Abstract
Noroviruses (NVs) cause many cases of oyster- or clam-associated gastroenteritis in various countries. We collected 191 samples from Japanese oysters intended for raw consumption that had been harvested from the sea in two different areas between December 2001 and February 2002. To detect, quantitate, and phylogenetically analyze the NV genome in purified concentrates from the stomachs and digestive diverticula of these oysters, we amplified the NV capsid gene by reverse transcription-PCR. Phylogenetic analysis was performed by using the neighbor-joining method. We detected the NV genome in 17 of 191 oysters (9%). Phylogenetic analysis indicated genogroup I (Norwalk virus type) in 3 of the 17 oysters and genogroup II (Snow Mountain virus type) in the other 14. Both genogroups showed wide genetic diversity. To quantify the NV capsid gene in these oysters, we performed real-time PCR using genogroup-specific probes. More than 102 copies of the NV genome were detected in 11 of 17 oysters. The results suggested that about 10% of Japanese oysters intended for raw consumption harbored NVs, and more than 50% of those oysters in which NVs were detected had a large amount.
Noroviruses (NVs), which belong to the family Caliciviridae, cause acute gastroenteritis (25). According to the Japanese 2001 Food Poisoning Surveillance Report (http://www.mhlw.go.jp/topics/syokuchu), NVs accounted for 28% (7,358 of 25,862) of cases of food poisoning overall and 99% (7,358 of 7,371) of purely viral cases. In addition, NVs reportedly cause gastroenteritis in large numbers of patients in many countries, suggesting that NVs are distributed worldwide (21).
NVs, enteroviruses, astroviruses, and hepatitis virus type A are likely to be transmitted by shellfish such as oysters and clams (19). The Japanese, other Asians, and the French eat large amounts of raw fish or shellfish. Raw consumption causes many cases of food poisoning or infectious gastroenteritis as well as hepatitis (10). Previous epidemiologic studies have linked many cases of NV gastroenteritis to the oyster harvesting season (20). These viruses, known to persist in the environment, are found to be concentrated in shellfish (9).
No conventional cell culture method has been developed for the propagation of NVs (2). Detection of NVs has relied mainly on reverse transcription-PCRs (RT-PCR), enzyme-linked immunosorbent assays (ELISA), and electron microscopy (2). NVs can be divided into two distinct genogroups, I (Norwalk virus type) and II (Snow Mountain virus type) (25). Previous studies that used stool specimens from patients with nonbacterial gastroenteritis have demonstrated the presence of broad genetic diversity in each genogroup of NVs (5, 7, 23). However, the epidemiologic and phylogenetic characteristics of NVs in oysters remain obscure. In addition, to our knowledge, few investigations have quantified NVs in patients' stool samples or in shellfish. This information is important for the prevention of food poisoning caused by NVs and for determining the infective dose and index of contamination for NVs. We therefore carried out detection, quantitation, and phylogenetic analysis of NV genomes in Japanese oysters.
MATERIALS AND METHODS
Samples and preparation of viral suspension.
