Abstract
The gene ere(A) of the plasmid pIP1100 is larger than originally reported and is organized as an integron gene cassette. The ere(A) gene cassette carries its own promoter and is propagated by a class 2 integron with an insertion sequence element, IS1, inserted upstream of the intI2 gene. The mobility of the ere(A) cassette has been demonstrated.
Integrons are natural cloning and expression systems (21). The integron platform codes for an integrase (IntI) that mediates recombination between a proximal primary recombination site (attI) and a target recombination sequence called an attC site (or 59-base element). The attC site is usually found associated with a single open reading frame in a structure termed a gene cassette (10, 25, 26). Insertion of the gene cassette at the attI site, which is located downstream of a resident promoter, Pc, internal to the intI gene, drives expression of the encoded proteins (12). More than 70 different antibiotic resistance genes have been characterized within integrons thus far (22). Multiresistance integrons (MRIs) evolved through successive integrations of antibiotic resistance loci within the integron platform. Five classes of MRIs have been reported based on the divergence of their integrase genes (22). Class 1 integrons are mostly found associated with the Tn21 transposon family, while the class 2 integrons are associated with the Tn7 transposon family (10).
Most of the attC sites of integron resistance gene cassettes identified to date share little homology. Their lengths and sequences vary considerably (lengths from 57 to 141 bp) and their sequence similarities are primarily restricted to their boundaries, which correspond to the inverse core site (ICS; RYYYAAC) and the core site (CS; G↓TTRRRY, where R is a purine, Y is a pyrimidine, and the arrow shows the recombination point) (7, 25).
Enterobacteria are intrinsically resistant to high levels of erythromycin (EM; MIC > 250 μg/ml) (2). However, oral EM is still prescribed to overcome intestinal infections due to antibiotic-resistant bacteria such as Vibrio cholerae (http://www.who.int/inf-fs/en/fact107.html), due in part to the high local concentrations of EM in the intestinal lumen (0.5 to 6 mg/g of feces). Several clinical isolates of Escherichia coli that are highly resistant to EM have been described, and their resistance mechanisms have been characterized to various extents. Interestingly, two types of EM-inactivating esterases have been identified among these E. coli strains. Further characterization has shown that these EM esterases were encoded by two genes, ere(A) (ereA from pIP1100 [14]) and ere(B) (ereB [3]), which share little similarity. Recently, a cassette carrying a gene showing about 90% identity with ere(A) was identified in a class 1 MRI carried on plasmid pLQ1723 in Providencia stuartii (16) and in other class 1 MRIs from different clinical bacterial isolates (6, 15, 28).
Determining the genetic context of the ere(A) gene in plasmid pIP1100.
The peculiar attC site carried by the ere(A) of the pLQ1723 cassette is related to the nucleotide sequence located immediately downstream of the ere(A) gene of pIP1100 (accession no. M11277) (14), suggesting that ere(A) of pIP1100 was also carried in an integron cassette. However, the region of similarity ended before an identifiable CS sequence could be found. Two features suggest that the deposited sequence was inaccurate: (i) the sequenced fragment was originally cloned from a Sau3A library, with the end of the attC site corresponding to a Sau3A site, and likely corresponding to a cloning artifact, and (ii) the annotated ere(A) gene of pIP1100 was shorter (345 codons) than the ere(A) gene of pLQ1723 (409 codons), but the homology between them extended a further 246 nucleotides (nt) upstream. Furthermore, the original investigators noted that even though the putative ere(A) gene product was a 38-kDa protein, when the protein was expressed in minicells its apparent Mr was about 43,000 (14). These observations led us to characterize the original ere(A) locus in pIP1100 in the original strain BM2195 (1). Since we suspected that the ere(A) gene was inside an integron, we screened the strain for the presence of the class 1, 2, and 3 integron-specific intI genes through PCR amplification using specific primers (Int1.F and Int1.R; I2UP and I2DW; and I3UP and I3DW, respectively; PCRs were performed with PCR Reddy mix (Abgene, Epsom, United Kingdom) following the manufacturer's instructions. Primer sequences can be found at http://www.pasteur.fr/recherche/unites/pmtg/integ/ere_primers). Primer sets specific for intI1 and for intI2 gave rise to amplified products, showing that BM2195 contained both class 1 and class 2 integrons. The association of ere(A) with the class 2 integron was established by PCR amplification using an ere(A)-specific primer (ereQ.L2) and the intI2 primer, I2DW. Finally, the entire class2 integron was obtained by PCR amplification with primers I2DW and orfX-Tn7, a primer that hybridizes just upstream of the Tn7 tnsE gene. The sole amplified fragment, about 5.4 kb long, was cloned (plasmid p1792) and sequenced. A list of the strains and plasmids used in this study is presented in Table 1.
