Abstract
Ceftriaxone and cefotaxime are extended-spectrum cephalosporins previously demonstrated to possess very similar in vitro activities against Streptococcus pneumoniae. Anecdotal reports of isolates with divergent in vitro susceptibilities to ceftriaxone and cefotaxime have been published. To determine the prevalence of pneumococcal isolates with divergent ceftriaxone and cefotaxime susceptibilities, we tested 1,000 clinical isolates collected by U.S. laboratories in 2001-2002 by broth microdilution and E-test. The percentages of isolates susceptible to ceftriaxone and cefotaxime were significantly different by both broth microdilution (98.6 and 96.6%, respectively; P < 0.05) and E-test (98.3 and 95.8%; P < 0.001). The differences observed were due solely to the activities of the two agents against penicillin-resistant isolates. Twenty-six of 188 penicillin-resistant isolates (13.8%) demonstrated different ceftriaxone and cefotaxime MIC interpretative phenotypes when tested by broth microdilution; 18 isolates were concurrently ceftriaxone susceptible and cefotaxime intermediate, 6 were ceftriaxone intermediate and cefotaxime resistant, and 2 were ceftriaxone susceptible and cefotaxime resistant (1.1% of penicillin-resistant isolates; 0.2% of all isolates tested). Sixteen of the 26 isolates (65%) were from southern U.S. states. The 26 isolates had serogroups and serotypes (6, 9, 14, 19, and 23) commonly associated with penicillin-resistant isolates; SmaI pulsed-field gel electrophoresis identified 18 isolates (69%) dispersed among five subtype groups and 8 isolates that were unrelated to any of the other isolates. We conclude that certain isolates of penicillin-resistant pneumococci are less susceptible to cefotaxime than to ceftriaxone and that these isolates are not the result of the spread of a single clone. Whether such isolates have increased in prevalence over time remains unknown.
Streptococcus pneumoniae is frequently identified as an etiologic agent in patients with community-acquired pneumonia, sinusitis, meningitis, and otitis media, and it is a common cause of invasive infections in the very young, the elderly, and in patients with serious underlying illnesses (17). Pneumococci isolated from patients in most countries were exquisitely susceptible to penicillin until the mid- to late 1990s. Epidemiologic evidence suggests that pneumococcal resistance to penicillin and other β-lactams emerged because of the development of strains with mosaic penicillin-binding proteins (PBPs). Molecular surveillance of penicillin-resistant and multidrug-resistant pneumococci from several countries has demonstrated that, in general, the majority of isolates circulating within a geographic area are derivatives of a relatively small number of clones (4, 7, 11, 12, 16, 20). Potential explanations for the rapid emergence of penicillin-resistant S. pneumoniae have been summarized previously (7, 16). In the United States, the rate of penicillin resistance among pneumococci now exceeds 20%, with >10% more isolates demonstrating intermediate levels of resistance to penicillin (8).
PBPs are cell membrane-associated serine peptidases that catalyze polymerization and cross-linking of peptidoglycan precursors in the assembly of bacterial cell walls. Penicillin and other β-lactams bind to different PBPs with varying avidities, leading to a decrease in cell wall synthesis, cell death, and cell lysis. Six PBPs have been identified in S. pneumoniae, and they include five high-molecular-mass PBPs (80 to 90 kDa; 1A, 1B, 2A, 2B, and 2X) and one low-molecular-mass PBP (45 kDa; PBP 3). PBP 2X and PBP 2B are the primary targets of penicillin. The development of high-level resistance to penicillin is a complex process that requires alterations of PBP 2X, PBP 2B, and PBP 1A and may also involve additional mechanisms (9, 22). Mosaic pbp genes are stably inherited in the presence or absence of selection, and strains carrying such genes serve as reservoirs for the horizontal transfer of β-lactam resistance to other pneumococcal strains (26).
