Abstract
The structure and thermal stability of empty and peptide-filled forms of the murine class II major histocompatibility complex (MHC) molecule I-Ek were studied at neutral and mildly acidic pH. The two forms have distinct circular dichroic spectra, suggesting that a conformational change may accompany peptide binding. Thermal stability profiles indicate that binding of peptide significantly increases the thermal stability of the empty heterodimers at both neutral and mildly acidic pH. Free energies calculated from these data provide a direct measure of this stabilization and show that the empty form of I-Ek is significantly more stable than that of class I MHC proteins. Furthermore, for the two MHC class II proteins that were analyzed (I-Ek and I-Ad), thermal stability was not significantly altered by acidification. In contrast, of four class I MHC molecules studied, three have shown a significant loss in complex stability at low pH. The marked stability exhibited by their empty form, as well as their resistance to low pH, as observed in this study, correlate well with the ability of class II MHC molecules to traverse and bind peptides in acidic endosomal vesicles.
Keywords: thermal stability, free energy
Class I and class II major histocompatibility complex (MHC) molecules are heterodimeric cell surface glycoproteins that bind antigenic peptides and display them for surveillance by T lymphocytes (1). The two MHC classes have a similar structure with two membrane-proximal immunoglobulin-like constant domains and a membrane-distal peptide-binding groove formed by two α-helices atop an eight-stranded β-sheet floor (2, 3). The interaction between the MHC molecule and the peptide not only forms the basis for the heterodimer function in antigen presentation but also plays a decisive role in its thermodynamic stability. For class I molecules, the presence of an appropriate peptide has been shown, in most circumstances, to be imperative for successful folding and surface expression at physiological temperature (1, 4–8). The free energy contributed by peptide binding to empty class I Kd heterodimers has been determined (9) and the binding energies contributed by peptide contacts at anchor positions (10–13) and at the peptide NH2 and COOH termini (8) have been reported for several alleles. The stability of class II molecules also depends on the peptide (1, 14), though apparently to a lesser extent than class I molecules (15). In addition, the binding of peptides to class II proteins is pH-dependent. For most class II alleles, peptide binding is enhanced at low pH (1, 16–20), consistent with the pH of the endosomal-like compartment (pH ≈5.0) where peptide loading takes place. At that pH, it has been suggested that the molecules adopt an alternate “open” conformation that facilitates peptide binding (21–24) and that has been proposed to be less stable than the form adopted at neutral pH (21, 23–25).
In spite of these observations, direct quantitative data regarding the energetic consequences of peptide binding or alteration in the pH on the stability of class II proteins has not been reported. Herein, we compare the thermal denaturation profiles exhibited by the murine class II protein I-Ek in the presence or absence of a specific peptide, moth cytochrome c. From these data, the free energy of the empty state, as well as that contributed by moth cytochrome c to the stabilization of the heterodimer, are derived. In addition, the thermal stability of two class II heterodimers, I-Ek and I-Ad, is analyzed at neutral and mildly acidic pH. To assess whether the differences in biosynthesis and localization of antigen binding between class I and class II proteins might be reflected in their resistance to low pH, the effect of the pH on the thermal stability of four class I proteins at the relevant pH range has also been studied.
EXPERIMENTAL PROCEDURES
Soluble Proteins.
Empty I-Ek heterodimers were expressed as glycosylphosphatidylinositol-anchored chimera in Chinese hamster ovary cells and were digested from the cell surface and purified as described (18, 19). Apparently, these GPI-linked chimeras do not traffic through the cell’s endosomal compartments and consequently lack bound peptides as evidenced by labeled peptide binding experiments (19), by the fact that no peptides can be eluted from the molecules (H. Schild, P. A. Reay, and M.M.D., unpublished results), by the observation that the I-Ek bearing cells were unable to present endogenously processed peptides to a specific T-cell line (18), and by their distinct unfolding behavior and stability as described herein. I-Ek molecules complexed with moth cytochrome c (residues 88–103) peptide were prepared and purified as reported (26, 27). Soluble I-Ad (mouse) with Eα peptide (residues 52–68) covalently linked to the heterodimer β chain was constructed by exchanging the peptide portion of I-Ad/OVA cDNA (28) by that of the Eα sequence obtained from an I-Ab/Eα construct (29). Baculovirus containing the I-Ad/Eα sequence was used to infect High Five insect cells (Invitrogen) as described (30) and the recombinant protein was immunoaffinity purified with the monoclonal antibody M5/114. Secreted forms of the following class I MHC molecules were obtained by following the procedures as outlined (9, 31): HLA-A2 [human, complexed with HIV pol (RT) peptide, residues 309–317], HLA-B27 (human, complexed with an endogenous mixture of CHO-derived peptides), H-2Kd [(mouse, complexed with human β2-microglobulin, β2m), complexed with a myeloma (MOPC21)-derived IgHV peptide, residues 49–58)], and HLA-B35 (human, complexed with HIV nef peptide, residues 73–82).
