Abstract
Lactic acid bacteria exhibiting activity against the gram-positive bacterium Bacillus subtilis were isolated from rice bran. One of the isolates, identified as Enterococcus faecalis RJ-11, exhibited a wide spectrum of growth inhibition with various gram-positive bacteria. A bacteriocin purified from culture fluid, designated enterocin RJ-11, was heat stable and was not sensitive to acid and alkaline conditions, but it was sensitive to several proteolytic enzymes. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis revealed that enterocin RJ-11 had a molecular weight of 5,000 in its monomeric form. The amino acid sequence determined for purified enterocin RJ-11 exhibited high levels of similarity to the sequences of enterocins produced by Enterococcus faecium.
Lactic acid bacteria (LAB) are widely used as starter cultures for dairy, meat, and vegetable fermentations (24, 38). Some LAB strains are known to produce various types of bacteriocins, which have bactericidal effects against gram-positive bacteria, including food-borne pathogens (17, 22, 23, 32). This beneficial trait has led to utilization of bacteriocins as food additives (9, 30). For example, nisin, a bacteriocin produced by Lactococcus lactis, has been used for food preservation in a large number of countries (7, 15).
Fermented soybean paste (miso) is a traditional food seasoning in Japan. Miso has been assumed to contribute to the health and lifespan of Japanese due to its favorable physiological effects, such as its antioxidative activity (3), its antimutagenic effect (18, 21, 29, 39), and its inhibitory effect on accumulation of cholesterol (1, 2). In the first step of the miso production process, called koji mold fermentation, a koji mold (Aspergillus oryzae) is grown on steam-cooked rice. During this process care must be taken to avoid contamination by spore-forming bacteria, such as Bacillus subtilis, since the conditions are favorable for growth of bacteria.
We previously tried to use LAB strains with antibacterial activity in the koji mold fermentation process to suppress the growth of contaminating bacteria. However, a preliminary test revealed that most LAB strains that are used industrially do not grow well on cooked rice. This poor growth may be ascribed to a deficiency of nutrients in rice, which are required by the LAB strains tested. In addition, the fairly low water activity of cooked rice seems to be unfavorable for the growth of LAB strains. We also attempted to isolate LAB strains which grow well on cooked rice and have an inhibitory effect on the growth of vegetative cells of B. subtilis, which is known to often contaminate preparations because of its spore-forming capacity. A large number of lots of rice bran were collected and used for screening LAB strains, since isolated strains are expected to grow well on steam-cooked rice. We found that rice bran is a good source for isolation of LAB strains that produce bacteriocins. In this paper, we describe purification and characterization of a bacteriocin produced by one of the isolates, Enterococcus faecalis strain RJ-11.
MATERIALS AND METHODS
Isolation of LAB strains with antibacterial activity.
We screened LAB strains from a total of 242 lots of rice bran, which were collected from various districts of Japan in 1999 and 2000. Ten grams of rice bran was suspended in 60 ml of 4 or 6.5% NaCl and incubated at 30°C for 11 days in a sealed plastic bag (Doi-Pack; Fujimori Kogyo Co., Tokyo, Japan) as a procedure for enrichment for LAB. The resulting suspension was serially diluted with 0.85% NaCl and spread onto plates (diameter, 90 mm) containing TGC agar (thioglycollate medium without dextrose; Difco). The plates were incubated at 30°C for 2 days to obtain an appropriate number of single bacterial colonies. Then 7 ml of soft nutrient agar (0.7% agar, 0.1% glucose, 0.25% yeast extract, 1% Polypeptone; pH 7.0) was added to 30 μl of an overnight culture of B. subtilis JCM1465T and was poured over the colonies grown on the TGC plates. After incubation at 30°C overnight, bacterial colonies that formed a growth inhibition zone around them were picked with a straight wire and purified twice on TGC agar plates by transferring single colonies. The resulting bacterial isolates (38 strains) were grown on GYP plates (1% glucose, 1% yeast extract, 0.5% Polypeptone, 0.2% beef extract, 0.025% sodium acetate, 0.02% MgSO4 · 7H2O, 0.001% FeSO4 · 7H2O, 0.001% MnSO4 · 4H2O, 0.001% NaCl; pH 6.8) with 0.5% CaCO3 powder to confirm the formation of clear zones caused by the production of lactic acid. The isolates that did not form clear zones were discarded at this step since they were not likely to be LAB strains. To investigate the antibacterial activity spectra of the LAB strains isolated, each strain was spotted and grown on TGC plates (diameter, 90 mm). Soft nutrient agar (7 ml) was added to 30 μl of a culture of one of the test bacteria (Table 1) grown to the early stationary phase in nutrient medium and then poured over the colonies of the LAB isolates grown on TGC plates. After incubation at 30°C, the diameters of the clear zones around the colonies of LAB isolates were measured. A cross-immunity test for the bacteriocin-producing strains isolated was performed in a similar way by using one of the other isolates as an indicator lawn.