Ceftriaxone and cefotaxime are extended-spectrum cephalosporins that have previously demonstrated very similar in vitro activities against pneumococci (25), although ceftriaxone does possess pharmacokinetic and pharmacodynamic advantages when compared to cefotaxime (5, 6). Virtually all strains of S. pneumoniae that are susceptible or intermediately resistant to penicillin are susceptible in vitro to both ceftriaxone and cefotaxime (21). Extended-spectrum cephalosporin resistance in pneumococci is conferred by changes within PBP 1A and PBP 2X. PBP 2B is not a target for extended-spectrum cephalosporins; consequently, resistance to extended-spectrum cephalosporins will not necessarily correlate with resistance to penicillin (16). Anecdotal reports of isolates with different levels of susceptibility and resistance to ceftriaxone and cefotaxime have appeared in the scientific literature (2, 3, 15; J. G. Gums, Progr. Abstr. 98th Am. Thoracic Soc., abstr. J70, 2002), but the prevalence of such phenotypes is largely unknown. As few recent studies have directly compared ceftriaxone and cefotaxime, the present study was undertaken to determine if isolates with discrepant ceftriaxone and cefotaxime MICs and MIC interpretations exist and, if so, to also determine their prevalence and the relatedness of isolates demonstrating these unanticipated phenotypes.
MATERIALS AND METHODS
Bacterial isolates studied.
One thousand clinical isolates of S. pneumoniae were tested. The isolates were collected from 118 microbiology laboratories in 18 states in the United States from April 2001 to May 2002. Each isolate was from a nonmeningeal specimen source and from a different patient. Isolates were considered to be etiologic agents of infection by individual laboratory algorithms and were collected without regard to patient age, gender, inpatient or outpatient status, or nonmeningeal specimen source quotas. The numbers of isolates collected from microbiology laboratories in each state were as follows: Alabama (75 isolates, 5 microbiology laboratories); Arizona (50 isolates, 3 labs); California (50 isolates, 14 labs); Florida (75 isolates, 8 labs); Georgia (50 isolates, 7 labs); Illinois (50 isolates, 12 labs); Iowa (50 isolates, 5 labs); Louisiana (75 isolates, 4 labs); Massachusetts (50 isolates, 6 labs); Minnesota (50 isolates, 3 labs); New Jersey (50 isolates, 7 labs); New York (50 isolates, 10 labs); Oklahoma (50 isolates, 2 labs); Pennsylvania (50 isolates, 15 labs); Tennessee (75 isolates, 5 labs); Utah (50 isolates, 3 labs); Washington (50 isolates, 3 labs); and Wisconsin (50 isolates, 6 labs). Of the 1,000 isolates studied, 23.1, 46.1, and 29.9% of isolates were from patients aged <18, 18 to 64, and >64 years, respectively, and 0.9% of isolates were from patients of unknown age; 59.5% of isolates were from males and 40.0% were from females, and 0.5% were from patients of unknown gender; 87.7% of isolates were from outpatients, 10.4% were from inpatients, and 1.9% were from patients of unknown location; 20.6% of isolates were from upper respiratory sources (nasopharynx, throat, nose, and sinus), 46.8% were from lower respiratory sources (sputum, bronchial washings, and tracheal aspirates), and 32.6% were from blood cultures. Clinical isolates were transported to a central laboratory (Focus Technologies, Herndon, Va.) where they were subcultured onto sheep blood agar and streaked for purity, and their identities were confirmed using standard clinical laboratory methods. The observation of alpha-hemolysis on blood agar and an optochin disk zone diameter of ≥14 mm were used as confirmatory tests; if necessary, a bile solubility test was also performed.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing of ceftriaxone, cefotaxime, and penicillin was performed in our laboratory using freeze-dried broth microdilution panels prepared by TREK Diagnostics (Cleveland, Ohio) in accordance with the recommended procedures of the National Committee for Clinical Laboratory Standards (NCCLS) (18). Ceftriaxone and cefotaxime were also tested by E-test (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions (incubation at 35°C in 5% CO2 for 18 to 24 h). MICs were interpreted as susceptible, intermediate, or resistant by using the M100-S13 NCCLS recommendations for nonmeningeal isolates of S. pneumoniae (19).
PFGE.
Twenty-six isolates that had discrepant ceftriaxone and cefotaxime MIC interpretations by broth microdilution, 26 (demographically matched) control isolates that were susceptible to both ceftriaxone and cefotaxime, and the 8 isolates that were resistant to both ceftriaxone and cefotaxime were analyzed by pulsed-field gel electrophoresis (PFGE). The 26 control isolates were all ceftriaxone susceptible and cefotaxime susceptible, but not all were penicillin susceptible; 18 of the 26 isolates were penicillin susceptible, 2 were penicillin intermediate, and 6 were penicillin resistant. PFGE of SmaI-restricted chromosomal DNA was performed according to a previously described method (14). DNA fragments were separated on a CHEF-DR III instrument (Bio-Rad Laboratories, Hercules, Calif.) for 20 h at 14°C on 1% agarose gels using ramped pulse times from 1 to 25 s (6 V/cm). Gels were examined by transillumination with UV light following ethidium bromide staining.