Circular Dichroism (CD) and Thermal Denaturation Studies.
CD spectra were recorded in a 0.1-cm path-length cell on an AVIV 62A DS spectropolarimeter (Aviv Associates, Lakewood, NJ) equipped with a thermoelectric cell holder, using a step size of 0.25 nm, a bandwidth of 1 nm, and a time constant of 1 sec. Protein solutions were made in 5 mM sodium phosphate/5 mM sodium acetate buffer and were brought to the indicated pH with concentrated stocks of HCl or NaOH. Spectra shown in the text represent the average of three measurements, each representing the average of five repetitive scans, and were smoothed by using a sliding window of nine (2.25 nm). Far-UV data are given as [θ]r, the mean residue ellipticity, and the near-UV data are expressed as molar ellipticity, [θ]. Thermal denaturation curves of the various MHC proteins were obtained by following the CD signal at 222 (class II) or 223 (class I) nm as a function of the temperature in a 1-cm path-length cell. The temperature was increased in a step-wise mode (2–2.5°C intervals) with each temperature jump being followed by a 100-sec equilibration time. Recording time was 50 sec. Each point in the melting curves shown in the text represents the average of three experiments. Reversibility of the thermal unfolding was demonstrated by using standard heating/cooling cycles in which samples were initially scanned at 25°C, heated to temperatures well above the midpoint temperature of the thermal unfolding transition Tm of the protein complex analyzed, immediately cooled back to 25°C, and scanned after an equilibration period of 1 h. The CD spectra at high temperatures were recorded separately to avoid the formation of kinetically driven irreversibly unfolded species due to long incubation times at high temperatures.
Free Energy Calculations.
Thermodynamic parameters were derived from the CD data presented in the text, assuming a two-state unfolding model. Derivation of the free energy change ΔG at physiological temperature (37°C), which lies below the transition region of the unfolding curves where the equilibrium constant K cannot be directly derived, was made by using the following form of the Gibbs–Helmholtz equation: ΔG(T) = ΔHm(1 − T/Tm) − ΔCp[(Tm − T) + T ln(T/Tm)], where T is the Kelvin temperature, Tm is the midpoint temperature of the thermal unfolding transition, ΔHm is the enthalpy change for unfolding measured at Tm, and ΔCp is the difference in heat capacity between the folded and unfolded conformations. Tm and ΔHm were obtained from the van’t Hoff equation: ΔH = RT2(δln K)/(δT), in which R is the gas constant. ΔCp values were assumed to be independent of temperature (32) and were estimated from: ΔCp = (δΔH)/(δT)p. Values of K(T) inside the transition region of the unfolding curves were derived, after sloping corrections, from the following relation: K(T) = (θN − θT)/(θT − θU), where (θN) and (θU) are the limiting ellipticity values representing the native and unfolded states, respectively, and (θT) is the observed ellipticity at T.
RESULTS AND DISCUSSION
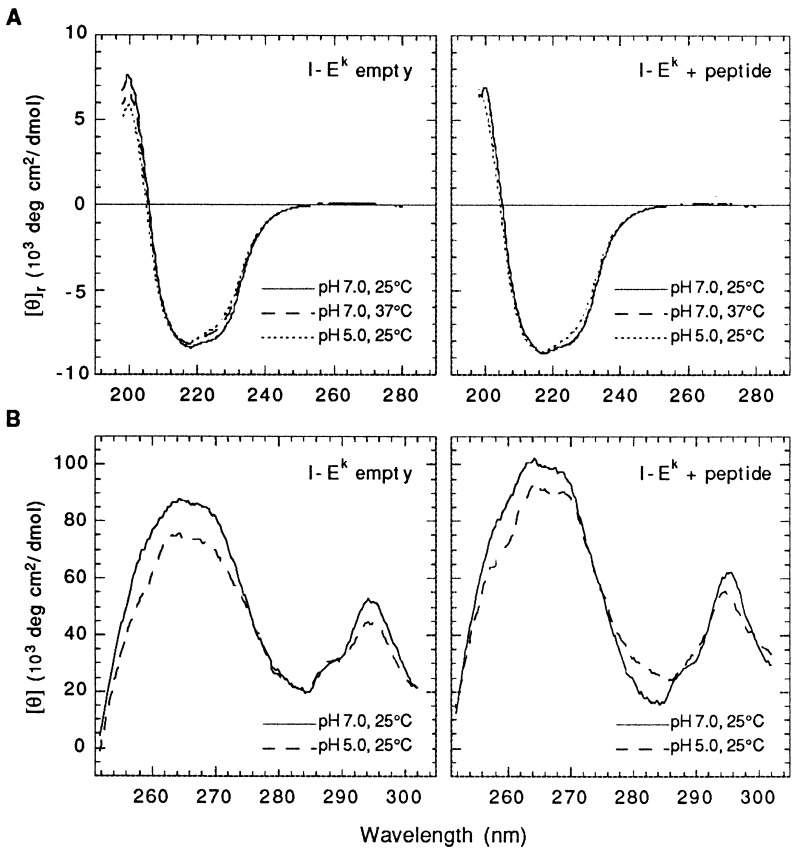