A total of 191 Japanese oysters (Crassosterea gigas or Crassosterea nippona) were collected from fish distributors who were handling the harvest from two areas of the Setouchi sea (areas A and B) that were about 500 km apart. Both areas are located in the offshore region west of the Honshu island. The sample numbers and harvesting months for these areas are shown in Table 1. All oysters had been approved for raw consumption by meeting the provisions of the Regulation of Food Sanitation Law (i.e., <50,000 bacteria per g of oyster by standard plate count with <230 representing the coliform group). The fresh oysters were shucked, and the stomachs and digestive diverticula of the oysters were removed by dissection on the day of harvest and then weighed and homogenized in nine times their weight of phosphate-buffered saline without magnesium and calcium (8). In brief, the stomachs and digestive diverticula were placed in a solution of phosphate-buffered saline with the addition of 0.1 ml of antifoam B (Sigma, St. Louis, Mo.) The samples were homogenized for two 30-s intervals at a maximum speed of 18,000 rpm with an Omni-mixer (OCI Instruments, Waterbury, Conn.). Six milliliters of chloroform-butanol (1:1, vol/vol) was added to the homogenate. Then, the mixture was homogenized for an additional 30 s, and 170 ml of Cat-Floc T (Calgon, Elwood, Pa.) was added to the homogenate (8). In addition, to test the adequacy of RNA extraction, we added poliovirus type II (Sabin strain, corresponding to 102 copies of viral genes/assay) to the homogenate samples. After homogenate samples were centrifuged at 3,000 × g for 30 min at 4°C, all of the supernatant was layered onto 3 ml of 30% sucrose solution and ultracentrifuged at 154,000 × g for 3 h at 4°C. Then, the pellet was resuspended in 300 μl of doubly distilled water containing 20 U of RNase inhibitor (Promega, Madison, Wis.) and stored at −80°C until use.
TABLE 1.
Oyster samples collected for this study
| Collection mo and yr | No. of samples from:
|
|
|---|---|---|
| Area A | Area B | |
| December 2001 | 20 | 16 |
| January 2002 | 40 | 20 |
| February 2002 | 80 | 15 |
| Total | 140 | 51 |
RNA extraction, RT-PCR, sequencing, and real-time PCR.
Viral RNA was extracted from 140 μl of viral suspension by using an available kit (QIAamp viral RNA mini kit; Qiagen GmbH, Hilden, Germany) and was finally suspended in 60 μl of doubly distilled water. Then, the RNA solution was treated with 5 U of DNase I (Takara, Tokyo, Japan). To amplify the partial capsid region of NVs by RT-PCR, we used genogroup-specific primers following reverse transcription as previously described (15). Primers for the first PCR were as follows: 5′-CGY TGG ATG CGN TTY CAT GA-3′ (COG1F; sense), 5′-CCA ACC CAR CCA TTR TAC A-3′ (G1-SKR; antisense), 5′-CAR GAR BCN ATG TTY AGR TGG ATG AG-3′ (COG2F; sense), and 5′-CCR CCN GCA TRH CCR TTR TAC AT-3′ (G2-SKR; antisense). Primers for the nested PCR were 5′-CTG CCC GAA TTY GTA AAT GA-3′ (G1-SKF; sense), 5′-CCA ACC CAR CCA TTR TAC A-3′ (G1-SKR; antisense), 5′-CNT GGG AGG GCG ATC GCA A-3′ (G2-SKF; sense), and 5′-CCR CCN GCA TRH CCR TTR TAC AT-3′ (G2-SKR; antisense) (12). We amplified the poliovirus type II VP1 gene by using the specific primers of 5′-AGC AAG CAC CGT ATT GAG CC-3′ (sense) and 