TABLE 1.
Classification of LAB isolates from rice bran on the basis of antibacterial spectra
| Indicator strain | Antibacterial activitya
|
|||||
|---|---|---|---|---|---|---|
| Group I strains (n = 18) | Group II strains (n = 2) | Group III strains (n = 6) | Group IV strains (n = 1) | Group V strains (n = 2) | Group VI strain (n = 1) | |
| Listeria monocytogenes SUB635 | +++ | +++ | +++ | +++ | ++ | ++ |
| Bacillus subtilis JCM1465T | +++ | ++ | ++ | + | + | + |
| Bacillus amyloliquefaciens B1 | ++ | ++ | ++ | − | + | + |
| Staphylococcus aureus SUB511 | ++ | ++ | ++ | ++ | + | + |
| Enterococcus sp. strain YJ-35 | +++ | +++ | ++ | +++ | ++ | + |
| Enterococcus durans KB-60 | ++ | +++ | ++ | +++ | ++ | ++ |
| Lactococcus lactis IFO12007 | + | ++ | ++ | +++ | + | + |
| Lactococcus lactis IO-1 | − | +++ | +++ | ++ | + | ++ |
| Escherichia coli JM109 | − | − | − | − | − | − |
Antibacterial activity was evaluated by measuring the size of a clear zone formed around colonies of an LAB isolate after a soft agar culture of indicator cells was overlaid and incubated overnight. Sizes of clear zones: +++, >22 mm; ++, 10 to 21 mm; +, <9 mm; −, no clear zone. n is the number of isolates belonging to the group. The representative group II, III, IV, V, and VI strains used for further study were RJ-11, RB-3, RJ-10, SJ-16, and YJ-35, respectively.
Assay of antibacterial activity.
The antibacterial activity was assayed by using a spot-on-lawn assay described by Mayr-Harting et al., with a slight modification (28). Cells of LAB strains were grown in MRS broth (Oxoid) supplemented with 0.5% CaCO3 at 30°C with shaking (120 strokes per min [spm]) to the early stationary phase; the initial pH of the medium was adjusted to pH 6.0. Culture fluid was obtained by removing the cells by centrifugation and was filtered with a cellulose acetate membrane filter (pore size, 0.2 μm; Advantec, Tokyo, Japan). The resulting sample was serially diluted twofold with MRS broth (pH 5.0). Soft nutrient agar (20 ml) was solidified in a sterile dish (140 by 100 mm) after addition of 60 μl of an overnight culture of Listeria monocytogenes SUB635. This strain was used as an indicator in the assay, since a clearer inhibitory zone was obtained with this organism than with B. subtilis. After 30 min of drying, 10 μl each diluted sample was spotted onto the plate. The resulting plates were incubated at 30°C overnight, and the titer was defined as the reciprocal of the highest dilution (2n) that resulted in inhibition of the indicator lawn. Thus, the arbitrary unit (AU) of antibacterial activity per milliliter was defined as 2n × 1,000 μl/10 μl. To examine the spectrum of activity of the purified bacteriocin against various bacterial strains, the test described above was performed with soft agar containing cells of test strains grown in a suitable medium to the early stationary phase. The abilities to inhibit a certain bacterial strain were compared by using the highest dilution that resulted in a growth inhibition zone.