Gels were normalized using S. pneumoniae R6 (ATCC 27336) restricted with SmaI. No standardized method for grouping of isolates into clonal groups has been developed. For the purpose of this study, isolates were defined as genetically indistinguishable, closely or possibly related, or genetically unrelated if their PFGE profiles differed by 0, 1 to 6, or ≥7 bands, respectively; this is a modification of the categories established by Tenover et al. (24). Isolates within each PFGE type with exactly the same PFGE profile were assigned the same subtype. Isolates differing by 1 to 6 bands from subtype 1 of each type were assigned a common type. Isolates with ≥7 band differences from subtype 1 of each type were considered unrelated isolates and were assigned a different PFGE type (10, 24). DNA banding patterns were also digitalized for analysis using Molecular Analyst software (Fingerprinting Plus, version 1.12; Bio-Rad Laboratories), and a dendrogram was calculated using the unweighted pair groups method with arithmetic averages and the Dice coefficient (the number of shared bands × 2 × 100/total number of bands in two samples). Dendrograms were generated to confirm the PFGE types and subtypes determined visually (10). Subtypes determined visually had ≥80% correlation in the dendrogram.
Serogrouping and serotyping.
Capsular serogroups and serotypes of the 60 isolates tested by PFGE were determined by slide coagglutination testing using group-specific antisera obtained from the Statens Seruminstitut (Copenhagen, Denmark).
Statistical analysis.
The assessment of statistical significance was made using χ2 testing with EpiInfo Statcalc, version 6 (Centers for Disease Control and Prevention). Uncorrected P values of <0.05 were considered statistically significant.
RESULTS
Among the 1,000 isolates of S. pneumoniae tested, 66.6% were penicillin susceptible, 14.6% were penicillin intermediate, and 18.8% were penicillin resistant by broth microdilution testing (Table 1). Penicillin-intermediate and -resistant isolates had higher MICs of ceftriaxone and cefotaxime than did penicillin-susceptible isolates. The percentages of all isolates susceptible to ceftriaxone and cefotaxime by both broth microdilution (98.6 and 96.6%, respectively; P < 0.05) and E-test (98.3 and 95.8%; P < 0.001) were significantly different. The differences observed were due solely to the differences in the activities of the two agents against penicillin-resistant isolates. By broth microdilution and E-test, the percentage of penicillin-resistant isolates susceptible to ceftriaxone (>90%) was >10% higher than that for cefotaxime (P < 0.001). Penicillin-resistant and -intermediate isolates demonstrated lower MICs at which 90% of isolates were inhibited (MIC90s) to ceftriaxone than to cefotaxime by broth microdilution (penicillin-resistant isolates, 1 versus 2 μg/ml; penicillin-intermediate isolates, 0.5 versus 1 μg/ml) and E-test (1 versus 1.5 μg/ml and 0.5 versus 0.75 μg/ml). For ceftriaxone alone and cefotaxime alone, less than 1% of all isolates demonstrated different MIC interpretative phenotypes (19) by broth microdilution and E-test.
TABLE 1.