The far-UV CD spectra of empty and peptide-filled I-Ek molecules at different temperatures and pHs are shown in Fig. 1A. At neutral pH and at 25°C, the spectra are very similar to each other and are consistent, as expected, with proteins containing a substantial amount of β-sheet and a considerable portion of α-helical structure (33). An increase in the temperature to 37°C, however, results in some loss of the CD signal exhibited by the empty but not by the peptide-filled molecules. Decreasing the pH to 5.0 has a similar effect on the CD spectrum of empty I-Ek but this time a slight decrease can also be observed in the spectrum exhibited by the peptide-filled heterodimers. The preferential decrease in the CD signal at the right side of the absorption band is consistent with a slight loss in helicity (33), suggesting that the lack of peptide and low pH are accompanied by limited melting in one or both of the α-helices that form the peptide-binding groove of the molecule. CD analysis in the near-UV range, and at 25°C, shows more pronounced differences between the two forms (Fig. 1B). Thus, these differences in the CD spectrum of the two forms suggest that the binding of peptide may be accompanied by a conformational change in the empty molecules. Changes observed in the near-UV spectra upon protonation may also reflect pH-induced structural rearrangements but can also be due to alterations in the polarity of the asymmetric environment around particular aromatic residues. As discussed below, for either form, changes in conformation induced by low pH should be of a fairly localized nature as they are not associated with a significant change in the free energy of the molecules.
Figure 1.
Far-UV (A) and near-UV (B) CD spectra of empty and peptide-loaded I-Ek heterodimers at neutral and mildly acidic pH. Protein concentrations were 0.3 mg/ml (A) and 1.5 mg/ml (B).
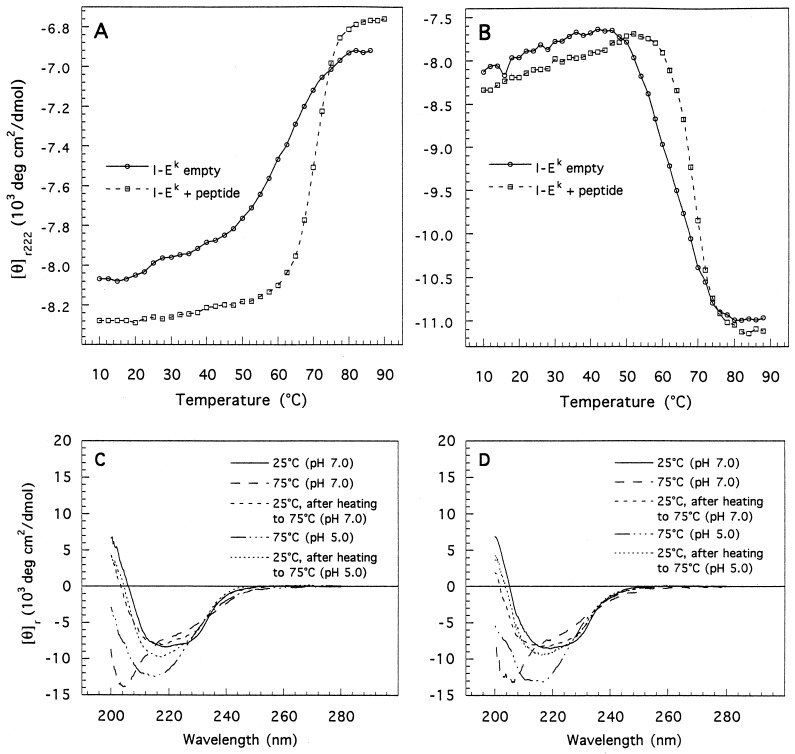
The thermal melting profiles of empty and peptide-bound I-Ek at neutral and mildly acidic pH are shown in Fig. 2A and the values for the midpoint of the thermal unfolding transitions Tm are summarized in Table 1. As shown (Fig. 2 C and D), at either pH, the thermal transitions of the two forms are largely reversible. At neutral pH, a sharp single transition with a Tm at 70°C is observed for the peptide-bound molecules. The presence of only a single transition suggests that chain dissociation and unfolding of the individual chains are coupled. In contrast, two comparatively broad transitions are evident for the empty heterodimers. The first is observed at low temperatures and may correlate with the limited loss in secondary structure observed for this form, going from 25° to 37°C (Fig. 1A). The second main transition is characterized by a Tm of 62°C. The pronounced broadening (low cooperativity) of the denaturation curve of empty I-Ek indicates that, compared with the peptide-bound conformation, this form is lacking in the strength and/or number of intramolecular contacts, giving rise to a ΔHm value as low as 45 kcal/mol (1 cal = 4.184 J). Structurally, such a low enthalpy of denaturation would be consistent with a “loose” or an extended conformation, an observation supported by hydrodynamic measurements (unpublished data). This loose structure could be similar to the “floppy” form observed during denaturation (21) and folding (34) of class II molecules and may also explain the tendency of these proteins to aggregate in the absence of peptide (14, 15). The relatively high Tm observed for this form, the far-UV CD data presented above, and the observation that empty class II molecules are recognized by conformational-sensitive antibodies (15) suggest, however, that the empty state still retains a high degree of the structure present in the peptide-filled conformation.
