Abstract
Amikacin has been very useful in the treatment of infections caused by multiresistant bacteria because it is refractory to the actions of most modifying enzymes. However, the spread of AAC(6′)-I-type acetyltransferases, enzymes capable of catalyzing inactivation of amikacin, has rendered this antibiotic all but useless in some parts of the world. The aminoglycoside 6′-N-acetyltransferase type Ib, which is coded for by the aac(6′)-Ib gene, mediates resistance to amikacin and other aminoglycosides. RNase H mapping and computer prediction of the secondary structure led to the identification of five regions accessible for interaction with antisense oligodeoxynucleotides in the aac(6′)-Ib mRNA. Oligodeoxynucleotides targeting these regions could bind to native mRNA with different efficiencies and mediated RNase H digestion. Selected oligodeoxynucleotides inhibited AAC(6′)-Ib synthesis in cell-free coupled transcription-translation assays. After their introduction into an Escherichia coli strain harboring aac(6′)-Ib by electroporation, some of these oligodeoxynucleotides decreased the level of resistance to amikacin. Our results indicate that use of antisense compounds could be a viable strategy to preserve the efficacies of existing antibiotics to which bacteria are becoming increasingly resistant.
Aminoglycoside antibiotics play an important role in the treatment of infections caused by aerobic gram-negative bacteria and some gram-positive bacteria. In the latter case they are generally used in combination with other antibiotics (36). They exert their biological action by binding to the decoding site of the 16S rRNA, which causes a decrease in translational fidelity (23). Bacterial resistance to aminoglycosides in the clinical setting is often mediated by modifying enzymes (20). Aminoglycoside N-acetyltransferases belonging to the GCN5-related N-acetyltransferase superfamily are an important group of enzymes that mediate inactivation of aminoglycosides by catalyzing the transfer of an acetyl group to a primary amine in the antibiotic molecule (27, 37). The aminoglycoside amikacin (AMK) has been very useful in the treatment of infections caused by multiresistant bacteria because it is refractory to the actions of most modifying enzymes (19, 33, 37). However, the spread of AAC(6′)-I-type acetyltransferases, enzymes capable of catalyzing the inactivation of AMK, has rendered this antibiotic all but useless in some parts of the world (1, 10, 11, 44). Although research is being conducted to generate new aminoglycoside antibiotics (15), AMK and netilmicin remain the only semisynthetic aminoglycosides available. The aac(6′)-Ib gene has been isolated from several mobile genetic elements such as integrons and transposons present in several bacterial species belonging to the families Enterobacteriaceae, Pseudomonadaceae, and Vibrionaceae (4, 26, 27, 35, 40, 41, 43, 44). Development of a method to control aac(6′)-Ib gene expression could help the usefulness of AMK to be regained. Antisense oligonucleotide technology has been used to control gene expression in eukaryotes and to a lesser extent in prokaryotes (6, 12, 14, 16, 17, 46, 50). Strategies based on the use of antisense oligonucleotides to fight antibiotic resistance have been researched before. The bactericidal action of norfloxacin was increased by the actions of antisense molecules that interfered with expression of the marRAB operon in Escherichia coli (50), and resistance to β-lactams was decreased by inhibition of bla expression in the presence of antisense peptide nucleic acids (12). The construction of plasmids coding for small oligoribonucleotides whose sequences are antisense to those of resistance genes mediated conversion to susceptibility by inducing RNase P digestion of the mRNA (14), and a plasmid containing a vanH promoter-vanA antisense gene cassette inhibited resistance to vancomycin in a clinical Enterococcus faecalis isolate (46). In this work we mapped the aac(6′)-Ib mRNA to identify single-stranded regions accessible for interaction with antisense oligodeoxynucleotides (ODNs). On the basis of this information we designed ODNs that interfere with the synthesis of AAC(6′)-Ib and that induce a reduction of the aac(6′)-Ib-mediated AMK resistance levels.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmid pNW1 is an F′ derivative that includes a copy of aac(6′)-Ib. Construction of this plasmid was carried out as follows. First, a recombinant plasmid, pNW0, was generated by insertion of a DNA fragment containing the aac(6′)-Ib gene into the EcoRI site of pFW11, a plasmid vector that codes for resistance to chloramphenicol (49). Next, pNW0 was introduced by transformation into E. coli CSH100 [F′ lac proA+ proB+ (lacIq lacPL8)] (28). Since the aac(6′)-Ib flanking regions in pNW0 share homology with a region of F′, homologous recombination in E. coli CSH100(pNW0) results in the integration of a fragment containing aac(6′)-Ib into F′. Conjugation of this strain with E. coli FW102 (a streptomycin-resistant derivative of E. coli CSH142) (49) and selection for colonies resistant to streptomycin and AMK but susceptible to chloramphenicol led to the isolation of E. coli FW102 harboring pNW1, an F′ derivative including aac(6′)-Ib. This plasmid was transferred by conjugation to E. coli TOP10; and the transconjugant, E. coli TOP10(pNW1), was used as a reporter strain for in vivo experiments. Plasmid pRS201 was generated by inserting an amplicon with the aac(6′)-Ib gene into pCR2.1. The amplicon was obtained by using primers 5′-GCCTCGTGATACGCCTATTTTT and 5′-GTTTAACGTTTGACATGAGGGC and recombinant plasmid pJHCT15.2 (45) as the template.
General DNA and RNA procedures.