Tricine-SDS-PAGE analysis of antibacterial compounds.
Cells of isolated LAB strains were grown in MRS broth at 30°C to the early stationary phase with vigorous shaking (120 spm). Culture supernatants were obtained by removing cells by centrifugation. Ammonium sulfate was dissolved in the supernatants to obtain 80% saturation. After the preparations stood at 4°C overnight, proteins were precipitated by centrifugation (3,000 × g, 20 min, 4°C) and dissolved in small volumes of 20 mM Na citrate buffer (pH 5.0). Then each solution was dialyzed against the same buffer at 4°C for several hours by using a cellulose ester membrane (molecular weight cutoff, 1,000; Spectrum Medical Industries Inc., Houston, Tex.). The resulting proteins were fractionated by Tricine-sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (16% polyacrylamide gel) by using the method of Schagger and Von Jagow (37). After electrophoresis, each gel was stained for proteins with Coomassie brilliant blue. To detect protein bands with antibacterial activity, the gel was washed three times in 0.1% Tween 80 (40 min each) at room temperature to remove SDS. Then soft nutrient agar medium (10 ml) containing cells of L. monocytogenes SUB635 (approximately 1 × 106 to 2 × 106 cells/ml) was overlaid to cover the whole gel (6 by 9 cm) by using the method of Martinez et al. (25). Clear zones caused by protein bands with antibacterial activity were detected after incubation at 30°C overnight.
Taxonomic analysis of isolated LAB strains.
For taxonomic identification isolated LAB strains were analyzed by using a VITEK2 kit (BioMerieux Inc., Hazelwood, Mo.) mainly composed of physiological tests. Moreover, a DNA fragment corresponding to the 16S ribosomal DNA region was PCR amplified by using chromosomal DNA as the template, which was prepared by the method of Pitcher et al. (34). The resulting amplified product was sequenced and compared to the sequences deposited in the DDBJ database by using the BLAST program.
Production of bacteriocin by E. faecalis RJ-11.
Cells of E. faecalis RJ-11 were grown under two different conditions, in a flask and in a jar fermentor, to determine the effects of changes in the pH of the medium on cell growth and the production of bacteriocin. In the case of the flask culture, 100 ml of MRS broth (initial pH, pH 6.0) was placed in a 500-ml Erlenmeyer flask and supplemented with 0.5% CaCO3 powder to neutralize the lactic acid produced during growth. In the case of the jar fermentor (type TBR-2-3-Z; vessel volume, 2 liters; Chiyoda Seisakusho Co., Ltd.), 700 ml of the same medium was placed in the fermentor, which was equipped with a pH controller to adjust the pH of the medium to 6.0 continuously by addition of 6 N NaOH. An overnight culture in MRS broth was inoculated into both media at a concentration of 5% (vol/vol) and cultured at 30°C with vigorous agitation (120 spm for the flask and 120 rpm for the jar fermentor). Aliquots were withdrawn from both cultures at appropriate times. Cell densities were monitored by measuring the turbidity (optical density at 660 nm), and antibacterial activity was assayed as described above after cells were removed by centrifugation and subsequently filtered through a membrane filter (pore size, 0.2 μm; Advantec).
Purification of bacteriocin from a culture fluid of E. faecalis RJ-11.