Figure 2.
Thermal denaturation of empty and peptide-loaded I-Ek heterodimers monitored by CD at pH 7.0 (A) and pH 5.0 (B). Protein concentrations were 40–60 μg/ml. (C and D) Far-UV CD scans of native, unfolded, and renatured empty (C) and peptide-filled (D) I-Ek at neutral and mildly acidic pH.
Table 1.
Thermal stability of class I and class II MHC molecules at neutral and mildly acidic pH
| Molecule | Peptide | pH | Tm, °C |
|---|---|---|---|
| I-Ek | — | 7.0 | 62 ± 1 |
| I-Ek | ANERADLIAYLKQATK | 7.0 | 70 ± 1 |
| I-Ek | — | 5.0 | 63 ± 1 |
| I-Ek | ANERADLIAYLKQATK | 5.0 | 68 ± 1 |
| I-Ad | ASFEAQGALANIAVDKA | 7.0 | 62 ± 1 |
| I-Ad | ASFEAQGALANIAVDKA | 5.0 | 61 ± 1 |
| HLA-A2 | ILKEQVHGV | 7.0 | 59 ± 1 |
| HLA-A2 | ILKEQVHGV | 5.5 | 45 ± 1 |
| HLA-B27 | Mixed | 7.0 | 66 ± 1 |
| HLA-B27 | Mixed | 7.0† | 63 ± 2 |
| HLA-B27 | Mixed | 5.0† | 52 ± 2 |
| Kd* | — | 7.0 | 45 ± 1 |
| Kd* | TYQRTRALV | 7.0 | 57 ± 1 |
| Kd | AYISSGSSTL | 7.0 | 54 ± 1 |
| Kd | AYISSGSSTL | 6.0 | 54 ± 1 |
| Kd | AYISSGSSTL | 5.5 | 54 ± 1 |
From ref. 9.
Measured in the presence of 300 mM urea.
With a Tm at 62°C, the empty form of I-Ek is substantially more stable than most class I molecules lacking peptides. Empty class I molecules usually fail to assemble at physiological temperature (1, 4–8) and, therefore, are likely to have Tm values lower than 37°C. Even when compared with empty H-2Kd heterodimers, which do assemble at physiological temperature (9), the empty form of I-Ek is still significantly more stable, with a Tm 17°C higher than that determined for that form of H-2Kd (Table 1).
To estimate the difference in conformational stability between the two forms of I-Ek, the CD melting curves of the two species (Fig. 2) were subjected to a thermodynamic analysis in which a two-state unfolding model was assumed. At physiological temperature and at neutral pH, we calculate changes in free energy of 2.7 and 6.1 kcal/mol for empty and peptide-filled I-Ek, respectively. Peptide binding thus confers an additional stability of 3.4 kcal/mol upon empty I-Ek heterodimers. This value is comparable, though somewhat lower than those calculated for the binding of specific peptides to class I Kd [4.2 kcal/mol (9)] and HLA-A2 [>5.8 kcal/mol (8)] molecules. The thermal stability we observe for empty I-Ek, though relatively low, is significant. At 37°C, more than 98% of the empty heterodimers would be expected to be folded on the basis of the equilibrium constant derived at this temperature. Moreover, this surprisingly high thermal stability of empty I-Ek may not be limited to I-E molecules. HLA-DR1 (human) heterodimers readily assemble in the absence of antigenic peptide and remain as such throughout a week-long incubation at 37°C (15).