Growth of bacteria was carried out by using Lennox Luria (L) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl). Transformation and electroporation were performed as described previously (34). Plasmid DNA preparations were carried out by using the QIAspin miniprep kit (Qiagen). A linear DNA template with aac(6′)-Ib under the control of the T7 promoter was generated by amplification by using the TaqPCR Master Mix kit (Qiagen), plasmid pRS201 DNA as the template, and primers 5′-TTGTAATACGACTCACTATAGGGAGACATGAGACAATAACCCTGATAAATGCTTC (the minimal T7 promoter is underlined, and the +1 nucleotide is double underlined (18) and 5′-GTTTAACGTTTGACATGAGGGC according to the recommendations of the supplier. This template was used to synthesize mRNA by using the MEGAscript T7 kit (Ambion) under the conditions specified by the supplier. To label the mRNA, about 5 μg was dephosphorylated by incubation with 5 U of alkaline phosphatase (New England Biolabs) in a buffer containing 100 mM NaCl, 50 mM Tris-HCl (pH 7.9), 10 mM MgCl2, and 1 mM dithiothreitol in a total volume of 10 μl. After phenol-chloroform extraction, the dephosphorylated mRNA was 5′ end labeled with 25 μCi of [γ-32P]ATP and 5 U of phage T4 polynucleotide kinase (New England Biolabs) in kinase buffer (50 mM Tris HCl [pH 7.5], 10 mM MgCl2, 5 mM dithiothreitol, 0.1 mM spermidine) in a final volume of 30 μl. The reaction mixture was incubated at 37°C for 1 h, and the unincorporated nucleotides were removed by using ProbeQuant G-50 microcolumns (Amersham Pharmacia Biotech). The ODNs were end labeled by a similar protocol, except that the dephosphorylation reaction was omitted. The computer-predicted secondary structure of the aac(6′)-Ib mRNA was generated by using the program mfold (version 3.1) (25, 52). Sequencing reactions were carried out by using the Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit (U.S. Biochemicals) with primer 5′-TCCGCTCATGAGACAATAACCCTG according to the recommendations of the supplier. Sequencing with this primer permits a comparison of the sequencing products with the fragments generated by RNase H cleavage of 5′-labeled mRNA.
RNase H reactions.
Single-stranded regions available for pairing to complementary ODNs were identified by RNase H mapping basically as described by Ho et al. (17). Solutions of random or semirandom (the 11th nucleotide is constrained) 20-mer libraries were heated for 3 min at 95°C and immediately placed on ice. Therafter, 1 μl of 10× RNase H buffer (400 mM Tris [pH 8.0], 40 mM MgCl2, 10 mM dithiothreitol) and 1 μl of labeled mRNA (about 0.1 μg) were added, and the mixture was incubated for 90 min at 25°C before addition of 1 U of RNase H (Epicenter) in a total volume of 10 μl. The final concentration of ODNs was 30 μM. After 10 min at 25°C, the reactions were stopped by addition of 2 volumes of Gel Loading Buffer II (Ambion), and the mixture was heated for 3 min at 95°C and analyzed on a 6% denaturing polyacrylamide gel by using a Sequi-Gen II nucleic acid sequencing cell (Bio-Rad). Radioactivity was detected with a phosphoimager (Cyclone Storage Phosphor system; Packard). For RNase H cleavage assays, the reactions were carried out by using the same protocol described above, but with replacement of the ODN libraries with the specific ODN (final concentration, 10 μM).
RNA binding assays.
Gel mobility shift assays were performed by mixing 5′-end-labeled ODNs (10 nM) with various concentrations of mRNA in a buffer containing 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. After 2 h at 25°C, 10 μl of 10% glycerol and 3 μl of gel loading buffer were added to the reaction mixture and the samples were analyzed on a 5% native polyacrylamide gel (acrylamide-bisacrylamide [19:1]).
In vitro activities of ODNs.
The in vitro activities of the ODNs were determined in cell-free coupled transcription-translation reactions in the presence or absence of ODNs. Coupled transcription-translation reactions were carried out with the EcoPro T7 system (Novagen) as recommended by the supplier. The reactions were performed in the presence of the same linear DNA template used to synthesize the mRNA and 40 μCi (specific activity, 1,000 Ci/mmol) of [35S]methionine (Perkin-Elmer), with the addition of 10 μM ODN when indicated. The products were electrophoresed in a sodium dodecyl sulfate-polyacrylamide gel as described previously (42). The gel was then fixed in a solution containing 30% methanol and 10% glacial acetic acid, immersed in fluorographic reagent (Amplify; Amersham) for 30 min with gentle agitation, dried, and exposed to a phosphoimager screen to detect radioactivity.
In vivo activities of ODNs.
The in vivo activities of the ODNs were determined essentially as described by White et al. (50), with slight modifications. ODNs were introduced into E. coli TOP10(pNW1) cells by electroporation. The cell suspensions (107 cells in a total volume of 50 μl) were placed on ice for 2 min and transferred to room temperature for 30 min to allow interaction between the mRNA and the ODNs. Then, 450 μl of L broth was added and the suspensions were incubated at 30°C for 30 min. At this point AMK was added (20 μg/ml) and the suspensions were incubated at 30°C for 1.5 h. The cells were then plated on L agar (no selection). The growing colonies represent cells that survived the exposure to AMK.
RESULTS
Mapping of aac(6′)-Ib mRNA.
The rational selection of antisense ODNs that can induce inhibition of gene expression requires knowledge of mRNA regions accessible for interaction with complementary ODNs. The mRNA molecules form complex secondary and tertiary structures that leave few regions available for pairing with antisense ODNs. Since it has been shown that computer-based models of the structures offer limited help in identifying these regions (21), the information provided by these models must be complemented with experimental data. We identified accessible single-stranded regions in aac(6′)-Ib mRNA by RNase H mapping in combination with computer-based prediction of the mRNA secondary structure.