Culture supernatant (400 ml) of E. faecalis RJ-11 cells grown in MRS medium was used as the bacteriocin source. After ammonium sulfate (Wako, Osaka, Japan) was dissolved in the culture supernatant to obtain 80% saturation, salted-out proteins were precipitated by centrifugation, dissolved in a small volume of 20 mM Na citrate buffer (pH 5.0) (buffer A), and dialyzed against buffer A at 4°C by using a cellulose ester membrane (molecular weight cutoff, 1,000; Spectrum Medical Industries Inc.). The dialysate was loaded onto a column of Sephadex G-50 (20 by 700 mm) that had previously been equilibrated with buffer A. Proteins were eluted with the same buffer A at a flow rate of 45 ml/h. Fractions (4.5 ml) were collected and tested for antibacterial activity by spotting aliquots (10 μl) on a lawn of L. monocytogenes. Active fractions were pooled and analyzed further. When dissociation of bacteriocins was needed, Tween 80 was added to buffer A at a concentration of 0.1%.
Sensitivity to proteolytic enzymes.
The purified bacteriocin was assessed to determine its sensitivity to various proteolytic enzymes. The enzymes used were protease from Streptomyces griseus (EC 3.4.24.4; 4 U/mg of protein; Sigma), α-chymotrypsin from bovine pancreas (EC 3.4.21.1; 41 to 60 U/mg; type I-S; Sigma), trypsin from bovine pancreas (EC 3.4.21.4; 10,000 U/mg; type I; Sigma), pepsin from porcine gastric mucosa (EC 3.4.23.1; 3,200 to 4,500 U/mg; Sigma), papain from Carica papaya (EC 3.4.22.2; 30,000 U/mg; Merck), and proteinase K from Tritirachium album (EC 3.4.21.14; 30 U/mg; Merck). A purified bacteriocin preparation (64,000 AU/ml) was incubated with each enzyme (final concentration, 2 mg/ml) for 7 h at the optimal temperature and pH. After incubation, the residual antibacterial activity was determined.
Amino acid sequencing of the purified bacteriocin from E. faecalis RJ-11.
Proteins corresponding to the bacteriocin were fractionated by Tricine-SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories Inc.) by using an electroblotting apparatus (Advantec EB-100; 1 mA/cm2; 2 h). The N-terminal amino acid sequence of the blotted protein was identified by automated Edman degradation by using a Shimadzu PSQ-21 protein sequencer. The sequence determined was used to retrieve similar sequences from a DDBJ database by using a BLAST search.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the DDBJ/GenBank/EMBL database under accession number AB100597.
RESULTS
Isolation of LAB strains with antibacterial activity from rice bran.
The nutrient requirement for growth of LAB is generally complicated, and we found that most of the LAB strains utilized in industry were unable to grow in steam-cooked rice, possibly because of a lack of some nutrients. Our aim is to utilize LAB to suppress the growth of contaminating bacteria (especially spore-forming bacteria, such as Bacillus strains) in the koji mold fermentation process in the production of miso (fermented soybean paste). We isolated novel LAB strains from rice bran since they were expected to grow well on steam-cooked rice. In the first step of the screening procedure, 38 colonies that produced clear zones on a lawn of B. subtilis were isolated on TGC plates. This medium was chosen in order to select strains that produce antibacterial compounds without an effect of lactic acid, which also has antibacterial activity, since a lack of sugar made it difficult for LAB strains to produce lactic acid. In the next step, the strains selected were tested for the formation of clear zones caused by lactic acid production on GYP plates containing CaCO3 powder, and the strains that formed no clear zones were discarded. During the course of this two-step screening procedure, a total of 30 LAB strains with antibacterial activity were isolated.
