Abstract
MexXY is an aminoglycoside-inducible multidrug transporter shown to contribute to intrinsic and acquired aminoglycoside resistance in laboratory isolates of Pseudomonas aeruginosa. To assess its contribution to aminoglycoside resistance in 14 clinical isolates demonstrating a panaminoglycoside resistance phenotype unlikely to be explained solely by aminoglycoside modification, expression of mexXY by these isolates was examined by reverse transcription-PCR. Elevated levels of mexXY expression were evident for most strains compared with those detected for an aminoglycoside-susceptible control strain, although there was no correlation between mexXY levels and the aminoglycoside MICs for the resistant strains, indicating that if MexXY was playing a role, other factors were also contributing. Deletion of mexXY from 9 of the 14 isolates resulted in enhanced susceptibilities to multiple aminoglycosides, confirming the contribution of this efflux system to the aminoglycoside resistance of these clinical isolates. Still, the impact of MexXY loss varied, with some strains clearly more or less dependent on MexXY for aminoglycoside resistance. Expression of mexXY also varied in these strains, with some showing high-level expression of the efflux genes independent of aminoglycoside exposure (aminoglycoside-independent hyperexpression) and others showing hyperexpression of the efflux genes that was to a greater or lesser degree aminoglycoside dependent. None of these strains carried mutations in mexZ, which encodes a negative regulator of mexXY expression, or in the mexZ-mexXY intergenic region. Thus, mexXY hyperexpression in aminoglycoside-resistant clinical isolates occurs via mutation in one or more as yet unidentified genes.
Pseudomonas aeruginosa is a major cause of opportunistic infections in immunocompromised patients, and the organism demonstrates a high level of intrinsic and acquired resistance to a variety of structurally unrelated antibiotics. While this organism causes a panoply of infections, one of the most frequent sites of P. aeruginosa infection remains the lungs of cystic fibrosis (CF) patients (10, 26). Long-term care of CF patients involves oral and parenteral administration of various groups of antibiotics, including the aminoglycosides, which are a family of related cationic antibiotics that find frequent use in the treatment of pseudomonal lung infections (7, 24). The recurrent use of aminoglycosides has unfortunately led to recalcitrant subpopulations that demonstrate elevated levels of resistance to this family of antimicrobials (10, 26). While in nonpseudomonal infections the development of aminoglycoside resistance is predominantly mediated through aminoglycoside-modifying enzymes (13, 14), the aminoglycoside resistance observed in P. aeruginosa is not due solely to inactivation of these antibiotics by the aminoglycoside-modifying enzymes. Although such enzymatic resistance mechanisms have been described in P. aeruginosa (4, 23, 28), they do not appear to be the predominant mechanism responsible for panaminoglycoside resistance in this organism (10, 26). Rather, aminoglycoside resistance, especially in clinical isolates from CF patients, is often caused by a poorly understood mechanism termed “impermeability resistance” (10, 26) that is defined by a general lack of aminoglycoside susceptibility (7, 24) as a result of reduced drug uptake and/or accumulation (2, 9, 17, 18). Transient, so-called adaptive resistance to aminoglycosides has also been reported in P. aeruginosa following exposure of susceptible strains to drugs (2, 6, 9, 17, 18). Although a mechanism was not invoked, the reversible nature of this resistance suggested that it was regulatory rather than mutational in nature.