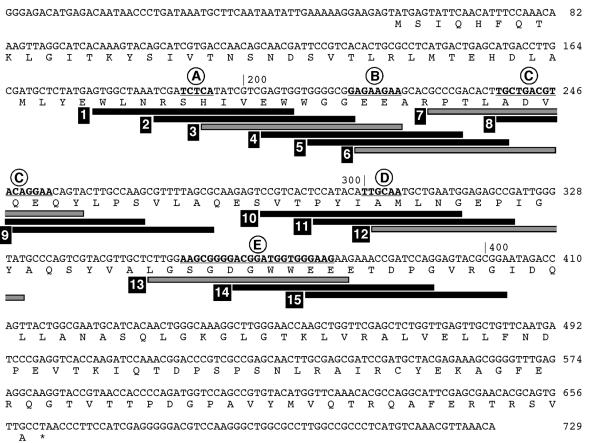
To identify accessible single-stranded regions by RNase H mapping, libraries of semirandom or random ODNs were mixed with radiolabeled aac(6′)-Ib mRNA, followed by treatment with RNase H. The semirandom libraries consisted of ODNs in which all but one of the nucleotides were randomized. The products of the reactions were analyzed in denaturing polyacrylamide gels (Fig. 1). We could detect five main regions (regions A to E) susceptible to digestion by RNase H. Two of them, regions A and D, produced an identical profile when any of the five ODN libraries was added to the reaction mixture (Fig. 1). Conversely, the cleavage fragments that defined regions B, C, and E were not identical when each of the ODN libraries was used (Fig. 1). Since RNase H digestion is not sequence dependent and the reactions generated discrete cleavage fragments, they must correspond to regions in the mRNA that were available to bind to the ODNs. Although other investigators (17) reported that experiments carried out with completely random libraries gave results less optimal than those obtained with semirandom libraries, we did not observe major differences in the qualities of the reactions when random or semirandom libraries were included in the reaction mixtures (Fig. 1). Controls for this experiment included reaction mixtures lacking ODNs or RNase H, or both. In all three cases, no mRNA cleavage was observed (Fig. 1). Another control included all reagents in the reaction mixture, but the mRNA was denatured by heating before addition of RNase H. As expected, the RNA was completely degraded under these conditions (Fig. 1). The gel in Fig. 1 also shows the products of a sequencing reaction with the DNA fragment corresponding to the mRNA and the locations of the DNA and RNA molecular size markers. The DNA and RNA 200-nucleotide fragments run with an apparent difference of about 3.5 nucleotides under the polyacrylamide gel electrophoresis conditions used. The difference in migration becomes smaller as the fragments increase in size, and the 300-nucleotide DNA and RNA fragments run practically at the same position (Fig. 1). The nucleotide sequences of the five main cleavage regions in the mRNA are shown in Fig. 2. These locations were determined by taking into account the positions of the molecular size markers and the DNA nucleotide sequence (Fig. 1).
FIG. 1.
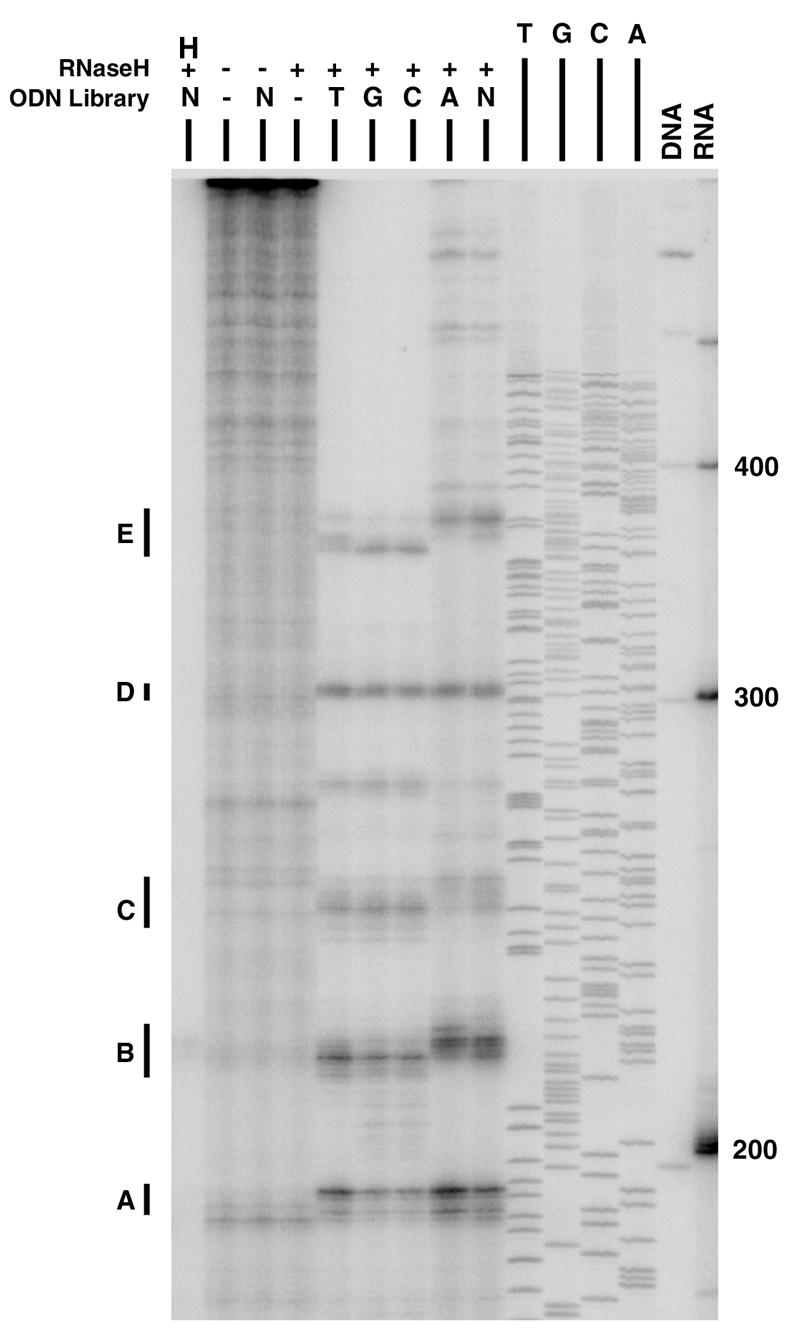
Determination of aac(6′)-Ib mRNA region accessible to antisense ODNs. RNase H mapping was carried out as described in Materials and Methods with a random library (N) or each of four semirandom libraries in which the 11th nucleotide (A, C, G, or T) was constrained. Control reactions were performed by leaving out ODNs or RNase H, or both, as indicated on top of the gel. Another control reaction mixture included all of the reagents, but the mRNA was denatured by heating before addition of RNase H (H). Size controls included DNA and RNA molecular weight standards (the number of nucleotides is shown to the right) as well as the products of a sequencing reaction. The regions (regions A through E) susceptible to RNase H digestion are indicated to the left of the gel.
FIG. 2.
Nucleotide sequences of the aac(6′)-Ib mRNA and ODNs. The complete sequence of the coding strand corresponding to the mRNA used in the mapping experiments is shown. The nucleotides in regions A to E are shown in boldface and are underlined. The bars represent the sequences of the ODNs assayed in RNase H cleavage and binding experiments (Fig. 4). Gray bars represent the ODNs that were selected for the binding and in vivo experiments (Fig. 5). The ODN sequences are complementary to those shown in the diagram.