In vitro susceptibilities of 1,000 isolates of S. pneumoniae to ceftriaxone and cefotaxime tested by broth microdilution and E-test
| Test method and antimicrobial agent | Isolate phenotypea | MIC (μg/ml)
|
MIC interpretationb
|
|||||
|---|---|---|---|---|---|---|---|---|
| Range | Mode | MIC50 | MIC90 | % Susceptible | % Intermediate | % Resistant | ||
| Broth microdilution | ||||||||
| Ceftriaxone | All isolates | ≤0.004-8 | 0.015 | 0.015 | 1 | 98.6 | 0.6 | 0.8 |
| Penicillin susceptible | ≤0.004-0.5 | 0.015 | 0.015 | 0.03 | 100 | 0 | 0 | |
| Penicillin intermediate | 0.015-1 | 0.5 | 0.25 | 0.5 | 100 | 0 | 0 | |
| Penicillin resistant | 0.12-8 | 1 | 1 | 1 | 92.6 | 3.2 | 4.3 | |
| Cefotaxime | All isolates | ≤0.004-8 | 0.015 | 0.015 | 1 | 96.6 | 1.8 | 1.6 |
| Penicillin susceptible | ≤0.004-1 | 0.015 | 0.015 | 0.03 | 100 | 0 | 0 | |
| Penicillin intermediate | 0.015-1 | 0.12, 0.5 | 0.25 | 1 | 100 | 0 | 0 | |
| Penicillin resistant | 0.25-8 | 1 | 1 | 2 | 81.9 | 9.6 | 8.5 | |
| E-test | ||||||||
| Ceftriaxone | All isolates | 0.004-4 | 0.012 | 0.016 | 0.5 | 98.3 | 0.8 | 0.9 |
| Penicillin susceptible | 0.004-0.75 | 0.012 | 0.012 | 0.032 | 100 | 0 | 0 | |
| Penicillin intermediate | 0.012-1 | 0.38 | 0.19 | 0.5 | 100 | 0 | 0 | |
| Penicillin resistant | 0.25-4 | 0.75 | 0.5 | 1 | 91.0 | 4.3 | 4.8 | |
| Cefotaxime | All isolates | 0.004-8 | 0.016 | 0.023 | 1 | 95.8 | 2.6 | 1.6 |
| Penicillin susceptible | 0.004-1 | 0.016 | 0.016 | 0.047 | 100 | 0 | 0 | |
| Penicillin intermediate | 0.016-1 | 0.38 | 0.19 | 0.75 | 100 | 0 | 0 | |
| Penicillin resistant | 0.38-8 | 1 | 1 | 1.5 | 77.7 | 13.8 | 8.5 | |
Of the 1,000 isolates tested, 666 were penicillin susceptible, 146 were penicillin intermediate, and 188 were penicillin resistant.
MICs interpreted using NCCLS 2003 guidelines (19).
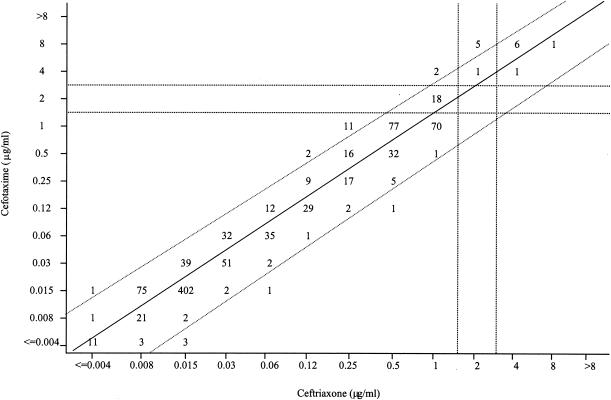
Identical ceftriaxone and cefotaxime MICs were observed for 670 of the 1,000 isolates; 974 isolates had MICs of ceftriaxone and cefotaxime within one doubling dilution of each other (Fig. 1). There was a propensity for nonidentical MICs within one doubling dilution to be one doubling dilution lower for ceftriaxone (n = 286 isolates) than for cefotaxime (n = 18). Twenty-one isolates had ceftriaxone MICs two doubling dilutions lower than for cefotaxime, compared with five isolates having cefotaxime MICs two doubling dilutions lower than for ceftriaxone.
FIG. 1.
Scatterplot of ceftriaxone and cefotaxime MICs determined by broth microdilution for 1,000 isolates of S. pneumoniae. The solid line is the linear regression line (r = 0.977). The area between the two diagonal dotted lines contains isolates with ceftriaxone MICs and cefotaxime MICs within ±1 doubling dilution of each other. The vertical dotted lines divide the isolates into ceftriaxone susceptible (MIC, ≤1 μg/ml), intermediate (MIC, 2 μg/ml), and resistant (MIC, ≥4 μg/ml) groups; the horizontal dotted lines divide the isolates into cefotaxime susceptible (MIC, ≤1 μg/ml), intermediate (MIC, 2 μg/ml), and resistant (MIC, ≥4 μg/ml) groups.