TABLE 1.
Bacterial strains and relevant plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44ΔlacU169 (φ80lacZ′ ΔM15) ΔargF hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| UB1637 | his lys trp recA56 rpsL | 13 |
| UB5201 | pro met recA56 gyrA | 13 |
| BM2195 | Wild-type, pIP1100, pIP1101, pIP1102 | 1 |
| ω165 | UB1637 R388 p2187 p112 | This work |
| ω171 | UB1637 R388 p2194 pTRC99A | This work |
| ω172 | UB1637 R388 p2194 p112 | This work |
| ω173 | UB1637 R388 p2195 pTRC99A | This work |
| ω174 | UB1637 R388 p2195 p112 | This work |
| P. stuartii | Wild-type, pLQ1723 [ere(A)] | P. H. Roy |
| Plasmids | ||
| R388 | Tpr Sur Tra+ IncW | 27 |
| pSU38 | Kanr oriP15A | 4 |
| pTRC99A | Apr oriColE1, expression vector | Pharmacia |
| p112 | pTRC99A::int11 | 20 |
| p2247 | 1,298 bp EcoRI-BamHI PCR fragment (ereA1 and ereA2) from pLQ1723 in pSU38, ere(A)pLQ1723 L101S, Kanr | This work (AY183454) |
| p1792 | 5.4-kb PCR fragment (I2DW and orfX-Tn7) from pIP1100 in pCR2.1, Kanr Apr Emr | This work (AY183453) |
| p1818 | 1,290-bp SacI-BamHI PCR fragment (ereAStQ.L2 and ereA2) from p1792 in pSU38, ere(A)pIP1100, Kanr Emr | This work |
| p1988 | 1,298-bp EcoRI-BamHI PCR fragment (ereA1 and ereA2) from pLQ1723 in pSU38, ere(A)pLQ1723, Kanr Emr | This work |
| p2187 | 1,492-bp BamHI PCR fragment (SatEnd and aadA1) from p1792 in pSU38, lacZα orientation, Kanr Emr | This work |
| p2188 | 1,492-bp BamHI PCR fragment (SatEnd and aadA1) from p1792 in pSU38, anti-lacZα orientation, Kanr Emr | This work |
| p2194 | EcoRI deletion in p2187, ere(A)(-aadA1) attC site, Kanr | This work |
| p2195 | PstI deletion in p2187, sat(-ere(A)) attC site, Kanr | This work |
The ere(A) gene cassette and its class 2 integron carrier.
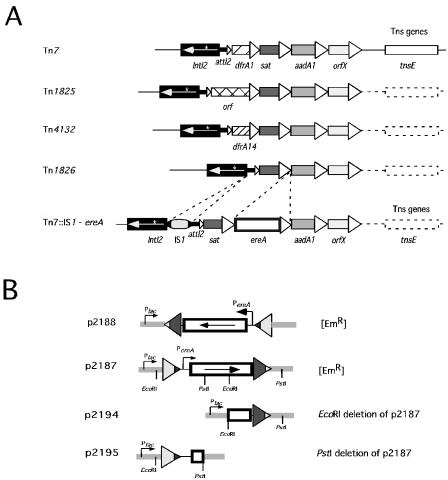
Sequence analysis of the p1792 insert revealed a unique class 2 integron, part of a Tn7 derivative, carrying four cassettes in the following order: sat, ere(A), aadA1, and orfX (Fig. 1). As for all previously described class 2 integrons, the intI2* gene was interrupted by a premature stop at codon 179. Compared to the four other class 2 integrons described so far, this integron is the only one showing disruption of the sat-aadA1-orfX cassette array (Fig. 1). Indeed, the only difference among the four other class 2 integrons is the presence of distinct cassettes in position 1 (dfrA1 in Tn7, orf in Tn1825, and dfrA14 in Tn4132) or the lack of cassettes upstream of sat (Tn1826) (11, 29, 30). A second exclusive feature of this integron resided in the presence of an IS1 element inserted 10 nt upstream of intI2* and 310 bp upstream of the attI2 recombination point. The rest of the sequence was identical to its Tn7 counterparts.
FIG. 1.