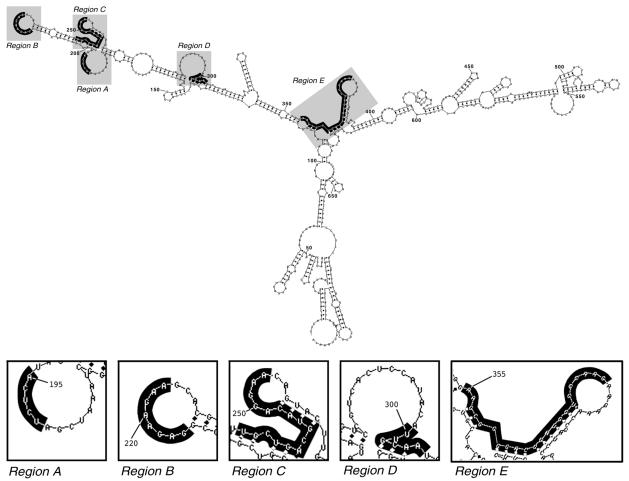
The single-stranded regions identified by RNase H mapping were compared to those predicted by generating a secondary structure of the aac(6′)-Ib mRNA by using the mfold program (52). Figure 3 shows the predicted secondary structure and the locations of the regions identified by RNase H mapping. Region A is located within a predicted loop. Although in region B the main products of digestion run at different positions depending on the ODN library used (Fig. 1), they were all located within a predicted single-stranded region. The main products of region C were also not identical when the reactions were carried out in the presence of different ODN libraries (Fig. 1). However, in this case the nucleotides encompassed by this region were not all located within a single-stranded loop. As shown in Fig. 3, this region has a more complex structure: a mismatch (C240), 3 nucleotides (U241, G242, A243) in a double-stranded structure, a mismatch (C244), and 4 nucleotides (G245, U246, A247, C248) in a double-stranded structure that is matched to a region in the RNA molecule different from that to which UGA hybridized, followed by nucleotides that are part of a loop (A249, G250, G251, A252, A253). The stronger bands within this region coincide with C244 and the nucleotides in the loop (Fig. 1). However, there are bands along the whole region, indicating that ODNs hybridized to nucleotides predicted to be in double-stranded form. It is possible that the predicted structure does not reflect exactly the real structure of the RNA molecule or that the complexity of the spatial structure at this region permits enough breathing at the short double-stranded stems to allow interaction with ODNs and, consequently, cleavage by the RNase H. A case like this has recently been described in which a stem RNA region was digested by a maxizyme due to a binding process that occurred during the breathing of the stem structure (22). The RNase H cleavage products that define region D were identical for all five ODN libraries (Fig. 1). Interestingly, although there was a big single-stranded loop, the products of digestion encompassed the end of the loop (U300, U301), 3 nucleotides (G302, C303, A304) predicted to exist as a double-stranded mismatch, and a nucleotide (A305) predicted to exist as a single-stranded mismatch (Fig. 3). It is possible that, unlike in the computer-generated structure, this region actually exists as a bigger single-stranded loop that includes the sequence GCAA. Conversely, as was discussed for region C, there may be enough breathing at the 3-nucleotide double-stranded sequence for the ODNs to be able to bind. However, for either of these two possibilities, we do not know why there was a preferential binding of the ODNs at these particular nucleotides of region D as opposed to the other nucleotides of the loop. The products of RNase H digestion that define region E are spread over a larger section of the mRNA molecule compared to the lengths of the products that define the other regions. The main products were located near the mismatched A355 and A356 nucleotides when the ODN libraries used were semirandom with G or C as the fixed nucleotide (Fig. 1). The main products in the case of semirandom libraries with A as the fixed nucleotide and the random library coincided with the loop that includes nucleotides G372 to G377. However, less intense but clear bands corresponding to nucleotides near these two locations were generated by the reactions containing either the semirandom libraries with T or A as the fixed nucleotide or the random library (Fig. 1). Examination of the predicted structure shows that it is a complex region, and as was discussed above for the other regions, the predicted structure may not be entirely correct or breathing at the region may result in hybridization and RNase H digestion. In Fig. 3 this region encompassing A355 to G377 is labeled, but RNase H digestion is more efficient at the nucleotides around A355 and A356 as well as G372, G373, and G374.
FIG. 3.
Secondary structure of aac(6′)-Ib mRNA generated with mfold software (52). The nucleotides over a black background indicate the regions identified by RNase H mapping.
RNase H cleavage of mRNA in the presence of ODNs.
A series of 15 ODNs encompassing regions A to E were designed for further analysis. Their nucleotide sequences are shown in Fig. 2. Their efficiencies for mediating RNase H degradation of the aac(6′)-Ib mRNA were determined by incubation of radiolabeled native mRNA with the antisense ODNs and RNase H. The products of these reactions were analyzed in denaturing polyacrylamide gels. Figure 4 shows that ODNs 1 to 3 (Fig. 4a) efficiently induced degradation of the mRNA. In the case of ODN 3, no undigested mRNA could be detected after the reaction, while there was a small amount of intact mRNA after incubation with ODNs 1 and 2. Conversely, inspection of the gel in Fig. 4b indicates that ODNs 4 to 6 were not equally efficient at inducing cleavage of mRNA. While ODN 4 practically did not induce mRNA digestion by RNase H, ODNs 5 and 6 efficiently mediated RNase H cleavage (Fig. 4b). The assay results shown in Fig. 4c indicate that while ODN 7 induced complete degradation of mRNA, in the presence of ODNs 8 and 9, not all of the mRNA was cleaved. The assay results shown in Fig. 4d and e indicate that with the exception of ODN 10, which did not mediate complete cleavage, all ODNs (ODNs 11 to 15) induced complete digestion of the mRNA. Inspection of the results shown in Fig. 4a through d indicates that the main reaction products are those expected on the basis of the location targeted by the ODNs. In the case of ODNs 13 to 15 (Fig. 4e), there were two main products in each reaction. One of them had the expected size, while the other one was the same size as the product obtained after incubation of the mRNA with ODN 6. It is possible that the presence of the repeat GAAGAA sequence found in region B and the region E loops (Fig. 2 and 3) is responsible for this result. However, the sole presence of the repeat does not seem to be enough for interaction with the heterologous ODN because ODN 6 did not mediate the cleavage of mRNA at region E.