Of the 1,000 isolates tested, 26 (2.6%) demonstrated different ceftriaxone and cefotaxime MIC interpretative phenotypes; 18 (1.8%) were concurrently ceftriaxone susceptible and cefotaxime intermediate, 6 (0.6%) were ceftriaxone intermediate and cefotaxime resistant, and 2 (0.2%) were ceftriaxone susceptible and cefotaxime resistant by broth microdilution testing (Fig. 1). All 26 isolates were penicillin resistant (Table 2). The differences in ceftriaxone and cefotaxime MICs were one doubling dilution for 19 of 26 isolates and two doubling dilutions for the other 7 isolates. E-test MICs were higher for cefotaxime than for ceftriaxone for 24 of the 26 isolates and identical for the other two isolates. Sixteen of the 26 isolates (65%) were from southern U.S. states (Alabama, Florida, Tennessee, Louisiana, Oklahoma, and Georgia). Eight other isolates were resistant to both cefotaxime and ceftriaxone by broth microdilution (Fig. 1); among these isolates, six of eight had MICs one doubling dilution lower for ceftriaxone (MIC range, 4 to 8 μg/ml) than for cefotaxime (MIC range, 4 to 8 μg/ml) by broth microdilution; by E-test, MICs for the eight isolates were lower for ceftriaxone (MIC range, 2 to 3 μg/ml) than for cefotaxime (MIC range, 4 to 8 μg/ml) (data not shown). Isolates with cefotaxime-susceptible and ceftriaxone-intermediate, cefotaxime-susceptible and ceftriaxone-resistant, and cefotaxime-intermediate and ceftriaxone-resistant phenotypes were not observed using broth microdilution testing or E-test.
TABLE 2.
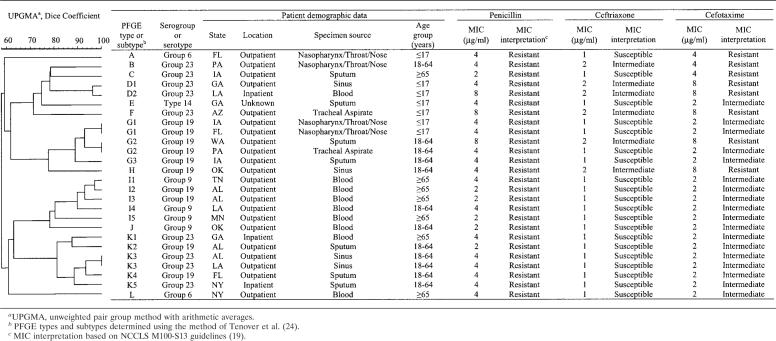
Genetic relatedness of 26 clinical isolates of S. pneumoniae with different MIC interpretative phenotypes for ceftriaxone and cefotaxime
By SmaI PFGE, 8 of the 26 isolates with disparate ceftriaxone and cefotaxime MIC interpretations were unrelated to any of the other isolates (PFGE types, A, B, C, E, F, H, J, and L) (Table 2) (24). Four groups of subtypes were observed, accounting for six, five, five, and two isolates, respectively. Three sets of two identical strains each were observed. Serogroups 19, 23, 9, and 6 and serotype 14 accounted for 10, 9, 4, 2, and 1 isolates, respectively. Of the eight ceftriaxone- and cefotaxime-resistant isolates, two strains were identified that were unrelated to any of the other isolates; we also identified one subtype with four isolates (including two isolates with identical PFGE profiles) and one subtype with two isolates (data not shown). All ceftriaxone- and cefotaxime-resistant isolates were serogroup 19 or 23. Fifteen of the 26 (57.7%) ceftriaxone-susceptible and cefotaxime-susceptible control isolates were unique isolates. Five subtypes were observed, including one subtype with three isolates and four subtypes with two isolates (including two isolates with identical PFGE profiles), respectively. Serogroups 6 and 19 accounted for five isolates each, there were two isolates each from serogroup 9, serogroup 23, serogroup 15, serogroup 11, serotype 14, and serotype 4, and one isolate each from serotype 3, serogroup 16, serogroup 18, and serotype 29/serogroup 35. Overall, 11 (42%) control isolates were contained in one of the identified subtypes compared to 18 (69%) of the isolates with disparate ceftriaxone and cefotaxime MIC interpretations and 6 of 8 (75%) ceftriaxone- and cefotaxime-resistant isolates. Serotypes were strongly correlated to clonal types identified using PFGE, as the majority of isolates within an identical clonal type belonged to the same serotype.