Increasingly, resistance to multiple antimicrobials in P. aeruginosa is explained by the operation of multidrug efflux systems, of which several have been identified in this organism to date (19, 22). One of these, encoded by mexXY (also known as amrAB), exports aminoglycosides and thus provides resistance to multiple aminoglycosides (1, 15, 29). This system has been implicated in both impermeability resistance (29) (in which efflux and not membrane impermeability explain the reduced levels of accumulation) and adaptive aminoglycoside resistance (6). The MexXY efflux pump is homologous to several previously studied pseudomonal multidrug efflux systems of the resistance-nodulation-division (RND) family, the family increasingly recognized as the most significant vis-à-vis the contribution to resistance to clinically relevant agents in gram-negative organisms (20). These pumps consist of an RND inner membrane drug-proton antiporter, a channel-forming outer membrane factor (OMF), and a periplasmic so-called membrane fusion protein (MFP) that is believed to facilitate assembly of the RND and OMF components into a functional efflux pump (21). The MexXY MFP-RND components are believed to associate with the previously identified OprM outer membrane component of the MexAB-OprM efflux system (1, 15); and this association is capable of extruding a variety of antibiotics in addition to aminoglycosides, including macrolides, tetracyclines, β-lactams, and quinolones (1, 12, 15). Still, a recent study (8) highlighting the involvement of outer membrane proteins OpmG, OpmI, and, to a lesser extent, OpmH in intrinsic aminoglycoside resistance in P. aeruginosa suggests that one or more of these may also function as the OMF for the MexXY pump. Expression of mexXY is negatively regulated by the product of the divergently transcribed mexZ (amrR) gene (1, 15, 29) and is inducible by substrate antibiotics, including aminoglycosides (11). The latter observation, in fact, explains the contribution of MexXY to adaptive aminoglycoside resistance (6, 10), although the role of MexZ in adaptive or mutational resistance that results from MexXY expression (29) remains undefined. In the study described in the present report, we examined the expression of this efflux system in clinical aminoglycoside-resistant isolates of P. aeruginosa and assessed its contribution to this broad-spectrum panaminoglycoside resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. All bacterial strains were grown in Luria-Bertani (LB) broth supplemented with 0.2% (wt/vol) sodium chloride and were incubated at 37°C for 18 h with shaking (90 rpm). For induction of mexXY, 100 μl of an overnight culture was inoculated into 10 ml of LB broth supplemented with kanamycin at one-quarter the MIC. Cultures of Escherichia coli S17-1 (27) carrying pCSV05-01 required the addition of tetracycline (10 μg/ml) to maintain the plasmid. In conjugations involving E. coli S17-1 (donor) and clinical strains of P. aeruginosa (recipients), the latter were incubated at 42°C without shaking. Solid media were prepared by addition of 1.5% (wt/vol) Bacto Agar and contained tetracycline (50 μg/ml), chloramphenicol (5 μg/ml), and sucrose (10% [wt/vol]), as required.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| K767 | PAO1 wild type | R. Hancock (University of British Columbia) |
| K258 | Clinical isolate | D. Speert (University of British Columbia) |
| K260 | Clinical isolate | D. Speert (University of British Columbia) |
| K2152 to K2156 | Clinical isolates | L. Tomalty (Kingston General Hospital) |
| K2157 to K2163 | Clinical isolates | D. Livermore (Antibiotic Resistance Monitoring and Reference Laboratory) |
| K1525 | K767 ΔmexXY | 3 |
| K2164 | K2152 ΔmexXY | Present study |
| K2165 | K2153 ΔmexXY | Present study |
| K2166 | K2155 ΔmexXY | Present study |
| K2167 | K2156 ΔmexXY | Present study |
| K2168 | K2160 ΔmexXY | Present study |
| K2169 | K2161 ΔmexXY | Present study |
| K2170 | K2162 ΔmexXY | Present study |
| K2171 | K258 ΔmexXY | Present study |
| K2172 | K2163 ΔmexXY | Present study |
| E. coli S17-1 | Donor strain used to promote transfer of pEX18Tc derivatives into P. aeruginosa; thi pro hsdR recA Tra+ | 27 |
| Plasmids | ||
| pEX18Tc | Gene replacement vector; sacB Tcr | 5 |
| pCSV05-01 | pEX18Tc::ΔmexXY | 3 |
Quantification of mexXY by RT-PCR.
Total bacterial RNA was isolated from 1.5 ml of late-log-phase P. aeruginosa cultures (with and without exposure to subinhibitory concentrations of kanamycin) by using the Qiagen RNeasy Mini kit (Qiagen Inc., Mississauga, Ontario, Canada) and treated with RNase-free DNase (Promega, Madison, Wis.) (1 U of enzyme/μg of RNA for 60 min at 37°C, followed by 15 min at 65°C). Reverse transcription (RT)-PCR was performed with the Qiagen OneStep RT-PCR kit (Qiagen Inc.) according to the instructions of the manufacturer. Primer pair rpsLF (5′-GCA ACT ATC AAC CAG CTG-3′) and rpsLR (5′-GCTGTG CTC TTG CAG GTT GTG-3′) and primer pair mexXF (5′-CAT CAG CGA ACG CGA GTA CAC-3′) and mexXR (5′-CAA TTC GCG ATG CGG ATT G-3′) were used to detect the rpsL and mexX messages, respectively. The reaction mixtures were incubated for 30 min at 50°C, followed by 15 min at 95°C and 18 or 20 cycles (for rpsL) or 27 or 29 cycles (for mexXY) of 1 min at 95°C, 1 min at 60°C and 1.5 min at 72°C, before finishing with a final 7 min of elongation at 72°C. The amount of product was assessed by gel electrophoresis in conjunction with densitometry measurements with gel analysis software (ONE-DSCAN; Scanalytics).