FIG. 4.
RNase H cleavage of aac(6′)-Ib mRNA. Radiolabeled mRNA was incubated with the ODNs specified on top (the lane numbers correspond to the ODN numbers shown in Fig. 2) and RNase H, as described in Materials and Methods. The products of the reaction were analyzed in denaturing polyacrylamide gels. RNA molecular size markers (MW; in nucleotides) are shown to the right. Reactions were performed with groups of three ODNs and control reaction mixtures that excluded ODN or RNase H, or both. Another control mixture contained an ODN with a sense sequence (S). For each group of reactions a gel was run with the controls. Since the controls behaved identically in all experiments, for clarity only the controls for the first group are shown.
mRNA binding of ODNs.
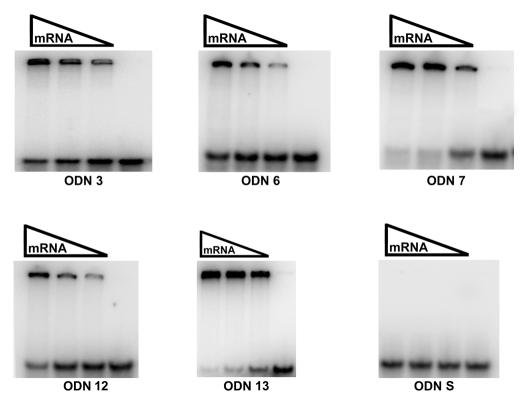
On the basis of the results shown in Fig. 4, we selected ODNs that include sequences in regions A to E and that were highly efficient in RNase H cleavage experiments. Their sequences are shown in Fig. 2 (gray bars). The selected ODNs were end labeled and used in mRNA binding experiments. Figure 5 shows that ODNs 3, 6, and 12, which target regions A, B, and D, respectively, bind to mRNA with less affinity than ODNs 7 and 13, which are antisense to the regions encompassing regions C and E, respectively. However, comparison of these results with those obtained in RNase H cleavage experiments indicates that while all five ODNs were very efficient in inducing mRNA digestion by RNase H, three of them (ODNs 3, 6, and 12) showed lower binding efficiencies compared to the binding properties of ODNs 7 and 13. An explanation for this apparent discrepancy may be that in the RNase H cleavage experiments a single ODN molecule can bind to an mRNA molecule, mediate RNase H digestion, and then bind to another one to repeat the process, while in the binding experiments, the products shifted must be present in a 1:1 stoichiometric ratio. Therefore, while the binding affinities of ODNs 3, 6, and 12 are lower than those exhibited by the other two ODNs, it may be sufficient to degrade most mRNA molecules under the conditions used to carry out the RNase H cleavage experiments.
FIG. 5.
ODN binding to aac(6′)-Ib mRNA. Various concentrations of mRNA (100, 50, or 10 nM) were incubated with 5′-end-labeled ODNs (10 nM) as described in Materials and Methods and analyzed in native polyacrylamide gels. A reaction without mRNA was also performed (the results are indicated in the rightmost lanes of the gels). The upper bands are ODNs bound to mRNA. The ODNs used are indicated below the gels (the numbers correspond to those shown in Fig. 2). ODN S, control mixture containing an ODN with a sense sequence.
In vitro activities of ODNs.
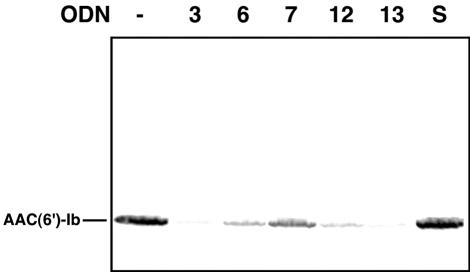
To study the in vitro effects of the five ODNs on expression of the aac(6′)-Ib gene, a DNA fragment consisting of the structural gene under the control of the T7 promoter was used as the template in cell-free coupled transcription-translation reactions containing radioactive methionine in the presence or absence of ODNs. Figure 6 shows that in the presence of any of the antisense ODNs there was a reduction in the amount of protein synthesized compared to the amount synthesized in reactions carried out without ODNs or with a control ODN with a sense sequence. ODN 7 showed less inhibitory activity than the other ODNs. These experiments demonstrated that the selected antisense ODNs can interfere with the expression of aac(6′)-Ib.
FIG. 6.
In vitro activities of ODNs. Cell-free coupled transcription-translation reactions were carried out as described in Materials and Methods in the absence (−) or presence of the indicated ODNs at 10 μM (the numbers correspond to those shown in Fig. 2). The products were run on sodium dodecyl sulfate- polyacrylamide gels, and the radioactivity was detected with a phosphoimager. S, control mixture containing an ODN with a sense sequence.
In vivo activities of ODNs.
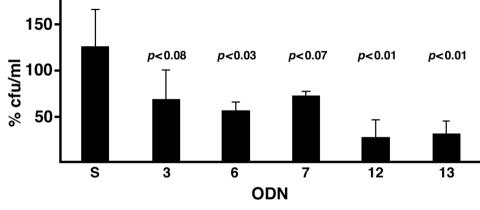
To evaluate the actions of the selected ODNs on the ability of AMK to exert a toxic effect on E. coli harboring the low-copy-number plasmid pNW1, electrocompetent cells were transformed with ODNs and exposed to the antibiotic before being plated on L agar without selection. The number of colonies in the plates reflected the number of cells that survived the treatment with AMK. Figure 7 shows the percentage of colonies formed by ODN-treated cells that were exposed to AMK compared with the number of cells that were subjected to electroporation without the addition of ODNs before being exposed to the antibiotic. A sample was treated with an ODN that is not antisense (Fig. 7, bar S). In this case the number of surviving cells is comparable to the number of surviving cells that were not transformed with an ODN. A comparison of the surviving cells that were treated with antisense ODN 3, 6, 7, 12, or 13 and the surviving cells that were treated with the control ODN (Fig. 7, bar S) indicates that in the presence of any of the five selected ODNs, there was a reduction in the number of surviving cells after exposure to AMK (Fig. 7). However, in the case of ODNs 3 and 7, the differences observed were not statistically significant (Fig. 7). These results strongly suggest that the appropriate antisense ODNs can interfere with the expression of AMK resistance.