DISCUSSION
The present study identified statistically significant differences in rates of susceptibility to ceftriaxone and cefotaxime for S. pneumoniae tested by both broth microdilution testing and E-test (Table 1). These differences were generally of one doubling dilution and were attributable to lower ceftriaxone MICs than cefotaxime MICs for penicillin-resistant isolates. Given that ceftriaxone and cefotaxime MICs and MIC interpretations are rarely reported together, the prevalence of such phenotypes is largely unknown (8, 13, 21, 25). Available data suggest that isolates with different ceftriaxone and cefotaxime MICs and MIC interpretative phenotypes do exist (13, 21), although their prevalence is low (2.6% in the present study [Fig. 1 and Table 2]). Data have not been published suggesting that the prevalence of these phenotypes may be increasing; however, phenotypes of pneumococci with different susceptibilities to ceftriaxone and cefotaxime have been reported previously (15). McDougal et al. identified 10 pneumococci isolated from 1991 and 1992 that were nonsusceptible to extended-spectrum cephalosporins (15). Of these 10 isolates, 8 had ceftriaxone MICs one doubling dilution lower than cefotaxime and 4 isolates would have had discrepant MIC interpretative phenotypes favoring ceftriaxone using current NCCLS breakpoints (19). The present study is the first to report the prevalence of these isolates in a large sample of pneumococci. Periodic studies to determine the prevalence of isolates with discrepant ceftriaxone and cefotaxime MICs and MIC interpretative phenotypes may be useful in the future to determine if disparate changes in the activities of these two agents are occurring.
Most pneumococcal isolates that are resistant to penicillin also have lowered susceptibility to extended-spectrum cephalosporins, including cefotaxime and ceftriaxone (Table 1). Among the 26 isolates with different ceftriaxone and cefotaxime MIC interpretative phenotypes all were penicillin resistant, and the serogroups or serotypes identified are known to include the six most prevalent serotypes associated with penicillin resistance (serotypes 6A, 6B, 9V, 14, 19F, and 23F) (16, 27). The test isolates with different ceftriaxone and cefotaxime MIC interpretations or those resistant to both extended-spectrum cephalosporins were more often associated with a clonal PFGE type or subtype and serotype (Table 2) than were control isolates susceptible to both ceftriaxone and cefotaxime (data not shown). However, isolates with different ceftriaxone and cefotaxime MIC interpretations can also arise independently. The identification of pneumococcal isolates with other unexpected phenotypes, such as isolates with higher amoxicillin MICs than penicillin MICs (9) and isolates with penicillin-susceptible and ceftriaxone- and cefotaxime-resistant phenotypes (1, 3, 23), have also been previously reported. Such phenotypes are generally attributed to PBP sequence changes, sometimes in combination with non-PBP-based mechanisms.
In conclusion, penicillin-resistant S. pneumoniae isolates with different MIC interpretative phenotypes for ceftriaxone and cefotaxime were identified by MIC testing. These phenotypes accounted for 13.8% (26 of 188) of penicillin-resistant isolates in the present study. Given that the MIC is a relatively crude method by which to measure differences in susceptibility, molecular techniques will need to be applied to such isolates to determine the mechanism(s) underlying the observed differences. The presence of isolates that are less susceptible to cefotaxime than ceftriaxone could potentially result from the use of cefotaxime once or twice daily as opposed to three times a day (5, 6, 19), and these phenotypes may be more common in certain geographic areas because of differences in local use of extended-spectrum cephalosporins. The emergence and global spread of penicillin-resistant pneumococci in a relatively short period of time suggests that isolates with disparate ceftriaxone and cefotaxime susceptibilities may also have the potential to spread, and therefore continued epidemiological surveillance for these phenotypes is important.
Acknowledgments
We thank Roche Laboratories, Inc., who financially supported this study.