Amplification of mexZ by PCR.
Total chromosomal DNA was prepared from overnight cultures as described by Sambrook and Russel (25), and mexZ was amplified by PCR with primers mexZF (5′-ATT GGA TGT GCA TGG GTG-3′) and mexZR (5′-TGG AGA TCG AAG GCA GC-3′). A 50-μl PCR mixture included 10 ng of chromosomal DNA, 0.6 μM each primer, 5% (vol/vol) dimethyl sulfoxide, 1× ThermoPol buffer (New England Biolabs), 0.2 mM deoxynucleoside triphosphate, and 2 U of Vent DNA polymerase (New England Biolabs). The mixture was incubated for 1 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final 5 min of elongation at 72°C. The PCR products were purified with the Qiagen PCR Purification kit and sequenced by Cortec DNA Service Laboratories Inc. (Kingston, Ontario, Canada) with the PCR primers.
Construction of mexXY deletion mutants.
Chromosomal deletions of mexXY were engineered into clinical isolates of P. aeruginosa by using mexXY gene replacement vector pCSV05-01 (3) following conjugal transfer of the plasmid from E. coli S17-1 (3). Briefly, 100 μl of a log-phase E. coli S17-1 LB broth culture was transferred to LB agar plates and immediately overlaid with an equal volume of a stationary-phase culture of a P. aeruginosa clinical isolate. Following incubation at 37°C for 18 h, the bacterial cells were resuspended in 1 ml of LB broth and diluted 10-fold before being plated onto LB agar plates containing 5 μg of chloramphenicol per ml (to counterselect E. coli S17-1) and 70 μg of tetracycline per ml. P. aeruginosa transconjugants harboring chromosomal inserts of pCSV05-01 were recovered from these plates and streaked onto LB agar containing sucrose (10% [wt/vol]). Sucrose-resistant colonies were then screened for deletion of mexXY by PCR with primers mexzbI (5′-AAG CTT AGG CTT GCG TTC GCA CTT GAG GTA GAG-3′) and mexhbI (5′-A CCG GAA TTC CAC CAG GAA GAA CAG CGG TAC-3′) as described before (3). The reaction mixtures were formulated as described above for the mexZ PCR and were incubated for 3 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 58°C, and 4 min at 72°C, with a final 5 min of elongation at 72°C.
Antimicrobial susceptibility testing.
The antimicrobial susceptibilities of the clinical P. aeruginosa strains and their ΔmexXY derivatives were assessed in microtiter trays by a twofold serial dilution technique (8). Briefly, 50-μl aliquots of log-phase cells grown in LB broth were added to an equal volume of LB broth containing serial twofold dilutions of antibiotic to yield a final cell concentration of 2.75 × 105 cells/ml. Following incubation at 37°C for 18 h, growth was assessed visually and the MIC was reported as the lowest concentration of antibiotic inhibiting visible growth.
RESULTS AND DISCUSSION
Aminoglycoside resistance in clinical strains of P. aeruginosa.
Fourteen clinical isolates and the designated PAO1 laboratory strain of P. aeruginosa (strain K767) were assessed for their susceptibilities to a range of aminoglycoside antibiotics. With few exceptions, the strains demonstrated panaminoglycoside resistance (Table 2), with MICs generally being above the accepted NCCLS breakpoint (16) for parenterally administered aminoglycosides used in the treatment of P. aeruginosa infections. Wild-type PAO1 reference strain K767 and two clinical isolates (isolates K2155 and K2159) were, however, substantially more susceptible to the aminoglycosides than the other strains (Table 2). Clinical isolates K2155 and K2159, particularly K2155, would nonetheless prove to be useful comparators in later assessments of mexXY expression and its contribution to aminoglycoside resistance in the other isolates. While most strains showed a range of susceptibilities to the various aminoglycosides, the MICs of spectinomycin, lividomycin, paromomycin, neomycin, and kanamycin were found to be universally elevated for all strains examined (Table 2). Given that P. aeruginosa is known to harbor a number of aminoglycoside-modifying enzymes, including APH(3′) (28), AAC(3) (4), AAC(6′) (4), and ANT(4′) (23), it is possible that enzymatic modification may contribute to the high level of resistance to these antibiotics that was observed. It is extremely unlikely, however, that aminoglycoside modification would satisfactorily explain the panaminoglycoside resistance observed in most of the clinical isolates, as shown in Table 2 (2, 6, 9). Indeed, this broad-range aminoglycoside resistance is reminiscent of the impermeability resistance that has previously been described in P. aeruginosa (7, 9, 10, 24).