Physical maps of the different class 2 integrons carried in Tn7-related transposons (A) and of the ere(A)pIP1100 cassette derivative constructions used in expression and recombination studies (B). (A) Black boxes symbolize the intI2 integrase genes, and the asterisk indicates the location of the premature stop codon; other boxes correspond to cassette genes. The attI2 recombination site is shown by a small open triangle; attC sites are represented by bigger open triangles. Transposition (Tns) genes are indicated on the right side of the different transposons. The class 2 integron investigated in this study is Tn7::IS1-ere(A) (see the text for details). The IS1 is shown by an oval with black boundaries. (B) The ere(A) gene is represented as an open box; the thin line represents the 5′ untranslated DNA region upstream of the ere(A) start codon. Plasmid constructions are described in Table 1. The sat and ere(A) attC sites are shown as triangles. Relevant EM resistance phenotypes conferred by plasmids are mentioned.
It is likely that the class 2 integron also carries a strong promoter, as it allows the phenotypic expression of all the cassettes in the array, as in class 1 integrons (12). Furthermore, the sat and aadA1 cassettes do not contain enough space 5′ of the genes to contain a promoter sequence. The class 2 cassette promoter has still to be precisely identified, but according to Hansen et al. and Levesque et al., it is located upstream of the intI2* gene (11) and not in the 5′ part of the intI gene as in the class 1 integron (12). The structure of this class 2 integron supports a promoter location within the 310-bp region between the IS1 insertion site and the attI2 recombination site, as no outward-facing promoter could be identified in the IS1 sequence itself.
Comparison of the ere(A) gene cassettes from pIP1100 and pLQ1723.
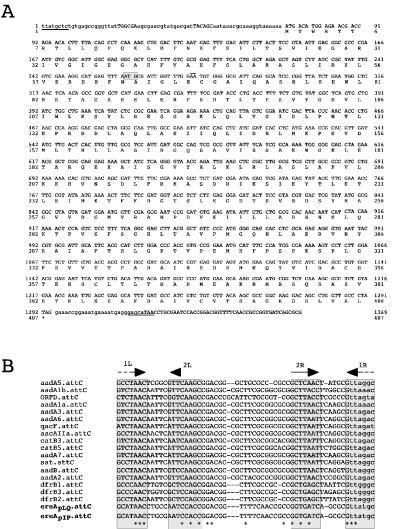
As indicated above, the ere(A) gene of pIP1100 gene was found to be the second cassette in this integron. Sequence analysis showed that this cassette was 1,369 nt long and coded for an ere(A) gene product of 406 amino acids, with a molecular mass of 44.5 kDa, similar to the data obtained from previous expression experiments (14). Comparison of the ere(A) cassette to the former ere(A) locus sequence showed that the differences between the two sequences essentially led to an extension of 62 codons at the 5′ end of the new sequence (Fig. 2). Furthermore, analysis of the ere(A) cassette boundaries revealed a 48-nt attC site (from the T of the ICS GCATAAC up to the G from the CS located at the junction with the aadA1 cassette), displaying all the characteristics of such recombination sites. Indeed, a canonical CS consensus could be identified at the recombination point, confirming that, as speculated, the original sequence was a chimera due to ligation between unrelated Sau3A fragments.
FIG. 2.
ere(A)pIP1100 cassette sequence and features (A) and comparison of the ere(A) attC sites to the short attC sites of other resistance cassettes (B). (A) The EreA EM esterase amino acid sequence is shown underneath the coding part of the cassette. The potential ribosome binding site is shown in bold, the proposed start codon in the former characterization of ere(A) is shown by a grey box (an additional A upstream of the overlined AA put this ATG in frame with the 3′ end of the gene), and the sequences that are complementary in the CS and ICS of the circularized ere(A) cassette are underlined. (B) CLUSTAL X (1.8) multiple sequence alignment of the attC sites from the following cassettes: aadA5 (GenBank no. AF137361), aadA1b (GenBank no. M95287), orfD (GenBank no. M95287), aadA1a (GenBank no. X12870), aadA3 (GenBank no. AF047479), aadA6 (GenBank no. AF140629), qacF (GenBank no. AF034958), aacAIIa (GenBank no. M29695), catB3 (GenBank no. U13880), catB5 (GenBank no. X82455), aadA7 (GenBank no. AF224733), sat (GenBank no. X15995), aadB (GenBank no. L06418), aadA2 (GenBank no. X68227), dfrB1 (GenBank no. U36276), dfrB3 (GenBank no. X72585), dfrB2 (GenBank no. J01773), ere(A)pLQ [ere(A) of pLQ1723; GenBank no. AF099140], ere(A)pIP [ere(A) of pIP1100; GenBank no. AY183453]. Putative IntI1 binding domains as defined by Stokes et al. (25) are boxed; conserved positions are indicated by stars.