REFERENCES
- 1.Centers for Disease Control and Prevention. 1994. Drug-resistant Streptococcus pneumoniae—Kentucky and Tennessee, 1993. Morb. Mortal. Wkly. Rep. 43:23-25. [PubMed] [Google Scholar]
- 2.Chiou, C. C., and M. C. McEllistrem. 2001. Novel penicillin-, cephalosporin-, and macrolide-resistant clones of Streptococcus pneumoniae serotypes 23F and 19F in Taiwan which differ from international epidemic clones. J. Clin. Microbiol. 39:1144-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corso, A., E. P. Severina, V. F. Petruk, Y. R. Mauriz, and A. Tomasz. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4:325-337. [DOI] [PubMed] [Google Scholar]
- 5.Craig, W. A. 1995. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagn. Microbiol. Infect. Dis. 22:89-96. [DOI] [PubMed] [Google Scholar]
- 6.Craig, W. A. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1-12. [DOI] [PubMed] [Google Scholar]
- 7.Doern, G. V., A. B. Brueggemann, M. Blocker, M. Dunne, H. P. Holley, Jr., K. S. Kehl, J. Duval, K. Kugler, S. Putnam, A. Rauch, and M. A. Pfaller. 1998. Clonal relationships among high-level penicillin-resistant Streptococcus pneumoniae in the United States. Clin. Infect. Dis. 27:757-761. [DOI] [PubMed] [Google Scholar]
- 8.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggeman. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Plessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Beall. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 11.Hall, L. M., R. A. Whiley, B. Duke, R. C. George, and A. Efstratiou. 1996. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J. Clin. Microbiol. 34:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ip, M., D. J. Lyon, W. Raymond, H. Yung, C. Chan, and A. F. B. Cheng. 1999. Evidence of clonal dissemination of multidrug-resistant Streptococcus pneumoniae in Hong Kong. J. Clin. Microbiol. 37:2834-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, R. N., A. H. Mutnick, and D. J. Varnam. 2002. Impact of modified nonmeningeal Streptococcus pneumoniae interpretative criteria (NCCLS M100-S12) on the susceptibility patterns of five parenteral cephalosporins: report from the SENTRY Antimicrobial Surveillance Program (1997 to 2001). J. Clin. Microbiol. 40:4332-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDougal, L. K., J. K. Rasheed, J. W. Biddle, and F. C. Tenover. 1995. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococccus pneumoniae isolates in the United States. Antimicrob. Agents Chemother. 39:2282-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munoz, R., J. M. Musser, M. Crain, D. E. Briles, A. Marton, A. J. Parkinson, U. Sorenson, and A. Tomasz. 1992. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin. Infect. Dis. 15:112-118. [DOI] [PubMed] [Google Scholar]
- 17.Musher, D. M. 2000. Streptococcus pneumoniae, p. 2128-2147. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. 18, no. 2. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing; 13th informational suppl., vol. 23, no. 1. Standard M100-S13. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Richter, S. S., K. P. Heilmann, S. L. Coffmann, H. K. Huynh, A. B. Bruggemann, M. A. Pfaller, and G. V. Doern. 2002. The molecular epidemiology of penicillin-resistant Streptococcus pneumoniae in the United States, 1994-2000. Clin. Infect. Dis. 34:330-339. [DOI] [PubMed] [Google Scholar]
- 21.Sahm, D. F., C. Thornsberry, D. C. Mayfield, M. E. Jones, and J. A. Karlowsky. 2002. In vitro activities of broad-spectrum cephalosporins against nonmeningeal isolates of Streptococcus pneumoniae: MIC interpretation using NCCLS M100-S12 recommendations. J. Clin. Microbiol. 40:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, A. M., and K. P. Klugman. 2001. Alterations in MurM, a cell wall muropeptide branching enzyme, increase high-level penicillin and cephalosporin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:2393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, A. M., R. F. Botha, H. J. Koornhof, and K. P. Klugman. 2001. Emergence of a pneumococcal clone with cephalosporin resistance and penicillin susceptibility. Antimicrob. Agents Chemother. 45:2648-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornsberry, C., P. T. Oglivie, H. P. Holley, Jr., and D. F. Sahm. 1999. Survey of susceptibilities of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolates to 26 antimicrobial agents: a prospective U.S. study. Antimicrob. Agents Chemother. 43:2612-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomasz, A. 1997. Antibiotic resistance in Streptococcus pneumoniae. Clin. Infect. Dis. 24:S85-S88. [DOI] [PubMed] [Google Scholar]
- 27.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, C. Lexau, A. Reingold, L. Lefkowitz, P. R. Cieslak, M. Cetron, E. R. Zell, J. H. Jorgensen, and A. Schuchat. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917-1924. [DOI] [PubMed] [Google Scholar]