TABLE 2.
Aminoglycoside susceptibilities of clinical P. aeruginosa strains
| Straina | MIC (μg/ml)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMI | GEN | KAN | LIV | NEO | PAR | SPC | STR | TOB | |
| K767 | 2 | 4 | 128 | 16 | 64 | 512 | 1,024 | 16 | 1 |
| K2152 (699) | 8 | 8 | 128 | 64 | 128 | 512 | 2,048 | 32 | 4 |
| K2153 (524) | 16 | 16 | 128 | 64 | 128 | 512 | 2,048 | 64 | 4 |
| K2154 (0006-3) | 16 | 512 | 512 | 256 | 256 | 1,024 | >2048 | 256 | 256 |
| K2155 (4420) | 1 | 4 | 16 | 16 | 8 | 32 | 512 | 16 | 1 |
| K2156 (8812-2) | 32 | 32 | 512 | 128 | 128 | 512 | 2,048 | 256 | 8 |
| K2157 (R68) | 64 | 64/128 | 1,024 | 512 | 512 | >2,048 | >2,048 | 256 | 128 |
| K2158 (R69) | 64 | 64 | 1,024 | 512 | 512 | >2,048 | >2,048 | 128 | 32 |
| K2159 (R92) | 8 | 8 | 128 | 16 | 64 | 64 | 64 | 4 | 8 |
| K2160 (R103) | 64 | 32 | 512 | 512 | 128 | 512 | 512 | 128 | 16 |
| K2161 (R109) | 64 | 64 | 1,024 | 256 | 512 | >2,048 | 256 | 64 | 16 |
| K2162 (R115) | 256 | 256 | >2,048 | >2,048 | 1,024 | >2,048 | >2,048 | 256 | 64 |
| K258 (C517M) | 32 | 16 | 512 | 128 | 256 | >2,048 | 2,048 | 64 | 16 |
| K260 (C510M) | 16 | 16 | 128 | 128 | 64 | 256 | 512 | 64 | 4 |
| K2163 (PS380) | 16 | 8 | 128 | 64 | 32 | 256 | 1,024 | 32 | 8 |
The designation of the laboratory of K. Poole is given, with the original hospital designation given in parentheses.
Abbreviations; AMI, amikacin; GEN, gentamicin; KAN, kanamycin; LIV, lividomycin; NEO, neomycin; PAR, paromomycin; SPC, spectinomycin; STR, streptomycin; TOB, tobramycin.
Expression of mexXY in aminoglycoside-resistant P. aeruginosa.
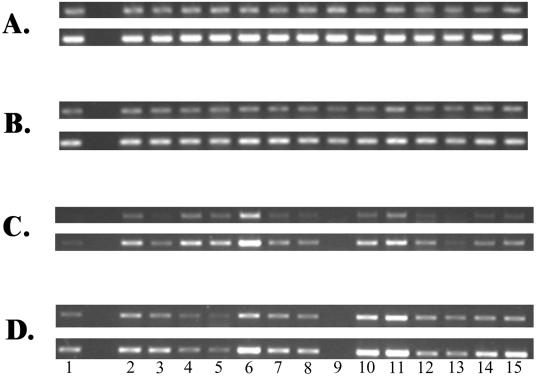
In light of suggestions that MexXY may contribute to aminoglycoside impermeability resistance, attempts were made initially to assess mexXY expression by RT-PCR and to correlate this with the resistance levels of the clinical isolates. RNA samples from the isolates grown in LB broth in the absence of aminoglycoside antibiotics were standardized by using rpsL gene expression (Fig. 1A), and although RNA was clearly added to the RT-PCR mixtures in equivalent amounts, a high degree of variation in the level of mexXY gene expression was observed among the aminoglycoside-resistant strains examined (Fig. 1C). Several isolates, including K2153, K2156, K2160, and K2161, showed high levels of mexXY expression relative to that for “susceptible” strain K2155 (Fig. 1C, top panel, lanes 4, 6, 10, and 11, respectively); and this was made more evident when the PCR was increased to 29 cycles (Fig. 1C, bottom panel, lanes 4, 6, 10, and 11, respectively). Westbrock-Wadman and colleagues (29) have reported similar increases in amrAB (mexXY) expression in strains from which amrR (mexZ) was deleted, although such mutants were not aminoglycoside resistant. PCR amplification and subsequent sequencing of mexZ and the mexZ-mexXY intergenic region failed to reveal any mutations in the mexZ genes of the resistant clinical strains described here. Similarly, in vitro-selected P. aeruginosa mutants that are resistant to tobramycin as a result of elevated levels of MexXY expression and that lack mexZ mutations have also been described (29). Two isolates, K2157 and K2158, did, however, carry the same mutation in the mexZ-mexXY intragenic region, a C-to-T transition at position −23 relative to the ATG start codon of mexX, although whether this would affect MexZ repression of mexXY is unclear. Interestingly, “susceptible” strain K2155 was the only strain that carried a mutation in mexZ, producing a L41Q substitution in the MexZ protein which is unlikely to affect its repressor activity, given the barely detectable level of mexX expression seen in this strain prior to antibiotic induction (Fig. 1C, top panel). Thus, despite the known role of MexZ as a negative regulator of mexXY expression, hyperexpression of this system in aminoglycoside-resistant clinical isolates is achieved by mutations in a gene(s) other than mexZ. Interestingly, K2159 failed to express detectable mexXY (Fig. 1C and D, lanes 9), and a mexZ gene could not be amplified from its genome, suggesting that a mexXY locus is absent or defective in this isolate, possibly explaining its general aminoglycoside susceptibility (Table 2).