5′-GTT TCA TGT CTG CTC CGT CTG-3′ (antisense) (unpublished data). The PCR protocol included incubation for 3 min at 94°C; then, 40 cycles of 94°C for 60 s, 50°C for 60 s, and 72°C for 2 min followed, with an additional 15 min for elongation at 72°C after the last cycle. This PCR procedure was repeated by using inner primers as a nested PCR (15). We used two types of positive controls and a viral gene-free negative control per five assays for amplification of NVs by PCR. Nested PCR is considered to be highly prone to false-positive results (2). Therefore, to check for conflicting PCR results, negative controls were included in each round of PCR. In addition, a first PCR round and a nested PCR round were performed independently. The size of the amplified DNA fragment was confirmed by electrophoresis on a 1.5% agarose gel. After purification of DNA fragments with a QIAquick PCR purification kit (Qiagen), the nucleotide sequence was determined with an automated DNA sequencer (ABI 310 DNA sequencer; Applied Biosystems, Foster City, Calif.) by using a dye terminator cycle sequencing ready reaction kit (Applied Biosystems) (15). We also quantified NV capsid genes by using real-time PCR as previously described (12). Fifteen microliters of DNase I-treated RNA solution was added to 15 μl of RT solution containing 100 mM Tris-HCl at pH 8.3, 150 mM KCl, 6 mM MgCl2, 1 mM deoxynucleoside triphosphate mixture, 10 mM dithiothreitol, 75 pmol of random hexamer (Takara), 30 U of RNase inhibitor (Takara), and 200 U of reverse transcriptase [Superscript II, RNaseH(−); Invitrogen, San Diego, Calif.]. The RT mixture was incubated at 42°C for 60 min (RT reaction) and then at 70°C for 15 min (for the denaturation of reverse transcriptase). The real-time PCR mixture contained 5 μl of cDNA (RT product), 17.5 μl of TaqMan universal PCR master mix (Applied Biosystems), a 400 nM concentration of each primer, and fluorogenic probes [probes for genogroup I, 10.5 pmol of RING1(a)-TP and 3.5 pmol of RING1(b)-TP; probe for genogroup II, 3.5 pmol of RING2-TP] (12). The fluorogenic probes for real-time PCR were as follows: 5′-AGA TYG CGA TCY CCT GTC CA-3′ [RING1(a)-TP], 5′-AGA TCG CGG TCT CCT GTC CA-3′ [RING1(b)-TP], and 5′-TGG GAG GGC GAT CGC AAT CT-3′ (RING2-TP) (12). The following PCR protocol was used: 2 min at 50°C and 10 min at 95°C, 50 cycles of 95°C for 15 s, and 56°C for 60 s (12). Data were corrected by using an internal standard as previously described (12). In addition, we tested recovery of the NV genome by using highly purified NV particles obtained by a sucrose gradient method from patients with gastroenteritis.
Phylogenetic analysis.
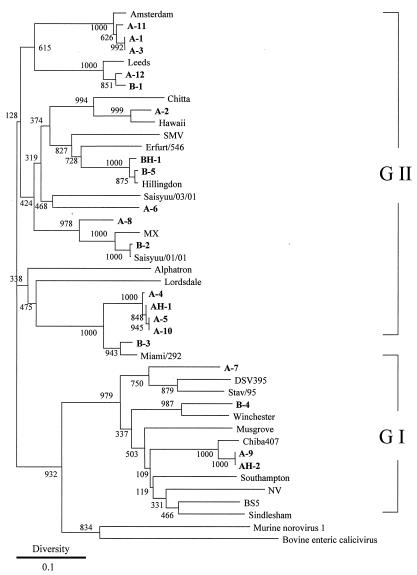
Capsid sequences of reference strains of NVs were obtained from GenBank (13). These strains and accession numbers are shown in the legend for Fig. 2. Phylogenetic analysis was performed as previously described (13). Briefly, all NV capsid region sequences (between 238 and 253 bases) were aligned by using Genetix-Win version 5.0 software (Software Development, Tokyo, Japan). A phylogenetic tree was constructed by using the neighbor-joining technique, specifically, Kimura's two-parameter method. Reliability of the tree was estimated by using 1,000 bootstrap replications.
FIG. 2.