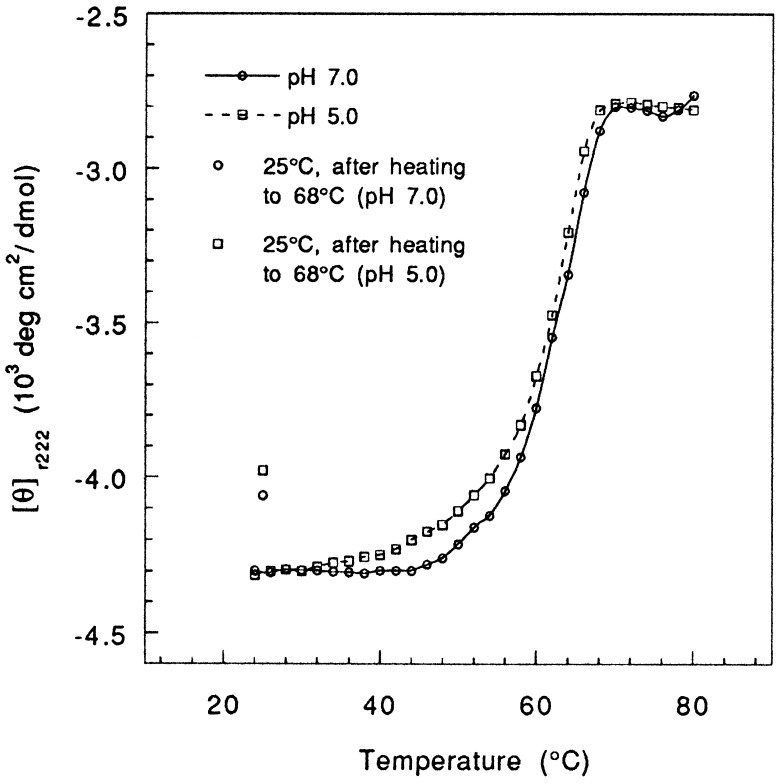
The thermal denaturation curves of empty and peptide-bound I-Ek at pH 5.0 are shown in Fig. 2B and the results are summarized in Table 1. Notably, these data show that protonation does not significantly affect the stability of either form of I-Ek. For both empty and peptide-filled I-Ek, we find melting temperature (63° and 68°C) and free energies (3.0 and 5.7 kcal/mol) that closely match those obtained at neutral pH. To assess whether the stability of I-Ek at low pH is unique to I-E molecules, we have also analyzed the thermal stability of another murine class II molecule, I-Ad, covalently linked to an Eα peptide (28–30). Consistent with previous observations (22, 24, 28), I-Ad is less stable than I-Ek as indicated by its lower transition midpoints at both pH 7.0 and 5.0 (Fig. 3 and Table 1). However, the melting profiles exhibited by the complex at neutral and mildly acidic pH deviate by only 1°C from each other and are virtually identical in appearance, thus indicating a similar degree of stabilization.
Figure 3.
Thermal denaturation of peptide-loaded I-Ad heterodimers (50 μg/ml) monitored by CD. The points shown at 25°C were taken from the full CD spectrum of the renatured protein.
The results presented above contrast with previous studies that suggested that protonation of class II proteins is associated with a substantial loss in complex stability as determined by their sensitivity to SDS-induced chain dissociation during SDS/ PAGE (21, 23–25). However, the significance of these assays is not clear. First, some of these studies (21, 25) involved pH levels (pH ≤4.5) well below those required for optimal peptide binding and may have triggered acid-derived denaturation of the proteins (35). In addition, the binding of SDS to class II molecules might be pH-dependent. Several studies have suggested that protonation of class II proteins may lead to an “opening up” of the protein structure resulting in an increase in the solvent accessibility of the molecules (21–24). Although the results presented herein and elsewhere (23, 36) indicate that such changes in the structure should be subtle, it is possible that under these conditions more SDS molecules can bind to the proteins, thus giving rise to their apparent SDS instability at low pH. Finally, a recent structural analysis of two I-Ek/peptide complexes has shown that the increase in peptide binding to class II molecules at low pH could be explained by small localized changes in the MHC structure that may actually stabilize the binary complex. Specifically, it was shown that the P6 pocket in I-E and DR class II molecules contains two acidic residues (Gluα11 and Aspα66) facing each other, whose protonation could be responsible for the observed enhancement in peptide binding to these molecules at low pH (37).