All 30 strains isolated were examined to determine their antibacterial activity spectra by using various gram-positive and gram-negative bacteria. As a result, they could be classified into six distinct groups on the basis of the spectra (Table 1). The antibacterial spectrum of the strains in group I, which was the major group and comprised 18 strains (60% of the total number of isolates), was very similar to that of nisin Z-producing strains. Taxonomic analysis with the VITEK2 kit revealed that one representative strain (SJ-9) was an L. lactis strain. Then all 18 of these strains were tested to determine their sensitivities to a nisin Z producer, L. lactis IO-1 (16, 26), and also to determine their growth-inhibitory effects against IO-1. Growth inhibition was not observed in any case, indicating that all 18 strains belonging to group I might produce nisin Z or a closely related bacteriocin. The other five groups (groups II to VI) had activity spectra different from that of group I. One representative strain was chosen from each group and examined for production of antibacterial compounds by using the culture fluid. The antibacterial activities of all five strains were completely eliminated when culture fluids were treated with proteinase K (data not shown). This result strongly suggests that the antibacterial compounds are composed of proteins and that the growth-inhibitory effects of lactate were negligible even if lactate was produced by the five LAB strains. When crude proteins in the culture fluids were fractionated by Tricine-SDS-PAGE and successively assayed for activity against L. monocytogenes, a single protein band exhibiting antibacterial activity was detected for all five strains tested (Fig. 1). The relative mobilities of the five protein bands were different, but the bands were distributed over a molecular weight range from 3,500 to 5,600. Moreover, the five strains were tested to determine their cross-immunities by checking their growth-inhibitory activities against each other and against the nisin Z producer L. lactis IO-1. All five strains inhibited the growth of the other four strains and L. lactis IO-1, indicating that the bacteriocins produced by these five strains were not nisin. The screening and characterization analysis revealed that LAB strains producing various types of bacteriocins could be easily isolated from rice bran. We chose strain RJ-11 (a representative strain of group II) for further study because this strain exhibited higher levels of activity against a wide range of gram-positive strains than strains belonging groups III to VI exhibited and it might be a promising organism for wide application in the food industry, as well as in the miso fermentation process.
FIG. 1.
Bioassay for proteins with antibacterial activity after Tricine-SDS-PAGE. Crude proteins in culture fluids of strains RJ-11 (lane 1), RB-3 (lane 2), RJ-10 (lane 3), SJ-16 (lane 4), and YJ-35 (lane 5) were fractionated by Tricine-SDS-PAGE (16% polyacrylamide gel). Proteins with antibacterial activity were visualized by overlaying soft nutrient agar containing cells of L. monocytogenes SUB635 on the gel and incubating it at 30°C overnight. The numbers on the left indicate the positions of molecular weight markers.
Taxonomic analysis of strain RJ-11.
Strain RJ-11 was taxonomically identified by using the VITEK2 kit. It was a gram-positive coccus that was nonmotile, nonsporing, and catalase negative, and other physiological tests revealed that the strain belonged to E. faecalis (8). Further determination of the nucleotide sequence (394 bp) of PCR-amplified partial 16S ribosomal DNA indicated that the sequence showed a high level of similarity (more than 99.5%) to the sequence of E. faecalis 16S ribosomal DNA deposited in the database (data not shown). Hence, we designated the new strain E. faecalis strain RJ-11. This taxonomic analysis also revealed that the other four representative strains (RB-3, RJ-10, SJ-16, and YJ-35) all belonged to the genus Enterococcus.
Conditions for bacteriocin production by E. faecalis RJ-11.