According to their similarity (94% nt identity), the ere(A) gene cassettes from pIP1100 and pLQ1723 should be considered different alleles of the same ere(A) cassette. Furthermore, their attC sites show only a single nucleotide difference (Fig. 2). Their ere(A)-encoded proteins shared 92.5% amino acid identity and are also related to a lesser extent (≈25% identity) to the ere(B) protein. The three EM esterases are of similar sizes, and the homology between them extends over the entire length of their sequences.
The high degree of similarity between the ere(A) cassettes from pIP1100 and pLQ1723 extended to the 73- and 75-nt sequences found upstream of the respective ere(A) genes. The presence of such a long 5′ untranslated region is in most cases connected to the presence of a promoter within this sequence.
In order to show the effective presence of such a promoter in the ere(A) cassette, we cloned the entire cassette either in the same orientation as or in the opposite orientation to the Plac promoter in the plasmid pSU38 (4). The ere(A) gene was generated by PCR amplification with the primers SatEnd and aadA1, digested with BamHI, and cloned in pSU38. The fragment was designed to include the complete attC site of the upstream sat cassette in order to reduce background expression due to the possible transcriptional read-through originating from plasmid promoters. Two plasmids, representing each of the two possible insert orientations, were selected: p2187, carrying ere(A) in the same orientation as the Plac promoter, and p2188, carrying ere(A) in the opposite orientation to the promoter. Both were found to confer the Emr phenotype to DH5α (Fig. 1B). This demonstrated that ere(A) expression was independent of the Plac promoter and relied on a promoter located in the cassette. The only putative promoter sequence we could identify by similarity to the E. coli σ70 −35 and −10 consensus sequences is shown in Fig. 2. Interestingly, the homology of this region with ere(A) of pLQ1723 suggests that it too carries its own promoter, as previously speculated by Peters and colleagues (15). This feature is rather unusual for integron cassettes, as only five resistance cassettes, cmlA, cmlA2/4/5, qacE, qacE2, and qacF/H (see Table 1 in reference 22), have been shown or suspected to contain their own promoters (5, 17, 24).
EreA from pIP1100 and pLQ1723 EM resistance phenotypes and characterization of the L101S mutation.
The EreA EM esterases from pIP1100 and pLQ1723 show 92.5% identity. In order to establish whether these differences affected the resistance against different macrolides they conferred, we determined and compared the MICs of several antibiotics for E. coli expressing either of the ere(A) alleles using the methods of Steers et al. (23) with ca. 105 CFU per spot. The EM MIC due to the ere(A)-encoded EM esterase in its original context (strain BM2195) had been previously found to be 2,048 μg/ml (1). In order to be able to see subtle differences between ere(A)pLQ1723 and ere(A)pIP1100 phenotypes, we cloned the two genes in the same vector and in identical expression contexts and we established the EM MICs for them in E. coli DB10, a strain that is sensitive to aminoglycosides and macrolide-lincosamide-streptogramin B molecules and especially to EM (MIC = 1 μg/ml) (9). The resistance phenotypes attributable to EreA from pIP1100 (plasmid p1818) and from pLQ1723 (plasmid p1988) for EM, clarithromycin, azithromycin, and telithromycin were determined in DB10, and the corresponding MICs are shown in Table 2. The two ere(A) alleles show identical resistance patterns, conferring resistance to both C14 macrolides (EM and clarithromycin), with the strains retaining antibacterial activity for azithromycin, a C15 macrolide, or telithromycin.
TABLE 2.
Macrolide resistance phenotype conferred by the different ere(A) alleles
| Strain | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Erythromycin | Clarithromycin | Azithromycin | Telithromycin | |
| DB10(pSU38) | 1 | 4 | 0.6 | 0.5 |
| DB10(pSU38::ereAL101S) | 1 | 4 | 0.6 | 0.5 |
| DB10(pSU38::ere(A)pLQ1723) | 25 | 20 | 0.6 | 0.5 |
| DB10(pSU38::ere(A)pIP1100) | 25 | 20 | 0.6 | 0.5 |
In these experiments, we isolated an ere(A) mutant of pLQ1723 that was sensitive to EM and clarithromycin in DB10 (Table 2). Characterization of this mutant (plasmid p2247) showed a single mutation in codon 101, a TTG-to-TCG transition, which led to the replacement of leucine 101 by a serine (GenBank AY183454). Interestingly, the amino acid position affected by the L101S mutation in this EreA mutant does not correspond to a residue conserved in the different esterases. Indeed, the corresponding amino acid is a valine in EreA of pIP1100 and a glutamate in EreB. Hence, further work will be necessary to understand the biochemical origin of this loss of activity.