FIG. 7.
Effects of antisense ODNs. Cells were treated with the indicated ODN (the numbers correspond to those shown in Fig. 2) before being exposed to AMK, as described in Materials and Methods. The numbers of CFU per milliliter were calculated from the numbers of colonies growing on plates with no selection and represent the number of cells that survived exposure to AMK after introduction of the ODNs. The percent CFU per milliliter represents the fraction of surviving cells compared to the number of cells in the sample that was subjected to electroporation without an ODN. Electroporation with a sense ODN (S) was used as a control. The numbers were obtained by averaging the results of three experiments. Error bars show standard deviations. P values were calculated with respect to the results for the control sense ODN. P values of <0.05 are statistically significant.
DISCUSSION
Pathogenic bacteria are becoming more resistant to antibiotics, and there have already been reports of multiple-drug-resistant pathogenic bacterial strains against which most available antibiotics are ineffective (39). Bacteria have developed diverse strategies to resist the actions of antibiotics. These strategies are modification of the target, removal of the antibiotic by efflux pumps that impede its accumulation inside the cell, or enzymatic modification of the antibiotic molecule (48). For aminoglycoside antibiotics, the last mechanism is the most common and involves three kinds of enzymes, N-acetyltransferases, O-adenylytransferases, O-phosphotransferases (27). To preserve the effectiveness of aminoglycoside antibiotics, it would be useful to generate compounds that can interfere with the modification mechanism. Other strategies involve the use of antisense ODNs to interfere with the expression of the modifying enzymes (14, 16, 17, 50).
Resistance to AMK in the clinical setting is most commonly mediated by N-acetyltransferases of the AAC(6′)-I type. The aac(6′)-Ib gene, responsible for resistance to several aminoglycosides, including AMK, has been found in a large number of bacterial species (4, 27, 35, 40, 41, 43, 44). Inhibition of expression of this gene would render these bacteria susceptible to AMK and other aminoglycosides. In this work we tested whether it is possible to interfere with the expression of aac(6′)-Ib using antisense ODNs, a mechanism that holds promise in the treatment of several diseases (21). We located regions of the mRNA available for interaction with ODNs using a combination of RNase H mapping and secondary structure prediction with mfold software. While the computer-generated structure shows several potential single-stranded regions that could be available for interacting with ODNs, the experimental results indicated that only some of them were capable of effectively binding to ODNs from the libraries. These results may reflect the facts that the mRNA molecule forms complex secondary and tertiary structures and only a limited number of the single-stranded regions are exposed and accessible for binding of the ODNs. Our experimental results identified five single-stranded regions that overlapped or that were very close to the single-stranded regions predicted with the mfold software. Regions A and B coincided with single-stranded regions predicted with the mfold software (Fig. 3). However, regions C, D, and E, as defined by RNase H mapping, encompass fragments predicted to exist as single- and double-stranded regions (Fig. 1 and 3). An explanation for the detection of fragments cleaved at these predicted double-stranded locations could be that the computer-generated structure is not completely correct or that there is enough breathing at the short double-stranded regions to allow cleavage by RNase H. A stem RNA region has recently been shown to be digested by a single-strand-specific enzyme due to a binding process that occurred during the breathing of the stem structure (22). Furthermore, in all five regions only parts of the loops were identified by RNase H mapping, which may reflect the complexities of the secondary and tertiary structures, which permit only a few nucleotides to be available for interaction with ODNs. The results of our mapping experiments and comparison to computer-based structure models coincide with the results of other studies that indicate that software prediction of the mRNA secondary structure is often not sufficient to predict the single-stranded regions available for interaction with ODNs (21).
Most ODNs among a group of ODNs designed on the basis of the mapping results were able to induce efficient mRNA cleavage by RNase H. However, when some of them were tested, a small percentage of the mRNA molecules were left undigested; and one of them (ODN 4) did not induce cleavage at all (Fig. 4). Binding experiments with ODNs that were selected among the ODNs that were more efficient at mediating RNase H cleavage showed that they bound to mRNA with different efficiencies (Fig. 5). It is our hypothesis that a possible explanation for this apparent inconsistency is the fact that, after binding and cleavage of an mRNA molecule, an individual ODN molecule can again be used to mediate cleavage of another mRNA molecule. All five ODNs selected interfered with synthesis of the protein in in vitro cell-free transcription translation experiments, although in one case there was a lesser degree of interference. Since no RNase H was added to the reaction mixtures, the mechanism of inhibition in these assays was most probably through steric hindrance. However, the extracts used may have other activities that play a role in the interactions between ODNs and mRNA. To determine the ability of the selected ODN to interfere with expression of AMK resistance in vivo, we performed survival experiments, as described by White et al. (50). The experimental results indicate that at least three of the ODNs (those targeting regions B, D, and E) were able to induce a statistically significant reduction in viability after exposure to AMK in the presence of the ODNs. The other two ODNs tested also showed a tendency to induce a reduction in viability, but the difference in the results compared to those obtained with the control ODN was not statistically significant. Different factors can affect the results of these experiments; among these are the number of molecules that reached the cytoplasmic compartment through the electroporation process and their stability. Since we did not use nonhydrolyzable analogs, ODNs that are hydrolyzed faster can be less able to interfere with synthesis of the protein, not because they are not appropriate but because they are degraded faster. Experiments with nonhydrolyzable ODN derivatives will permit us to determine if stability is responsible for the differences in the efficiencies of the inhibitory activities of the ODNs. Unmodified ODNs are also rapidly degraded by nucleases present in blood and other body fluids (8, 21). Their half-lives have been determined to be on the order of minutes in blood and somewhat longer in cerebrospinal fluid (47). Therefore, the use of nonhydrolyzable ODN derivatives will take us a step closer to the practical use of the antisense strategy to regain the usefulness of antibiotics to which bacteria have become resistant. We do not know the mechanism by which the ODNs reduced the levels of resistance to AMK; the most probable ones are steric hindrance after binding to the mRNA or induction of cleavage through the cell's RNase H. Although the results of the in vitro experiments suggest that under those conditions steric hindrance may play an important role, the mechanism in vivo may be different. Experiments with RNase H-deficient E. coli mutants will help clarify this question.