FIG. 1.
Expression of rpsL (A and B) and mexX (C and D) in P. aeruginosa clinical isolates grown without (A and C) and with (B and D) kanamycin (at one-quarter the MIC) as determined by semiquantitative RT-PCR. Lanes 1, K2155; lanes 2, PAO1 strain K767; lanes 3, K2152; lanes 4, K2153; lanes 5, K2154; lanes 6, K2156; lanes 7, K2157; lanes 8, K2158; lanes 9, K2159; lanes 10, K2160; lanes 11, K2161; lanes 12, K2162; lanes 13, K258; lanes 14, K260; lanes 15, K2163. The rpsL reaction served as an internal control that ensured that equal amounts of RNA were used in all of the RT-PCRs shown. The PCR portion of the rpsL reactions was carried out for 18 cycles (upper panels) or 20 cycles (lower panels). The PCR portion of the mexX reactions was carried out for 27 cycles (upper panels) or 29 cycles (lower panels).
Despite the variability of mexXY expression observed in the clinical strains and although many of the aminoglycoside-resistant strains expressed mexXY at levels above the level of expression by K2155, there was no clear correlation between mexXY expression and aminoglycoside resistance. Indeed, strain K2162 (Fig. 1C, lane 12) expressed substantially less mexXY than several isolates (e.g., K2153 and K2156; Fig. 1C lanes 4 and 6, respectively), yet the MICs of a range of aminoglycosides were 4- to 16-fold higher for K2162 than for these isolates (Table 2). Still, mexXY expression is typically aminoglycoside inducible (6, 11), and as MIC determinations clearly involve drug exposure, it was possible that aminoglycoside MICs would correlate better with aminoglycoside-induced mexXY levels. Thus, mexXY expression by the clinical isolates following aminoglycoside (i.e., kanamycin) exposure was examined (Fig. 1D). Not unexpectedly, many of the isolates, including generally susceptible strain K2155, showed increased levels of mexXY expression in the presence of kanamycin compared to the levels of expression by their counterparts not exposed to kanamycin (compare Fig. 1C and D). Again, too, most of the aminoglycoside-resistant clinical isolates showed higher levels of mexXY expression than K2155 (Fig. 1D). Still, no good correlation between mexXY levels and aminoglycoside resistance was observed, with many highly resistant isolates (e.g., K2154, K2157, K2158, and K2162; Fig. 1D lanes 5, 7, 8, and 12, respectively) expressing mexXY at levels comparable to the levels expressed by K2155 (Fig. 1D, lane 1) and much less than the levels expressed by more susceptible isolates (e.g., K2156, K2160, K2161, and K2163; Fig. 1D lanes 6, 10, 11, and 15, respectively). Thus, while mexXY is clearly expressed at high levels in a number of clinical aminoglycoside-resistant P. aeruginosa isolates, at this point we are uncertain about its contribution to resistance.
Contribution of MexXY to aminoglycoside resistance.