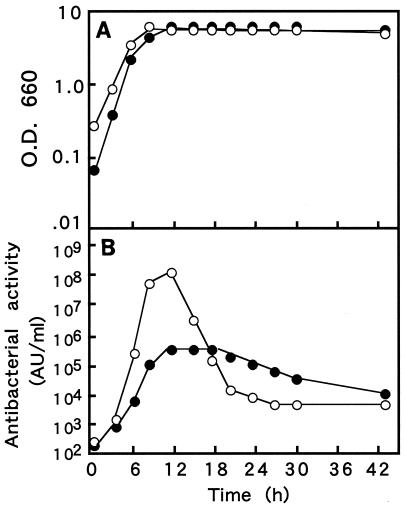
The effects of two different culture conditions on cell growth and production of bacteriocin were examined. First, cells of E. faecalis RJ-11 were grown at 30°C in a flask containing MRS broth (initial pH, pH 6.0) to which CaCO3 powder was added to neutralize the lactic acid produced during growth. The amount of bacteriocin in the culture fluid reached the maximum level (4 × 105 AU/ml) in the early stationary phase and then decreased gradually (Fig. 2). The pH of the medium decreased to 5.0 to 5.5 in the stationary phase, possibly because of an accumulation of lactic acid. Second, we examined the production of bacteriocin in a jar fermentor in which the pH of the medium was constantly adjusted to 6.0 by addition of NaOH. Under these culture conditions, a markedly higher concentration of bacteriocin (1 × 108 AU/ml, 250-fold higher than the concentration in the flask culture) was observed in the early stationary phase, although the generation time and cell density in the stationary phase were almost the same under the two culture conditions (Fig. 2).
FIG. 2.
Production of bacteriocin by E. faecalis RJ-11. Cells were cultured in a flask (•) or in a jar fermentor (○). At appropriate intervals, cell densities (A) and antibacterial activities in the culture fluid (B) were determined. O.D 660, optical density at 660 nm.
Purification of the bacteriocin produced by E. faecalis RJ-11.
We attempted to purify the bacteriocin by using crude proteins in the culture fluid as a starting point. A combination of Tricine-SDS-PAGE and a bioassay for antibacterial activity indicated that the molecular weight of the bacteriocin is around 5,000 (Fig. 1, lane 1). Thus, crude proteins salted out from culture fluid by addition of ammonium sulfate were subjected to Sephadex G-50 gel permeation chromatography. When citrate buffer was used throughout the chromatographic procedure, all proteins with antibacterial activity eluted in the void volume of the column, in which proteins whose molecular weights were more than 30,000 were expected to elute (Fig. 3A). This result led us to assume that bacteriocins might aggregate and form large assemblages. Therefore, we performed chromatography with Tween 80 added to the citrate buffer at a concentration of 0.1%. As a result, a second active peak eluted in addition to the original peak (Fig. 3B). The molecular weight of the protein in the second peak was estimated to be approximately 4,000 when the elution volume was compared with the elution volume of standard proteins in the same column. Analysis of the proteins in the two active fractions by Tricine-SDS-PAGE clearly indicated that a major band corresponding to a molecular weight of 5,000 appeared for both fractions (Fig. 4A). We also confirmed that both bands exhibited activity against L. monocytogenes in the bioassay (Fig. 4B). The proteins in the second peak were apparently homogeneous, as judged by electrophoresis (Fig. 4A, lane 3), and were used as purified bacteriocin in further examinations. We tentatively designated this bacteriocin enterocin RJ-11.
FIG. 3.
Elution profiles of proteins with antibacterial activity. Crude proteins in culture fluid were eluted from a Sephadex G-50 column (20 by 700 mm) by using citrate buffer in the absence (A) or in the presence (B) of 0.1% Tween 80. Each fraction contained 4.5 ml of the eluate.
FIG. 4.
Tricine-SDS-PAGE analysis of purified enterocin RJ-11. Proteins were fractionated by Tricine-SDS-PAGE (16% polyacrylamide gel). The resulting gel was stained with Coomassie brilliant blue (A) or assayed for activity against L. monocytogenes (B). The samples used were crude proteins salted out from culture fluid and then dialyzed (lane 1) and proteins in the fraction of the first active peak (lane 2) and the second active peak (lane 3) eluted by Sephadex G-50 gel permeation chromatography.
Characterization of enterocin RJ-11.