While in this work we show that antisense ODNs could, in theory, be developed to preserve the efficacy of AMK against the rising number of resistant strains isolated in the clinical setting in several parts of the world, many problems remain to be solved. Aminoglycoside 6′-N-acetyltransferase type Ib variants that exhibit variability at the N-terminal region have been isolated (4). These differences could result in changes in the tertiary structure of the mRNA, making some of the variants resistant to ODNs that can efficiently inhibit the expression of other variants. We generated secondary structures of the mRNAs coded for by these variants using the mfold program, and we found that the single-stranded regions identified in this work are still present (unpublished results). We are now performing RNase H mapping and testing the effects of selected ODNs to determine if they are effective against all or most variants. In addition, the aac(6′)-Ib gene is often found in high-copy-number plasmids (51), which may make it very difficult to reach inhibition levels high enough for phenotypic conversion to susceptibility. A solution to this problem could be the finding of inhibitors that interfere with the residual enzymatic activity. Although aminoglycoside 6′-N-acetyltransferases have recently been characterized (2, 4, 5, 7, 24, 30-32, 37, 38), no inhibitors have reached the market to date. However, the recent discovery of inhibitors of aminogylcoside-modifying enzymes (1a, 3) and the known inhibitors of β-lactamases (29) suggest that if enough research efforts are devoted to finding them, appropriate inhibitors of N-acetyltransferases will be developed. Thus, antisense ODNs or ODN analogs and enzyme inhibitors could have a synergistic activity that results in the phenotypic conversion to AMK susceptibility. Finally, methods to ensure that the compounds reach the cell's cytoplasm must be developed. Although in general bacteria do not take up most ODNs or ODN analogs (13), some encouraging results have already been reported. ODN analogs have been found to reach the cytoplasm of Mycobacterium (16), and attachment of cell-permeabilizing peptides to peptide nucleic acids dramatically improved cell uptake (9). We are developing additional strategies to improve cell uptake of ODNs and ODN analogs.
Other research groups have also obtained promising results using antisense strategies to deal with antibiotic resistance. White et al. (50) demonstrated an increase in the bactericidal action of norfloxacin by the actions of antisense molecules selected on the basis of the results of RNase H mapping of mRNA. These antisense molecules interfered with expression of the E. coli marRAB operon. Resistance to β-lactams was decreased by inhibition of expression of bla in the presence of antisense peptide nucleic acids, which are DNA mimic compounds with a pseudopeptide backbone that are very stable in body fluids (12). A different strategy consisted of the use of endogenous the RNase P enzyme to degrade the cat mRNA and induce susceptibility to chloramphenicol by transformation of E. coli cells with recombinant plasmids coding for small oligoribonucleotides that can form the appropriate stemlike structure to induce RNase P digestion of the mRNA (14). Resistance to vancomycin could be inhibited when a clinical E. faecalis isolate harbored a recombinant plasmid containing a vanH promoter-vanA antisense gene cassette (46). Our results as well as these alternative antisense approaches indicate that antisense ODNs or ODN analogs could be used as part of viable strategies to preserve the efficacies of existing antibiotics to which bacteria are becoming increasingly resistant.
Acknowledgments
This work was supported by Public Health Service grant AI47115 (to M.E.T.) from the National Institutes of Health. R.S. and N.W. were supported in part by MSD grant R25 GM56820 from the National Institutes of Health.
We are indebted to Fred Whipple for generously providing plasmid pFW11 and E. coli FW102. We thank Ramona Chavideh for excellent technical support.
REFERENCES
- 1.The Aminoglycoside Resistance Study Groups. 1995. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J. Chemother. 7(Suppl. 2):17-30. [PubMed] [Google Scholar]
- 1a.Boehr, D. D., K. Draker, K. Koteva, M. Bains, R. E. Hancock, and G. D. Wright. 2003. Broad-spectrum peptide inhibitors of aminoglycoside antibiotic resistance enzymes. Chem. Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 2.Boehr, D. D., S. I. Jenkins, and G. D. Wright. 2003. The molecular basis of the expansive substrate specificity of the antibiotic resistance enzyme aminoglycoside acetyltransferase-6′-aminoglycoside phosphotransferase-2". The role of ASP-99 as an active site base important for acetyl transfer. J. Biol. Chem. 278:12873-12880. [DOI] [PubMed] [Google Scholar]
- 3.Burk, D. L., and A. M. Berghuis. 2002. Protein kinase inhibitors and antibiotic resistance. Pharmacol. Ther. 93:283-292. [DOI] [PubMed] [Google Scholar]
- 4.Casin, I., B. Hanau-Bercot, I. Podglajen, H. Vahaboglu, and E. Collatz. 2003. Salmonella enterica serovar Typhimurium bla(PER-1)-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavideh, R., S. Sholly, D. Panaite, and M. E. Tolmasky. 1999. Effects of F171 mutations in the 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] enzyme on susceptibility to aminoglycosides. Antimicrob. Agents Chemother. 43:2811-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallabrida, S. M., M. A. De Sousa, and D. H. Farrell. 2000. Expression of antisense to integrin subunit beta 3 inhibits microvascular endothelial cell capillary tube formation in fibrin. J. Biol. Chem. 275:32281-32288. [DOI] [PubMed] [Google Scholar]
- 7.Dery, K., B. Soballe, M. Witherspoon, D. Bui, R. Koch, D. J. Sherratt, and M. E. Tolmasky. 2003. The aminogylcoside 6′-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell's cytoplasm. Antimicrob. Agents Chemother. 47:2897-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dove, A. 2002. Antisense and sensibility. Nat. Biotechnol. 20:121-124. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson, M., P. E. Nielsen, and L. Good. 2002. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277:7144-7147. [DOI] [PubMed] [Google Scholar]
- 10.Flidel-Rimon, O., E. Leibovitz, A. Juster-Reicher, M. Amitay, A. Miskin, Y. Barak, and B. Mogilner. 1996. An outbreak of antibiotic multiresistant Klebsiella at the Neonatal Intensive Care Unit, Kaplan Hospital, Rehovot, Israel, November 1991 to April 1992. Am. J. Perinatol. 13:99-102. [DOI] [PubMed] [Google Scholar]