To better assess the contribution of MexXY to aminoglycoside resistance in clinical isolates, the genes were deleted from these isolates and the impact on aminoglycoside resistance was assessed. Despite numerous attempts, deletion of mexXY was achieved in only nine strains (Table 3), although for each of these strains the aminoglycoside MICs decreased a minimum of twofold, indicating that MexXY does, in fact, contribute to aminoglycoside resistance in clinical isolates. Interestingly, the biggest decline in MICs upon deletion of mexXY (8- to 64-fold) was found for strain K2156, which generally showed the highest levels of mexXY expression (see K2167 in Table 3), indicating that MexXY is a major determinant of aminoglycoside resistance in this isolate. In two other isolates that also showed substantial levels of mexXY expression (i.e., K2160 and K2161), the loss of mexXY had a substantial impact (2- to 32-fold declines in MICs; see K2168 and K2169 in Table 3) on the panaminoglycoside resistance of these strains. Still, K2163 also showed substantial levels of mexXY expression, and the loss of mexXY had only a modest impact on susceptibility (two- to eightfold declines in MICs; see K2172 in Table 3). In this instance, another resistance determinant(s) is likely playing an important role and, for some aminoglycosides, at least can compensate for the loss of this efflux system, although the presence of mutations affecting MexXY expression or activity cannot be ruled out. In contrast, some strains expressing modest levels of mexXY (e.g., K2162) still showed substantial increases in aminoglycoside susceptibilities upon the loss of mexXY (up to 32-fold decreases in MICs; see K2170 in Table 3). In this instance, mexXY is clearly a major determinant of aminoglycoside resistance in this isolate. It is interesting, however, that even in strains in which MexXY is clearly having a major impact (e.g., K2160, K2161, and K2162), the loss of mexXY yields strains that are still substantially aminoglycoside resistant (the MICs for K2168, K2169, and K2170 are much greater than those for the other ΔmexXY derivatives). Clearly, then, another factor(s) is also contributing to the high-level aminoglycoside resistance in these isolates.
TABLE 3.
Aminoglycoside susceptibilities of ΔmexXY derivatives of clinical P. aeruginosa strains
| Strain | MIC (μg/ml)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMI | GEN | KAN | LIV | NEO | PAR | SPC | STR | TOB | ERY | TET | NOR | |
| K767 | 2 | 4 | 128 | 16 | 64 | 512 | 1,024 | 16 | 1 | 512 | 16 | 0.5 |
| K1525 | 1 | 2 | 64 | 4 | 16 | 32 | 128 | 2 | 1 | 128 | 16 | 0.5 |
| ΔMICb | 2 | 2 | 2 | 4 | 4 | 16 | 8 | 8 | 1 | 4 | 1 | 1 |
| K2152 | 8 | 8 | 128 | 64 | 128 | 512 | 2,048 | 32 | 4 | 512 | 32 | 1 |
| K2164 | 4 | 4 | 64 | 8 | 32 | 32 | 128 | 4 | 2 | 128 | 32 | 1 |
| ΔMIC | 2 | 2 | 2 | 8 | 4 | 16 | 16 | 8 | 2 | 4 | 1 | 1 |
| K2153 | 16 | 16 | 128 | 64 | 128 | 512 | 2,048 | 64 | 4 | 512 | 32 | 1 |
| K2165 | 2 | 4 | 64 | 16 | 16 | 16 | 128 | 8 | 4 | 128 | 128 | 1 |
| ΔMIC | 8 | 4 | 2 | 4 | 8 | 32 | 16 | 8 | 1 | 4 | 0.25 | 1 |
| K2155 | 1 | 4 | 16 | 16 | 8 | 32 | 512 | 16 | 1 | 256 | 32 | 4 |
| K2166 | 0.5 | 2 | 16 | 4 | 8 | 8 | 128 | 4 | 1 | 32 | 32 | 4 |
| ΔMIC | 2 | 2 | 1 | 4 | 1 | 4 | 4 | 4 | 1 | 8 | 1 | 1 |
| K2156 | 32 | 32 | 512 | 128 | 128 | 512 | 2,048 | 256 | 8 | 512 | 16 | 8 |
| K2167 | 1 | 4 | 64 | 4 | 16 | 32 | 64 | 4 | 1 | 256 | 16 | 4 |
| ΔMIC | 32 | 8 | 8 | 32 | 8 | 16 | 32 | 64 | 8 | 2 | 1 | 2 |
| K2160 | 64 | 32 | 512 | 512 | 128 | 512 | 512 | 128 | 16 | 256 | 16 | 1 |
| K2168 | 16 | 8 | 256 | 32 | 64 | 64 | 64 | 16 | 8 | 32 | 16 | 1 |
| ΔMIC | 4 | 4 | 2 | 16 | 2 | 8 | 8 | 8 | 2 | 8 | 1 | 1 |
| K2161 | 64 | 64 | 1,024 | 256 | 512 | >2,048 | 256 | 64 | 16 | 512 | 16 | 0.5 |
| K2169 | 8 | 8 | 256 | 32 | 64 | 64 | 64 | 8 | 8 | 256 | 16 | 0.5 |
| ΔMIC | 8 | 8 | 4 | 8 | 8 | 32 | 4 | 8 | 2 | 2 | 1 | 1 |
| K2162 | 256 | 256 | 2,048 | 2,048 | 1,024 | >2,048 | 2,048 | 256 | 64 | 512 | 32 | 2 |
| K2170 | 32 | 16 | 512 | 64 | 128 | 256 | 128 | 16 | 16 | 512 | 32 | 2 |
| ΔMIC | 8 | 16 | 4 | 32 | 8 | 8 | 16 | 16 | 4 | 1 | 1 | 1 |
| K258 | 32 | 16 | 512 | 128 | 256 | 2,048 | 2,048 | 64 | 16 | 1,024 | 64 | 1 |