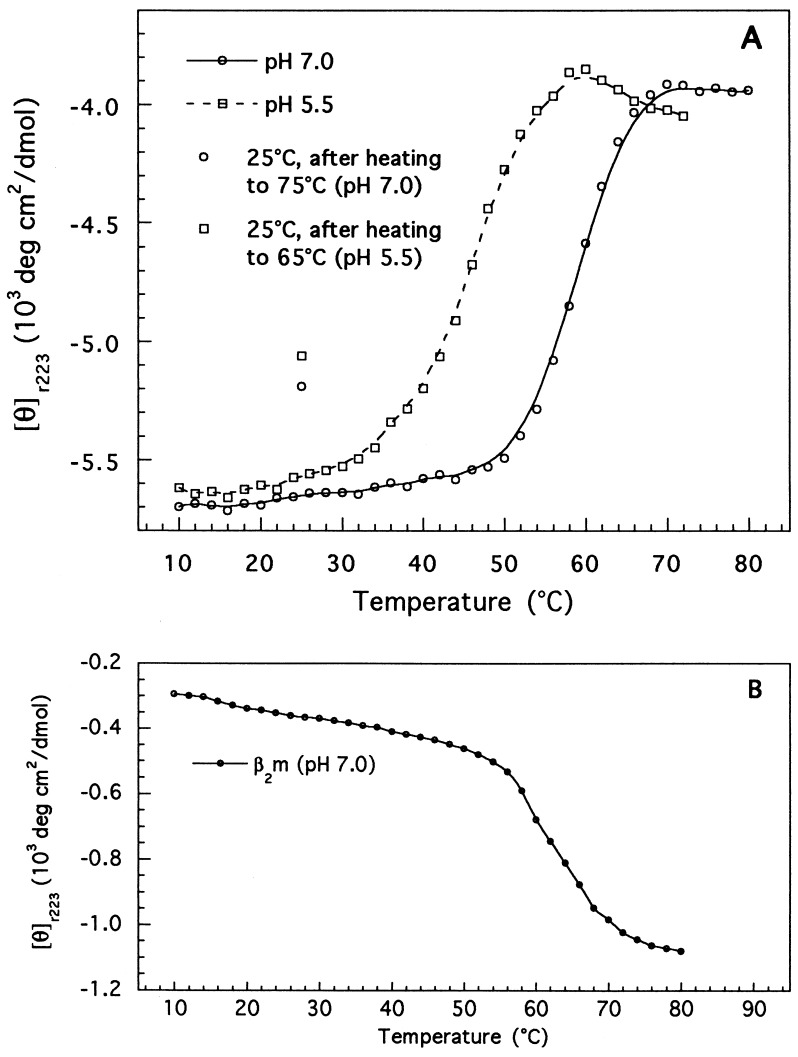
As opposed to the results observed herein for class II molecules, a significant number of class I proteins become less stable at low pH. In particular, it has been shown that the dissociation rate of the β2m subunit of the class I protein, HLA-B7, increases by almost an order of magnitude going from pH 8.0 to 6.0 (38). Similarly, peptide binding studies conducted on three different class I molecules showed that protonation was accompanied by a decrease in complex stability (39). Herein, we have analyzed the effect of the pH on the thermal stability of four class I molecules. Shown in Fig. 4 are the results obtained for one of these molecules, HLA-A2, complexed with the HIV pol (RT) peptide. At neutral pH, HLA-A2 unfolds through a single transition with a Tm at 59°C. The lack of a clear transition of the β2m subunit (Fig. 4B) strongly suggests that peptide dissociation and unfolding of the protein heavy and β2m chains have all occurred simultaneously (8, 9). We next determined the stability of HLA-A2 at mildly acidic pH (pH 5.5). At that pH, the HLA-A2 heavy chain unfolds with a Tm at 45°C, 14°C lower than that measured at neutral pH. Also note that, under these conditions, a second transition that closely matches the melting profile of free β2m is also present, indicating that the two protein chains unfold in a relatively independent manner. Similar profiles and low Tm values have been observed before for HLA-A2 molecules complexed with peptides whose NH2 or COOH terminus was substituted by methyl groups (8) and for empty H-2Kd heterodimers (9). These data suggest then that the apparent instability of HLA-A2 at low pH originates from a pH-induced peptide-dissociation concomitant with chain separation and denaturation.
Figure 4.
Thermal denaturation of peptide-loaded HLA-A2 heterodimers (50 μg/ml) (A) and free β2m (50 μg/ml) (B) monitored by CD. The curve for free β2m was factored by 0.25 to represent its relative contribution to the mean residue ellipticity of the heterodimer. The points shown at 25°C were taken from the full CD spectrum of the renatured protein.
Three additional class I molecules were also analyzed. The first, HLA-B27, unfolded at neutral pH through a single transition with a Tm at 66°C (Table 1). Initial attempts to unfold this molecule at mildly acidic pH (5.0–5.8) resulted in poorly reversible transitions due to aggregation of the protein sample during the thermal cycles. To overcome this problem, melting was performed in the presence of subcritical concentrations (0.3 M) of urea, a chaotrope. At neutral pH, this resulted in only a minor reduction in the thermal stability of the protein, lowering the Tm by only 3°C (to 63°C, Table 1). At pH 5.0, however, the Tm decreased to 52°C, a degree of destabilization similar to that observed for HLA-A2. Similar problems were also encountered when another class I molecule, HLA-B35, was analyzed. In this case, however, complete melting profiles could not be obtained at low pH, even when urea concentrations as high as 0.75 M were used. However, within the range of pH 5.0–5.8, the partial melting curves observed showed a consistent and progressive shift toward lower temperatures compared with those obtained at neutral pH, suggesting that acidification was accompanied by a loss of thermal stability. Interestingly, the murine class I heavy chain H-2Kd complexed with human β2m subunit, was the only class I molecule not destabilized by low pH (Table 1). As mentioned above, this form of H-2Kd is unusual among class I MHCs in that its empty state exhibits an exceptionally high thermal stability.