Phylogenetic tree construct based on partial sequences of the capsid gene of NVs. The distance was calculated by Kimura's two-parameter method, and the tree was plotted by using the neighbor-joining method. Numbers at each branch indicate bootstrap values for the clusters supported by that branch. Accession numbers of our strains are indicated. A and B refer to the areas of harvest, and positive samples from this study are designated in boldface type. AH and BH refer to human NV strains detected in patients with gastroenteritis living near sites A and B. GI, genogroup I; GII, genogroup II. Strains and GenBank accession numbers were as follows: DSV395/90/Saudi Arabia, U04469; NLV/Stav/95/Nor, AF145709; Hu/NLV/Winchester/94/UK, AJ277609; Hu/NLV/Sindlesham/95/UK, AJ277615; BS5/Germany, AF093797; NV/8FiiA/68/US, M87661; Hu/NLV/Musgrove/89/UK, AJ277614; Hu/NLV/Chiba407/1987/JP, AB042808; Southampton/91/UK, L07418; Hu/NLV/Leeds/90/UK, AJ277608; Hu/NLV/Amsterdam/98-18/1998/NET, AF195848; NLV/Miami/292/1994/US, AF414410; HV/NLV/Hillingdon/90/UK, AJ277607; NLV/Erfurt/546/00/DE, AF427118; SMV/76/US (Snow Mountain virus), U70059; Hu/NLV/GII/Hawaii virus/1971/US, U07611; Aichi 124-89, AB031013; NV/Saisyuu/03/01, AB091399; NV/Saisyuu/01/01, AB091398; MX/89/Mexico, U22498; Hu/NLV/Alphatron/98-2/1998/NET, AF195847; Lordsdale/93/UK, X86557; AH-1, AB111894; BH-1, AB111896; BH-2, AB111895; murine norovirus 1, AY228235; bovine enteric calicivirus, AJ11099.
RESULTS
Detection and phylogenetic analysis of NVs in Japanese oysters.
The NV capsid gene was amplified and detected in 17 of 191 oysters (9%). Amplicons from oysters were electrophoresed for comparison with corresponding NV protostrains (Fig. 1). All amplicons of NVs from oysters were sequenced successfully. As shown in Table 2, NV genomes were detected in 13 of 60 oysters in January 2002 (9 of 40 from area A and 4 of 20 from area B). NV genomes were detected in 4 of 95 oysters in February 2002 (3 of 80 from area A and 1 of 15 from area B). No NV genome was detected in oysters harvested in December 2001. A phylogenetic tree constructed by the neighbor-joining method is shown in Fig. 2. Of 17 amplicons, 3 were classified into genogroup I (Norwalk virus type), while 14 were classified into genogroup II (Snow Mountain virus type). These strains could be subdivided into five clusters of genogroup II and two clusters of genogroup I. Both genogroups were detected in areas A and B. Several amplicons harvested in area A (A-4, A-5, and A-10; A-1, A-3, and A-11) were closely related genetically (i.e., exhibiting less than 1% genetic diversity). Interestingly, the sequences of amplicons B-2 Saisyuu/01/01 and (derived from a Korean oyster) were closely related genetically, although we only partially sequenced the capsid gene of these strains. The two areas of harvest were about 500 km apart, but no regionally distinctive distribution of an NV genogroup was evident in our phylogenetic tree. Nucleotide diversity among these amplicons exceeded 10% (see Fig. 2), representing wide genetic diversity. Some human NVs found in stools from gastroenteritis patients who lived near oyster sampling sites (within about 30 km) during the investigation period between December 2001 and February 2002 were genetically related to our strains from oysters. Approximately 30 to 40% diversity was noted between some animal calicivirus strains and some of our strains from oysters.
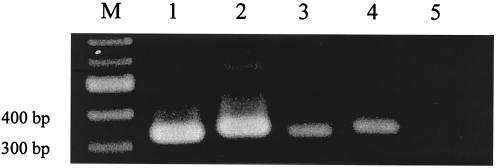
FIG. 1.
Detection of NV capsid gene from oysters. Nested PCR products were electrophoresed on a 1.5% agarose gel. M, marker; lane 1, positive control for genogroup I (DSV395/90/Saudi Arabia, accession no. U04469); lane 2, positive control for genogroup II (Arg320, accession no. AF190817); lane 3, genogroup I amplicon from an oyster (A-9 strain in Fig. 2); lane 4, genogroup II amplicon from an oyster (B-3 strain in Fig. 2); lane 5, negative control (free of viral RNA). RT-PCR, nested PCR, and electrophoresis procedures are described in detail in Materials and Methods.
TABLE 2.
