler, H. M., and S. M. Finegold. 1991. Antibacterial susceptibility tests: anaerobic bacteria, p. 1133-1137. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 40.Zhao, X., C. Xu, J. Domagala, and K. Drlica. 1997. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc. Natl. Acad. Sci. USA 94:13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]