Physiologically, the differences in stability between the two MHC classes correlate well with their roles in the antigen-presenting cell. Class I MHC molecules assemble and bind peptides inside the endoplasmic reticulum (ER), a situation that allows these two events to be coupled and that provides a means for regulating the peptides to be presented; heterodimers that fail to bind the right peptides are likely to unfold and, subsequently, be discarded. This is consistent with a molecule that is heavily dependent on peptide binding for being properly folded and assembled. In contrast, for class II molecules, coupling protein folding and peptide binding would be mechanistically not feasible since these two processes take place in different cellular compartments, the ER and the endosome, respectively. This, in turn, would necessitate a structure that is relatively independent of peptide for its assembly (1). In this regard, while the invariant chain protein is present and associates with class II MHCs in the ER, it is not required for the folding and assembly of most class II alleles (1, 15), although it may act to keep the nascent molecules in a disaggregated form, ensuring a proper trafficking of the proteins (14, 15).
Similarly, the ability of class II molecules to maintain their stability in a low pH environment is also expected. Otherwise, they would be subjected to a pH-induced unfolding and subsequent proteolytic degradation inside the acidic endosomes in which they reside for several hours (40). The price paid for such a pH resistance is that once bound, some peptides are slow to exchange from class II proteins even at low pH. This includes the class II invariant chain-derived peptide (CLIP) that occupies the molecules’ binding site upon their entrance to the endosomal site (41). Indeed, in vitro, class II–CLIP complexes can be efficiently loaded with antigenic peptides at low pH only in the presence of some detergents (42, 43), and the half-life measured for CLIP bound to HLA-DR3 molecules (at pH 4.5 and 37°C) is significantly longer than that required for class II molecules to mature inside the endosome (44). In vivo, this problem is solved by the action of MHC-like catalysts, such as HLA-DM, that promote the release of the bound CLIP and, thus, allow for a timely exchange by high-affinity peptides (45, 46).
While the rationale behind the apparent resistance of class II molecules to low pH is clear, the significance, if any, of the apparent sensitivity of most class I molecules to low pH is less obvious. One possibility is that this might be important in preventing internalized class I molecules (47) from binding endocytosed “class II-restricted” peptides during recycling through acidic compartments. This, in turn, would act to increase segregation of the two presentation pathways. However, as pointed out above, some variation in pH resistance exist within this MHC class.
The difference in stability between empty class I and class II molecules may also affect the peptide repertoire available for the two MHC classes. Because they have an inherently higher thermal stability, empty class II molecules would be expected to bind and present low-affinity peptides better than class I molecules, in which case complexes are likely to have a half-life that is too short to be physiologically significant. An extended peptide repertoire for class II molecules has been noted (25) and is consistent with observations suggesting a larger set of protein determinants for class II compared with class I-restricted T lymphocytes (48).
Acknowledgments
We thank Jack Aviv and his colleagues in Aviv Associates (Lakewood, NJ) for the use of the 62A DS spectropolarimeter and for their helpful advice and assistance and Dr. Harden McConnell for critical reading of the manuscript. HLA-B27 used in this study was a generous gift from José A. Lebron and Pamela J. Bjorkman. Z.R. was supported by a postdoctoral fellowship from the Rothschild Foundation, J.D.A. was supported by an American Cancer Society postdoctoral fellowship, J.J.B. was supported by a National Institute of Health training grant (AI 19512) and by a fellowship from the Irvington Institute for Medical Research, and D.S.L. is supported by a Howard Hughes Medical Institute predoctoral fellowship. Funding for this work was from the Howard Hughes Medical Institute.