Genogroups of and NV genome copy numbers in Japanese oysters
| Collection mo and yr | Area | Strain | Genogroup | NV genome copy no.a | GenBank accession no. |
|---|---|---|---|---|---|
| January 2002 | A | A-1 | II | 2.7 × 103 | AB097905 |
| A | A-2 | II | 2.9 × 103 | AB097906 | |
| A | A-3 | II | NDb | AB097907 | |
| A | A-4 | II | ND | AB097908 | |
| A | A-5 | II | 7.5 × 103 | AB097909 | |
| A | A-6 | II | 1.6 × 102 | AB097910 | |
| A | A-7 | I | 2.9 × 104 | AB097911 | |
| A | A-8 | II | ND | AB097912 | |
| A | A-9 | I | 2.2 × 102 | AB097913 | |
| B | B-1 | II | ND | AB097917 | |
| B | B-2 | II | 6.2 × 102 | AB097918 | |
| B | B-3 | II | 1.5 × 102 | AB097919 | |
| B | B-4 | I | 2.4 × 102 | AB097920 | |
| February 2002 | A | A-10 | II | ND | AB097914 |
| A | A-11 | II | 1.4 × 102 | AB097915 | |
| A | A-12 | II | ND | AB097916 | |
| B | B-5 | II | 4.8 × 102 | AB097921 |
Units are copy numbers of NV genome per oyster.
ND, not detected (<102 copies/oyster).
Quantitation of NV capsid gene by real-time PCR.
As shown in Table 2, more than 102 copies of the NV genome were found in 11 of 191 oysters (6%) (3 from genogroup I and 8 from genogroup II). More than 103 copies of the NV genome were detected in four oysters harvested in area A. In January 2002, about 15% of oysters harvested in both areas (6 of 40 oysters from area A and 3 of 20 oysters from area B) had large amounts of the NV genome. In February 2002, 3 of 80 oysters harvested in area A (4%) and 1 of 15 oysters harvested in area B (7%) had relatively large amounts of the NV genome. Thus, more than 5% of oysters harvested in the two areas during January and February of 2002 carried large numbers of NVs. In a recovery test, about 85 to 90% of NV genomes were recovered (data not shown). In this study, a greater amount of NV genome was detected in oysters harvested from site A than in oysters from site B. However, no plausible sources of contamination with NVs, including wastewater treatment plants and combined sewer overflows, were present in the vicinity of either site. During the investigation period, no significant difference in seawater quality (defined as <70 coliform groups/100 ml of seawater by the Japanese Enforcement Regulation of the Food Sanitation Law for farming of oysters for raw consumption) was observed between sites (data not shown).
DISCUSSION
We demonstrated that about 10% of oysters harvested from two areas in January and February 2002 for raw consumption had NV genomes, while about 5% had large numbers of NVs (more than 102 copies). More than 10% nucleotide diversity was observed in our strains. No region-specific preponderance of any given genogroup was evident in our phylogenetic tree. The results suggested that Japanese oysters harvested in winter in both areas were highly contaminated with a genetically diverse population of NVs.
NV genomes have been detected in shellfish such as oysters and clams in the United States (9, 14, 22), United Kingdom (17), Canada (16), and Japan (23), and these viruses are associated with gastroenteritis in humans. NVs can be divided into two genogroups, each of which can be further divided into four to six clusters (4). Fankhauser et al. demonstrated that the polymerase gene of NVs detected during outbreaks of gastroenteritis in the United States can be divided into two clusters within genogroup I and four clusters within genogroup II (4). Capsid genes of NVs from stool specimens from patients during outbreaks of gastroenteritis in Japan can be divided into two clusters in genogroup I and four clusters in genogroup II (13). Our phylogenetic analysis revealed that NVs in Japanese oysters can be divided into two clusters in genogroup I and five clusters in genogroup II. This suggests wide genetic diversity, as has been noted in NVs from stool specimens (4, 5, 13). Transmission of NVs can occur via contaminated shellfish, especially oysters, and by person-to-person contact (10). Many reports have implicated NVs and enteroviruses in shellfish-associated outbreaks and demonstrated the presence of NV gene sequences in shellfish and in specimens from patients. Various types of NVs have been shown to cause oyster-associated gastroenteritis in Japan (23).