- 11.Galani, I., E. Xirouchaki, K. Kanellakopoulou, G. Petrikkos, and H. Giamarellou. 2002. Transferable plasmid mediating resistance to multiple antimicrobial agents in Klebsiella pneumoniae isolates in Greece. Clin. Microbiol. Infect. 8:579-588. [DOI] [PubMed] [Google Scholar]
- 12.Good, L., and P. E. Nielsen. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355-358. [DOI] [PubMed] [Google Scholar]
- 13.Good, L., R. Sandberg, O. Larsson, P. E. Nielsen, and C. Wahlestedt. 2000. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiology 146(Pt 10):2665-2670. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada, C., R. Salavati, and S. Altman. 1997. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc. Natl. Acad. Sci. USA 94:8468-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddad, J., L. P. Kotra, B. Llano-Sotelo, C. Kim, E. Azucena, M. Liu, S. Vakulenko, C. Chow, and S. Mobasheri. 2002. Design of novel antibiotics that bind to the ribosomal acyltransfer site. J. Am. Chem. Soc. 124:3229-3237. [DOI] [PubMed] [Google Scholar]
- 16.Harth, G., P. C. Zamecnik, J. Y. Tang, D. Tabatadze, and M. A. Horwitz. 2000. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-l-glutamate/glutamine cell wall structure, and bacterial replication. Proc. Natl. Acad. Sci. USA 97:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, S. P., D. H. Britton, B. A. Stone, D. L. Behrens, L. M. Leffet, F. W. Hobbs, J. A. Miller, and G. L. Trainor. 1996. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 24:1901-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imburgio, D., M. Rong, K. Ma, and W. T. McAllister. 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 39:10419-10430. [DOI] [PubMed] [Google Scholar]
- 19.Karlowsky, J. A., M. E. Jones, C. Thornsberry, I. R. Friedland, and D. F. Sahm. 2003. Trends in antimicrobial susceptibilities among Enterobacteriaceae isolated from hospitalized patients in the United States from 1998 to 2001. Antimicrob. Agents Chemother. 47:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurreck, J. 2003. Antisense technologies. Eur. J. Biochem. 270:1628-1644. [DOI] [PubMed] [Google Scholar]
- 22.Kuwabara, T., M. Warashina, and K. Taira. 2002. Cleavage of an inaccessible site by the maxizyme with two independent binding arms: an alternative approach to the recruitment of RNA helicases. J. Biochem. 132:149-155. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, S. R., R. L. Gonzalez, and J. D. Puglisi. 2003. Comparison of X-ray crystal structure of the 30S subunit-antibiotic complex with NMR structure of decoding site oligonucleotide-paromomycin complex. Structure 11:43-53. [DOI] [PubMed] [Google Scholar]
- 24.Magnet, S., T. A. Smith, R. Zheng, P. Nordmann, and J. S. Blanchard. 2003. Aminoglycoside resistance resulting from tight drug binding to an altered aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 47:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 26.Melano, R., A. Corso, A. Petroni, D. Centron, B. Orman, A. Pereyra, N. Moreno, and M. Galas. 2003. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J. Antimicrob. Chemother. 52:36-42. [DOI] [PubMed] [Google Scholar]
- 27.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, and K. J. Shaw. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H., D. Ganem, P. Lu, and A. Schmitz. 1977. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J. Mol. Biol. 109:275-298. [DOI] [PubMed] [Google Scholar]
- 29.Page, M. G. 2000. β-Lactamase inhibitors. Drug Resist. Update 3:109-125. [DOI] [PubMed] [Google Scholar]
- 30.Panaite, D. M., and M. E. Tolmasky. 1998. Characterization of mutants of the 6′-N-acetyltransferase encoded by the multiresistance transposon Tn1331: effect of Phen171-to-Leu171 and Tyr80-to-Cys80 substitutions. Plasmid 39:123-133. [DOI] [PubMed] [Google Scholar]
- 31.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reish, O., S. Ashkenazi, N. Naor, Z. Samra, and P. Merlob. 1993. An outbreak of multiresistant Klebsiella in a neonatal intensive care unit. J. Hosp. Infect. 25:287-291. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, vol. 1, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scaglione, F., S. Dugnani, G. Demartini, M. M. Arcidiacono, C. E. Cocuzza, and F. Fraschini. 1995. Bactericidal kinetics of an in vitro infection model of once-daily ceftriaxone plus amikacin against gram-positive and gram-negative bacteria. Chemotherapy 41:239-246. [DOI] [PubMed] [Google Scholar]
- 37.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shmara, A., N. Weinsetel, K. J. Dery, R. Chavideh, and M. E. Tolmasky. 2001. Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob. Agents Chemother. 45:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover, F. C. 2001. Development and spread of bacterial resistance to antimicrobial agents: an overview. Clin. Infect. Dis. 33(Suppl. 3):S108-S115. [DOI] [PubMed] [Google Scholar]
- 40.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolmasky, M. E. 2000. Bacterial resistance to aminoglycosides and β-lactams: the Tn1331 transposon paradigm. Front. Biosci. 5:D20-D9. [DOI] [PubMed] [Google Scholar]
- 42.Tolmasky, M. E. 1990. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24:218-226. [DOI] [PubMed] [Google Scholar]
- 43.Tolmasky, M. E., R. M. Chamorro, J. H. Crosa, and P. M. Marini. 1988. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 32:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolmasky, M. E., and J. H. Crosa. 1987. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 31:1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolmasky, M. E., M. Roberts, M. Woloj, and J. H. Crosa. 1986. Molecular cloning of amikacin resistance determinants from a Klebsiella pneumoniae plasmid. Antimicrob. Agents Chemother. 30:315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres Viera, C., S. Tsiodras, H. S. Gold, E. P. Coakley, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., and R. T. Inouye. 2001. Restoration of vancomycin susceptibility in Enterococcus faecalis by antiresistance determinant gene transfer. Antimicrob. Agents Chemother. 45:973-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlestedt, C. 1994. Antisense oligonucleotide strategies in neuropharmacology. Trends Pharmacol. Sci. 15:42-46. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775-781. [DOI] [PubMed] [Google Scholar]
- 49.Whipple, F. W. 1998. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 26:3700-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woloj, M., M. E. Tolmasky, M. C. Roberts, and J. H. Crosa. 1986. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob. Agents Chemother. 29:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.