Recombination properties of the ere(A) cassette.
The ere(A) cassette attC site is unique among the attC sites identified thus far in integron and superintegron cassettes (18, 25). Its closest structural relatives are the attC sites of the dfrB1, -B2, and -B3 cassettes, which share about 65% identity with it. Together with the ere(A) recombination site, these sites correspond to the shortest attC sites characterized so far (57 bp). Their primary sequences, apart from the conserved ICS and CS sequences, are fairly different from the other “short” attC sites found in 14 resistance cassettes (Fig. 2). These short attC sites are all related and resemble the XCRs and XSRs, the attC sites of the Xanthomonas superintegron (SI) cassettes (18).
The recombination proficiency of the ere(A) cassette was demonstrated from its original context between the sat and aadA1 cassettes and its ability to be integrated in a class 1 integron through IntI1 catalysis using the conduction assay developed by Martinez and de la Cruz (13). Strain ω165 was constructed by successive transformations of strain UB1637 by R388, a natural conjugative IncW plasmid harboring a class 1 integron conferring resistance to trimethoprim through its dfrB2 cassette (13), by plasmid p2187, and by p112, a plasmid expressing the intI1 integrase gene under the control of the Ptrc promoter (Table 1). The plasmid p2187 insert encompassed the complete attC site of the sat cassette, the entire ere(A) cassette, and the beginning of the aadA1 cassette (see above; Table 1). Induction of intI1 expression in ω165 was then followed by conjugation using strain UB5201 as a recipient. Recombination and integration of the ere(A) cassette alone at the attI1 site of the R388 integron was found to occur at a frequency of 11.6 × 10−2 ± 3 × 10−2.
In order to precisely determine the contribution of each of the two attC sites [the sat(-ere(A)) and ere(A)(-aadA1) attC sites] in ere(A) cassette recombination from p2187, we conducted the same experiments using plasmids p2194 (strain ω172) and p2195 (strain ω174) instead of p2187 as the substrate for IntI1. Plasmids p2194 and p2195 are p2187 derivatives which, respectively, carry either only the ere(A)(-aadA1) attC site or only the sat(-ere(A)) attC site (Table 1). The exconjugants from the different experiments were then plated on different selective media in order to establish the rates of integration through recombination of the different attC sites carried by p2194 or p2195 into the attI1 or attC sites of the R388 class 1 integron. Since integration of the entire plasmid at the attI site would be expected to generate Kanr Tps transconjugants while integration at the attC site would generate Kanr Tpr transconjugants, the integration frequency was calculated as the ratio of Kanr transconjugants to R388 UB5201 transconjugants, where the number of R388 transconjugants was the sum of the number of the Tpr UB5201 clones and Tps clones among the Kanr UB5201 transconjugants. The precise location of the recombination event was established by PCR using primer attI1.1, which hybridizes to the attI1 site of R388, and primer M13fd, which hybridizes in pSU38 downstream of the attC site recombination point. Twenty random clones per test set were mapped.
The sat attC site belongs to the family of related cassette recombination sites mentioned above, and their recombination properties have been extensively studied (see for examples (8, 11, 13). As shown in Table 3, in identical conditions, the ere(A) attC × R388 attI1 site recombination frequency was found repeatedly to be only one-ninth of the sat attC × R388 attI1 frequency. Therefore, as these sites show all the structural features common to the various attC sites of MRI and SI cassettes (19, 25), differences in structures should help identify the factors governing the variations in recombination efficiencies.
TABLE 3.
Recombination frequencies
| Strain | Relevant characteristics | Integration frequencya |
|---|---|---|
| ω171 | ere(A)(-aadA1) attC; IntI1 not overexpressed | <7 × 10−9 |
| ω172 | ere(A)(-aadA1) attC; IntI1 overexpressed | (2.4 ± 0.5) × 10−2 [56/3] |
| ω173 | sat(-ere(A)) attC; IntI1 not overexpressed | <2 × 10−9 |
| ω174 | sat(-ere(A)) attC; IntI1 overexpressed | (22.6 ± 6.1) × 10−2 [57/3] |
Values are averages of three independent trials; values in brackets are attI1/dfrB2 attC integrations.