Although no conventional culture method for NVs is available at present (2), other NV detection methods, such as electron microscopy (EM), ELISA, and RT-PCR, can be used (2). EM and ELISA are practical for the detection of NVs in diarrheal stool specimens that contain a large amount of virus (105 to 106 particles/ml) (3). Determination of copy numbers of NV genome in foods is important, although these numbers do not exactly correspond to infective doses of NVs. However, our results and those of previous reports suggested that various foods, including shellfish, contain much smaller numbers of NVs than the number of NVs found in stool specimens (11). This hinders the detection of NVs in foods by EM and ELISA, since their sensitivity is limited (6). In contrast, RT-PCR is not quantitative but is highly sensitive, being able to detect a few copies of the NV genome in a sample (1). Recently, a quantitative real-time PCR method has been developed (12). We applied this method to determine the copy numbers of NV genomes in Japanese oysters. Some of our oyster-derived strains were found to be genetically related to strains infecting patients with gastroenteritis during the period of our investigation (Fig. 2). The results suggested that genetically similar types of NVs from oysters might be associated with NV-associated outbreaks during our investigation period. Some genetic similarities exist between animal calicivirus and human calicivirus (18, 24), and there are many types of animal caliciviruses (2). Interestingly, some of our strains had unique sequences that were genetically related to some animal calicivirus strains (30 to 40% diversity; Fig. 2). Thus, we could not rule out the inclusion of animal calicivirus strains in our study. In addition, the NV genome was detected in abundance in oysters harvested in January and February of 2002 but not those harvested in December of 2001. Japanese epidemiologic data for the interval between October and December of 2001 (http://www.mhlw.go.jp/topics/syokuchu) suggest that the incidence of oyster- or clam-associated gastroenteritis outbreaks was low at that time, although these data are not directly linked to our own. The population of NV-contaminated Japanese oysters from both areas in December of 2001 may indeed have been low. Yearlong month-to-month studies regarding the prevalence of NVs in oysters may be needed.
Fecally excreted NVs, enteroviruses, astroviruses, and hepatitis virus type A from patients with gastroenteritis or hepatitis enter sewage and seawater, where they may remain infective for long periods of time (9). In many countries, oyster beds are located near shore, in an estuary or a bay. It is possible that various shellfish (especially oysters and clams) contain high concentrations of viral pathogens, such as NV, enterovirus, and hepatitis A virus (19). In the course of this study, relatively large amounts of NV genes in oysters from site A were detected. However, no significant difference in bacteriologic quality of seawater was noted between the two sites, and no likely sources of NV contamination were nearby. Why relatively large numbers of the NV genome were detected in oysters harvested from site A is not known. To prevent food poisoning by microorganisms in raw oysters, the Japanese government regulates the oyster industry under the Enforcement Regulation of Food Sanitation Law. However, this regulation focuses mainly on bacteriology. We detected NVs in 10% of government-approved oysters. Additional regulation based on viral sanitation protocols may be needed.
Further molecular epidemiologic studies of causative viral agents (NVs, enteroviruses, astroviruses, and hepatitis virus type A) carried out by using phylogenetic analysis and quantitative real-time PCR methods will contribute to an understanding of the epidemiology of these viruses and provide a more accurate assessment of the risk factors for shellfish-associated illnesses.
Acknowledgments
We thank Mitsuaki Oseto (Ehime Prefectural Institute of Public Health and Environmental Science), Masaaki Sugieda (Shizuoka Prefectural Institute of Public Health and Environmental Science), and Yukio Morita (Gunma Prefectural Institute of Public Health and Environmental Sciences) for constructive discussions. We also thank Miyuki Ikeda and Kyoko Ikejima (Gunma Prefectural Institute of Public Health and Environmental Sciences) for their skillful assistance.