ABBREVIATIONS
- MHC
major histocompatability complex
- β2m
β2-microglobulin
References
- 1.Germain R N, Margulies D H. Annu Rev Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkman P J, Saper M A, Samraoui B, Bennet W S, Strominger J L, Wiley D C. Nature (London) 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 4.Ljunggren H-G, Stam N J, Öhlém C, Neefjes J J, Höglund P, Heemels M T, Bastin J, Schumacher T N, Townsend A, Kärre K, Ploegh H L. Nature (London) 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 5.Kvist S, Hamann U. Nature (London) 1990;348:446–448. doi: 10.1038/348446a0. [DOI] [PubMed] [Google Scholar]
- 6.Hosken N A, Bevan M J. Science. 1990;248:367–370. doi: 10.1126/science.2326647. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz-Navarrete V, Hämmerling G J. Proc Natl Acad Sci USA. 1991;88:3594–3597. doi: 10.1073/pnas.88.9.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvier M, Wiley D C. Science. 1994;265:398–402. doi: 10.1126/science.8023162. [DOI] [PubMed] [Google Scholar]
- 9.Fanestock M L, Tamir I, Narhi L, Bjorkman P J. Science. 1992;258:1658–1662. doi: 10.1126/science.1360705. [DOI] [PubMed] [Google Scholar]
- 10.Guo H-C, Madden D R, Silver M L, Jardetzky T S, Gorga J C, Strominger J L, Wiley D C. Proc Natl Acad Sci USA. 1993;90:8053–8057. doi: 10.1073/pnas.90.17.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker K C, Bednarek M A, Hull L K, Utz U, Cunningham B, Zweerink H J, Biddison W E, Coligan J E. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 12.Ruppert J, Sidney J, Celis E, Kubo R T, Grey H M, Sette A. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 13.Saito Y, Peterson P A, Matsumura M. J Biol Chem. 1993;268:21309–21317. [PubMed] [Google Scholar]
- 14.Germain R N, Rinker A G. Nature (London) 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 15.Stern L J, Wiley D C. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 16.Jensen P E. J Exp Med. 1990;171:1779–1784. doi: 10.1084/jem.171.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sette A, Southwood S, O’Sullivan D, Gaeta F C A, Sidney J, Grey H M. J Immunol. 1992;148:844–851. [PubMed] [Google Scholar]
- 18.Wettstein D A, Boniface J J, Reay P A, Schild H, Davis M M. J Exp Med. 1991;174:219–228. doi: 10.1084/jem.174.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reay P A, Wettstein D A, Davis M M. EMBO J. 1992;11:2829–2839. doi: 10.1002/j.1460-2075.1992.tb05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen P E. J Exp Med. 1991;174:1111–1120. doi: 10.1084/jem.174.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dornmair K, Rothenhausler B, McConnell H M. Cold Spring Harbor Symp Quant Biol. 1989;54:409–416. doi: 10.1101/sqb.1989.054.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Lee J M, Kay C M, Watts T H. Int Immunol. 1992;4:889–897. doi: 10.1093/intimm/4.8.889. [DOI] [PubMed] [Google Scholar]
- 23.Boniface J J, Lyons D S, Wettstein D A, Allbritton N L, Davis M M. J Exp Med. 1996;183:119–126. doi: 10.1084/jem.183.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runnels H A, Moore J C, Jensen P E. J Exp Med. 1996;183:127–136. doi: 10.1084/jem.183.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadegh-Nasseri S, Germain R N. Nature (London) 1991;353:167–170. doi: 10.1038/353167a0. [DOI] [PubMed] [Google Scholar]
- 26.Matsui K, Boniface J J, Reay P A, Schild H, Fazekas de St. Groth B, Davis M M. Science. 1991;254:1788–1791. doi: 10.1126/science.1763329. [DOI] [PubMed] [Google Scholar]
- 27.Boniface J J, Allbritton N L, Reay P A, Kantor R M, Stryer L, Davis M M. Biochemistry. 1993;32:11761–11768. doi: 10.1021/bi00095a003. [DOI] [PubMed] [Google Scholar]
- 28.Kozono H, White J, Clements J, Marrack P, Kappler J. Nature (London) 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 29.Ignatowicz L, Winslow G, Bill J, Kappler J, Marrack P. J Immunol. 1995;154:3852–3862. [PubMed] [Google Scholar]
- 30.Kozono H, Parker D, White J, Marrack P, Kappler J. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 31.Garboczi D N, Hung D T, Wiley D C. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Privalov P L, Gill S J. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 33.Greenfield N, Fasman G D. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 34.Dornmair K, McConnell H M. Proc Natl Acad Sci USA. 1990;87:4134–4138. doi: 10.1073/pnas.87.11.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creighton T E. Proteins. New York: Freeman; 1993. p. 293. [Google Scholar]
- 36.Driscoll P C, Altman J D, Boniface J J, Sakaguchi K, Reay P A, Omichinski J G, Appella E, Davis M M. J Mol Biol. 1993;232:342–350. doi: 10.1006/jmbi.1993.1394. [DOI] [PubMed] [Google Scholar]
- 37.Fremont D H, Hendrickson W A, Marrack P, Kappler J. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 38.Parker K C, Strominger J L. Biochemistry. 1985;24:5540–5550. doi: 10.1021/bi00341a039. [DOI] [PubMed] [Google Scholar]
- 39.Stryhn A, Pedersen L Ø, Romme T, Olsen A C, Nissen M H, Thorpe C J, Buus S. J Immunol. 1996;156:4191–4197. [PubMed] [Google Scholar]
- 40.Tulp A, Verwoerd D, Dobberstein B, Ploegh H L, Pieters J. Nature (London) 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 41.Cresswell P. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 42.Riberdy J M, Newcomb J R, Surman M J, Barbosa J A, Cresswell P. Nature (London) 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 43.Avva R R, Cresswell P. Immunity. 1994;1:763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh P, Amaya M, Mellins E, Wiley D C. Nature (London) 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 45.Weber D A, Evavold B D, Jensen P E. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 46.Sloan S V, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller D M. Nature (London) 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 47.Yewdell J W, Bennink J R. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 48.Bevan M J. Nature (London) 1989;342:478–479. doi: 10.1038/342478a0. [DOI] [PubMed] [Google Scholar]