The antibacterial activity of purified enterocin RJ-11 was examined for sensitivity to various proteolytic enzymes and heat treatment. Under the conditions tested, the activity of enterocin RJ-11 was completely eliminated by proteinase K and S. griseus protease. The activity was also partially eliminated by either α-chymotrypsin, trypsin, or pepsin but not by papain (Table 2). The activity was extremely heat stable, since no decrease in activity was observed after boiling for 30 min (Table 2). Similarly, the activity did not decrease when enterocin RJ-11 was kept at 30°C for 6 h at pH 2 or 12. The antibacterial spectrum of enterocin RJ-11 was investigated by using various bacterial strains (Table 3). Enterocin RJ-11 was very active against B. subtilis and Bacillus amyloliquefaciens, which are occasionally detected as contaminants in the koji mold fermentation stage of miso production. It was also effective against the four other Enterococcus strains isolated from rice bran, in accordance with the cross-immunity test described above. Enterocin RJ-11 was not active against gram-negative bacteria, such as Escherichia coli, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium, like most bacteriocins obtained from other LAB strains (17, 22, 23).
TABLE 2.
Sensitivity of purified enterocin RJ-11 to various proteases and heat treatment
| Treatmenta | Residual activity (AU/ml)b |
|---|---|
| Control | 64,000 |
| Protease (pH 7.5, 37°C)c | 0 |
| Proteinase K (pH 7.5, 37°C) | 0 |
| α-Chymotrypsin (pH 7.5, 25°C) | 8,000 |
| Trypsin (pH 7.5, 25°C) | 32,000 |
| Papain (pH 6.0, 37°C) | 64,000 |
| Pepsin (pH 2.0, 37°C) | 32,000 |
| Control | 102,400 |
| 100°C, 15 min | 102,400 |
| 100°C, 30 min | 102,400 |
| pH 2, 30°C, 6 h | 102,400 |
| pH 12, 30°C, 6 h | 102,400 |
In each protease treatment,the sample was incubated for 7 h under the optimum pH and temperature condition indicated in parenthesis.
L. monocytogenes was used as the indicator strain.
A product of S. griseus.
TABLE 3.
Antibacterial spectra of purified enterocin RJ-11
| Strains | Antibacterial activity (AU/ml)a |
|---|---|
| Enterococcus faecalis RJ-11b | 400 |
| Enterococcus sp. strain RB-3b | 51,200 |
| Enterococcus sp. strain RJ-10b | 51,200 |
| Enterococcus sp. strain SJ-16b | 3,200 |
| Enterococcus sp. strain YJ-35b | 409,600 |
| Enterococcus durans KB-60 | 1,638,400 |
| Lactococcus lactis IFO12007 | 12,800 |
| Leuconostoc mesenteroides IFO3426 | 204,800 |
| Bacillus subtilis JCM1465T | 819,200 |
| Bacillus amyloliquefaciens B1 | 409,600 |
| Bacillus cereus B3 | 3,200 |
| Listeria monocytogenes SUB635 | 204,800 |
| Staphylococcus aureus SUB511 | 1,600 |
| Escherichia coli JM109 | 0 |
| Pseudomonas aeruginosa ATCC 10145 | 0 |
| Salmonella typhimurium SUB636 | 0 |
Antibacterial activity was determined by using various bacteria after serial dilutions.
LAB strain isolated in this study.
Amino acid sequence of enterocin RJ-11.
The N-terminal amino acid sequence of the purified enterocin RJ-11 protein was determined by using a peptide sequencer. A sequence comprised of 44 amino acid residues was determined (Fig. 5). Judging from the calculated molecular weight of this polypeptide (5,049), the sequence may correspond to the entire polypeptide of enterocin RJ-11.
FIG. 5.
Amino acid sequence of enterocin RJ-11 and comparison with the sequences of enterocins L50A and L50B. The sequence containing 44 amino acid residues determined for purified enterocin RJ-11 was deduced from four sets of data analyzed independently. The sequences of enterocins L50A and L50B were obtained from the DDBJ database (accession numbers of AJ223633 and Y16413, respectively). Residues identical to residues of enterocin L50A and/or enterocin L50B are indicated by a black background.