| K2171 | 8 | 4 | 256 | 32 | 64 | 64 | 512 | 16 | 8 | 1,024 | 64 | 1 |
| ΔMIC | 4 | 4 | 2 | 4 | 4 | 32 | 4 | 4 | 2 | 1 | 1 | 1 |
| K2163 | 16 | 8 | 128 | 64 | 32 | 256 | 1,024 | 32 | 8 | 256 | 32 | 1 |
| K2172 | 8 | 2 | 64 | 16 | 16 | 32 | 256 | 8 | 4 | 128 | 32 | 1 |
| ΔMIC | 2 | 4 | 2 | 4 | 2 | 8 | 4 | 4 | 2 | 2 | 1 | 1 |
Abbreviations: AMI, amikacin; GEN, gentamicin; KAN, kanamycin; LIV, lividomycin; NEO, neomycin; PAR, paromomycin; SPC, spectinomycin; STR, streptomycin; TOB, tobramycin; ERY, erythromycin; TET, tetracycline; NOR, norfloxacin.
ΔMIC, ratio of MIC for the parent strain and the MIC for the corresponding ΔmexXY derivative.
While MexXY was initially identified as an aminoglycoside efflux system, this pump is also capable of extruding unrelated antibiotics, including erythromycin, tetracycline, and norfloxacin (1, 15, 29). We therefore assessed the impact of mexXY deletion on the susceptibilities of the clinical strains to these nonaminoglycoside antimicrobials. While the loss of MexXY had a modest impact on erythromycin resistance, with MICs declining less than or equal to eightfold for all strains, the tetracycline and norfloxacin MICs did not change at all (Table 3). This is reminiscent of earlier studies in which the loss of mexXY in otherwise wild-type laboratory strains had a negligible impact on susceptibilities to these agents, apparently because they could be exported by other efflux systems (e.g., MexAB-OprM), which could thus compensate for the loss of MexXY (15, 29).
mexXY expression patterns in aminoglycoside-resistant clinical isolates.
While no correlation between aminoglycoside resistance and mexXY expression in the presence or absence of an added aminoglycoside antibiotic was found to exist, clear patterns of mexXY expression were evident among those aminoglycoside-resistant isolates in which MexXY was a contributing factor. In some instances, the best example being K2156, mexXY was expressed at levels markedly higher than the level of expression seen by K2155 in the absence of antibiotic (Fig. 1C, lane 6); and no increase in mexXY expression was observed following drug (i.e., kanamycin) exposure (Fig. 1D, lane 6). A similar pattern of drug-independent hyperexpression was observable for clinical strains K2153 (Fig. 1C and D, lanes 4) and K2154 (Fig. 1C and D, lanes 5), although a contribution of MexXY to aminoglycoside resistance was not confirmed for K2154. In most instances, however, maximal mexXY expression by the clinical isolates was achieved upon aminoglycoside exposure, although two distinct patterns of drug-dependent mexXY hyperexpression were evident. These two patterns were differentiated by the levels of mexXY expressed prior to aminoglycoside exposure. One group produced very modest levels of mexXY in the absence of aminoglycoside (e.g., K260 and K2163; Fig. 1C, lanes 14 and 15, respectively), while the other group expressed substantial mexXY even in the absence of an aminoglycoside (e.g., K2160 and K2161; Fig. 1C, lanes 10 and 11, respectively). These different patterns of expression seen in the clinical aminoglycoside-resistant isolates suggest that different mutations are responsible for the enhanced mexXY expression observed and that MexXY-mediated aminoglycoside resistance can arise in P. aeruginosa in several ways. While the drug-independent mexXY hyperexpression seen in, for example, K2156 is somewhat reminiscent of hyperexpression of the mexAB-oprM and mexCD-oprJ multidrug efflux systems by nalB and nfxB mutants, respectively, as a result of mutations in linked repressor genes and is consistent with a mutation in mexZ, such mutations, as mentioned previously, were absent from all aminoglycoside-resistant isolates examined. Thus, not only is mexXY hyperexpression alone insufficient for aminoglycoside resistance, but mutations in mexZ are not associated with the mexXY hyperexpression that clearly occurs in and that contributes to the aminoglycoside resistance of at least some clinical isolates. Thus, additional genes contribute both to MexXY-dependent aminoglycoside resistance in P. aeruginosa and to mexXY hyperexpression in aminoglycoside-resistant strains.