This work was supported by Research on Food and Chemical Safety for Health, Labour, and Welfare Programs from the Ministry of Health, Labour, and Welfare, Japan.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentesch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atomer, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danoe, F. W. 1994. Electron microscopy for the detection of gastroenteritis viruses, p. 101-130. In A. Z. Kapikian (ed.), Viral infections of gastrointestinal tract. Marcel Dekker, Inc., New York, N.Y.
- 4.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 5.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 6.Glass, R. I., J. Noel, T. Ando, R. L. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 7.Green, S. M., K. E. Dingle, P. R. Lambden, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1994. Human enteric Caliciviridae: a new prevalent small round-structured virus group defined by RNA-dependent RNA polymerase and capsid diversity. J. Gen. Virol. 75:1883-1888. [DOI] [PubMed] [Google Scholar]
- 8.Guyader, F. L., F. H. Neill, M. K. Estes, S. S. Monroe, T. Ando, and R. L. Atmer. 1996. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl. Environ. Microbiol. 62:4268-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyader, F. L., L. Haugarreau, L. Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye, S., K. Yamashita, S. Yamadera, M. Yoshikawa, N. Kato, and N. Okabe. 2000. Surveillance of viral gastroenteritis in Japan: pediatric cases and outbreak incidents. J. Infect. Dis. 181:S270-S274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaykus, L. A., R. de Leon, and M. D. Sobsey. 1996. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 62:2074-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, B. F. Hoshino, N. Takeda, and K. Katayama. 2003. A broadly reactive and highly sensitive assay for Norwalk-like viruses on real-time quantitative RT-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, K., H. S. Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225-239. [DOI] [PubMed] [Google Scholar]
- 14.Kingsley, D. H., G. K. Meade, and G. P. Richards. 2002. Detection of both hepatitis A virus and Norwalk-like virus in imported clams associated with food-borne illness. Appl. Environ. Microbiol. 68:3914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 16.Leers, W. D., G. Kasupski, R. Fralick, S. Wartman, J. Garcia, and W. Gary. 1987. Norwalk-like gastroenteritis epidemic in a Toronto hospital. Am. J. Public Health. 77:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lees, D. N., K. Henshilwood, J. Green, C. I. Gallimore, and D. W. G. Brown. 1995. Detection of small round-structured viruses in shellfish by reverse transcription-PCR. Appl. Environ. Microbiol. 61:4418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to Norwalk-like virus. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalf, T. G., J. L. Melnick, and M. K. Estes. 1995. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 20.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181:S284-S287. [DOI] [PubMed] [Google Scholar]
- 21.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 22.Shieh, Y., S. S. Monroe, R. L. Fankhauser, G. W. Langlois, W. Burkhardt III, and R. S. Baric. 2000. Detection of Norwalk-like virus in shellfish implicated in illness. J. Infect. Dis. 181:S360-S366. [DOI] [PubMed] [Google Scholar]
- 23.Sugieda, M., K. Nakajima, and S. Nakajima. 1996. Outbreaks of Norwalk-like virus-associated gastroenteritis traced to shellfish: coexistence of two genogroups in one specimen. Epidemiol. Infect. 116:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugieda, M., and S. Nakajima. 2002. Viruses detected in the caecum contents of healthy pigs representing a new genetic cluster in genogroup II of the genus “Norwalk-like virus.” Virus Res. 87:165-172. [DOI] [PubMed] [Google Scholar]
- 25.Wang, J., X. I. Jiang, H. P. Madore, J. Gray, U. Desselberger, T. Ando, Y. Seto, I. Oishi, J. F. Lew, K. Y. Green, and M. K. Estes. 1994. Sequence diversity of small round-structured viruses in the Norwalk virus group. J. Virol. 68:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]