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in GenBank under accession no. AY183453.
Acknowledgments
We thank G. Gerbaud and P. Courvalin for kindly providing E. coli BM2195 and P. H. Roy for P. stuartii 1723. We thank Dean Rowe-Magnus for helpful discussions and for critical reading of the manuscript. L.B. is a CROUS (programme de cooperation Algéro-Française) doctoral fellow.
This work was supported by the Institut Pasteur, the CNRS, and the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires from the MENRT and the DGA (contract 0134020).
REFERENCES
- 1.Andremont, A., G. Gerbaud, and P. Courvalin. 1986. Plasmid-mediated high-level resistance to erythromycin in Escherichia coli. Antimicrob. Agents Chemother. 29:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andremont, A., and C. Tancrede. 1981. Reduction of the aerobic Gram negative bacterial flora of the gastro-intestinal tract and prevention of traveller's diarrhea using oral erythromycin. Ann. Microbiol. (Paris) 132B:419-427. [PubMed] [Google Scholar]
- 3.Arthur, M., D. Autissier, and P. Courvalin. 1986. Analysis of the nucleotide sequence of the ereB gene encoding the erythromycin esterase type II. Nucleic Acids Res. 14:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C. Y., L. L. Chang, Y. H. Chang, T. M. Lee, and S. F. Chang. 2000. Characterisation of drug resistance gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J. Med. Microbiol. 49:1097-1102. [DOI] [PubMed] [Google Scholar]
- 7.Collis, C. M., M. J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase Intl1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 8.Collis, C. M., G. D. Recchia, M. J. Kim, H. W. Stokes, and R. M. Hall. 2001. Efficiency of recombination reactions catalyzed by class 1 integron integrase IntI1. J. Bacteriol. 183:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta, N., R. W. Hedges, D. Becker, and J. Davies. 1974. Plasmid-determined fusidic acid resistance in the Enterobacteriaceae. J. Gen. Microbiol. 83:191-196. [DOI] [PubMed] [Google Scholar]
- 10.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 11.Hansson, K., L. Sundstrom, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 13.Martinez, E., and F. de la Cruz. 1990. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 9:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ounissi, H., and P. Courvalin. 1985. Nucleotide sequence of the gene ereA encoding the erythromycin esterase in Escherichia coli. Gene 35:271-278. [DOI] [PubMed] [Google Scholar]
- 15.Peters, E. D., M. A. Leverstein-van Hall, A. T. Box, J. Verhoef, and A. C. Fluit. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plante, I., D. Centron, and P. H. Roy. 2003. An integron cassette encoding erythromycin esterase, ere(A), from Providencia stuartii. J. Antimicrob. Chemother. 51:787-790. [DOI] [PubMed] [Google Scholar]
- 17.Ploy, M. C., P. Courvalin, and T. Lambert. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob. Agents Chemother. 42:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe-Magnus, D. A., A.-M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multi-resistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe-Magnus, D. A., A. M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowe-Magnus, D. A., A. M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 21.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 22.Rowe-Magnus, D. A., and D. Mazel. 2002. The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 292:115-125. [DOI] [PubMed] [Google Scholar]
- 23.Steers, E., E. L. Foltz, B. S. Graves, and J. Riden. 1959. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot. Chemother. 9:307-311. [PubMed] [Google Scholar]
- 24.Stokes, H. W., and R. M. Hall. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26:10-19. [DOI] [PubMed] [Google Scholar]
- 25.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 26.Sundstrom, L. 1998. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS Suppl. 84:37-42. [DOI] [PubMed] [Google Scholar]
- 27.Swift, G., B. J. McCarthy, and F. Heffron. 1981. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol. Gen. Genet. 181:441-447. [DOI] [PubMed] [Google Scholar]
- 28.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tietze, E., J. Brevet, H. Tschape, and W. Voigt. 1988. Cloning and preliminary characterization of the streptothricin resistance determinants of the transposons Tn1825 and Tn1826. J. Basic Microbiol. 28:129-136. [DOI] [PubMed] [Google Scholar]
- 30.Young, H. K., M. J. Qumsieh, and M. L. McIntosh. 1994. Nucleotide sequence and genetic analysis of the type Ib trimethoprim-resistant, Tn4132-encoded dihydrofolate reductase. J. Antimicrob. Chemother. 34:715-725. [DOI] [PubMed] [Google Scholar]