Acknowledgments
This work was supported by funding from the Canadian Bacterial Diseases Network (one of the Networks of Centers of Excellence). K.P. is a Canadian Cystic Fibrosis Foundation Scholar. M.L.S. is the recipient of a Natural Sciences and Engineering Research Council graduate student fellowship.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daikos, G. L., G. G. Jackson, V. T. Lolans, and D. M. Livermore. 1990. Adaptive resistance to aminoglycoside antibiotics from first-exposure down-regulation. J. Infect. Dis. 162:414-420. [DOI] [PubMed] [Google Scholar]
- 3.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 6.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plesiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley, J. C., G. H. Miller, and A. L. Smith. 1995. Mechanism of amikacin resistance in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Diagn. Microbiol. Infect. Dis. 22:331-336. [DOI] [PubMed] [Google Scholar]
- 8.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlowsky, J. A., M. H. Saunders, G. A. Harding, D. J. Hoban, and G. G. Zhanel. 1996. In vitro characterization of aminoglycoside adaptive resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:1387-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLeod, D. L., L. E. Nelson, R. M. Shawar, B. B. Lin, L. G. Lockwood, J. E. Dirk, G. H. Miller, J. L. Burns, and R. L. Garber. 2000. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J. Infect. Dis. 181:1180-1184. [DOI] [PubMed] [Google Scholar]
- 11.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, K. J. Shaw, et al. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 14.Miller, G. H., F. J. Sabatelli, L. Naples, R. S. Hare, K. J. Shaw, et al. 1995. The changing nature of aminoglycoside resistance mechanisms and the role of isepamicin—a new broad-spectrum aminoglycoside. J. Chemother. 7(Suppl. 2):31-44. [PubMed] [Google Scholar]
- 15.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Eighth informational supplement. M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Parr, T. R., Jr., and A. S. Bayer. 1988. Mechanisms of aminoglycoside resistance in variants of Pseudomonas aeruginosa isolated during treatment of experimental endocarditis in rabbits. J. Infect. Dis. 158:1003-1010. [DOI] [PubMed] [Google Scholar]
- 18.Perlin, M. H., and S. A. Lerner. 1986. High-level amikacin resistance in Escherichia coli due to phosphorylation and impaired aminoglycoside uptake. Antimicrob. Agents Chemother. 29:216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 20.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 21.Poole, K. 2002. Outer membranes and efflux: the path to multidrug resistance in gram-negative bacteria. Curr. Pharm. Biotechnol. 3:77-98. [DOI] [PubMed] [Google Scholar]
- 22.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 23.Sabtcheva, S., M. Galimand, G. Gerbaud, P. Courvalin, and T. Lambert. 2003. Aminoglycoside resistance gene ant(4′)-IIb of Pseudomonas aeruginosa BM4492, a clinical isolate from Bulgaria. Antimicrob. Agents Chemother. 47:1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiman, L., F. Mehar, W. W. Niu, H. C. Neu, K. J. Shaw, G. Miller, and A. Prince. 1996. Antibiotic susceptibility of multiply resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis, including candidates for transplantation. Clin. Infect. Dis. 23:532-537. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Shawar, R. M., D. L. MacLeod, R. L. Garber, J. L. Burns, J. R. Stapp, C. R. Clausen, and S. K. Tanaka. 1999. Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 28.Torres, C., M. H. Perlin, F. Baquero, D. L. Lerner, and S. A. Lerner. 2000. High-level amikacin resistance in Pseudomonas aeruginosa associated with a 3′-phosphotransferase with high affinity for amikacin. Int. J. Antimicrob. Agents 15:257-263. [DOI] [PubMed] [Google Scholar]
- 29.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]