DISCUSSION
We isolated LAB strains with antibacterial activity from a large number of rice bran samples collected from all over Japan. A total of 30 strains were isolated during this screening procedure, and it should be noted that rice bran is a good source for isolation of LAB strains. Some of the strains isolated grew well on steam-cooked rice and produced antibacterial compounds (data will be presented elsewhere). This characteristic is promising for application of the strains in the koji mold fermentation process of miso production to suppress the growth of contaminating bacteria, such as Bacillus. Kato et al. tried to suppress the growth of contaminating Bacillus by using a LAB strain producing nisin during miso production (19, 20). Their method consisted of a lactic acid fermentation process in which a LAB strain was grown solely on steam-cooked rice supplemented with soybean extract for 24 h before addition of the koji mold. Utilization of LAB strains capable of growing well on steam-cooked rice described in this work should make it possible to suppress contaminating bacteria at the beginning of koji mold fermentation without an additional lactic acid fermentation step and thus should be a more practical method. The potency of these strains in the miso production process is now being examined to assess the effectiveness of the organisms at the industrial level.
Thirty LAB strains isolated in this work could be classified into six groups on the basis of their antibacterial activity spectra (Table 1). It is noteworthy that 60% of the strains (18 of 30 strains), which comprised the major group (group I), seemed to produce a nisin-like bacteriocin and should belong to the genus Lactococcus. It was reported previously that nisin Z-producing strains belonging to the genus Lactococcus were isolated from well-aged Nukadoko (a bed of fermented rice bran used for making Japanese pickles) (10). It can be postulated that rice bran is a suitable source for isolation of LAB strains that produce nisin-like bacteriocins. The other 12 strains were classified into groups II to VI. Further analysis of the five representative strains of these groups showed that all of them produced a bacteriocin with a low molecular weight (3,500 to 5,500), at least in the monomeric form (Fig. 1). Taxonomic analysis suggested that all five of the strains belong to the genus Enterococcus. We concluded that these five strains produce different types of bacteriocins, since the cross-immunity test indicated that all five strains clearly exhibited activity against the other four strains. From these results, it can be postulated that two major genera of bacteriocin-producing LAB strains (Lactococcus and Enterococcus) are present in rice bran, although we cannot rule out the possibility that other genera of LAB exist in rice bran and could be isolated by a different screening procedure. We chose one strain belonging to group II, E. faecalis strain RJ-11, for further characterization of the bacteriocin produced, since this strain had a significant inhibitory effect against a wide range of gram-positive bacteria (Table 1) and is a promising candidate for application in food-manufacturing processes.
The effects of the culture conditions on cell growth and on the production of enterocin RJ-11 were examined from the viewpoint of accumulation of lactate in cultures (Fig. 2). In a batch culture in an Erlenmeyer flask in which lactate was neutralized with CaCO3 powder, the antibacterial activity in the culture fluid increased in proportion to the cell growth and reached the maximum level in the early stationary phase. On the other hand, a 250-fold-higher level of activity was observed in a jar fermentor in which the pH of the medium was kept at 6.0, although the generation time and cell density in the stationary phase were almost the same as those in the batch culture. The high productivity may be partly ascribed to optimization of the physiological state of the cells due to efficient pH adjustment by neutralizing lactate (27). This result indicates that pH control is critical for achieving high levels of production of enterocin RJ-11 for application of this bacteriocin in the food industry. In both cultures, the activity markedly decreased after it reached the maximum level in the early stationary phase. The reason for this has not been determined, but some proteolytic enzyme(s) produced by strain RJ-11 might cause degradation and inactivation of enterocin RJ-11 (33).
Enterocin RJ-11 was purified from the culture fluid to apparent homogeneity, as judged by SDS-PAGE analysis (Fig. 4A). The molecular weight of enterocin RJ-11 in the monomeric form was estimated to be 5,000, and the monomeric form was confirmed to have antibacterial activity by a bioassay (Fig. 4B). The elution profile for antibacterial peptides following Sephadex G-50 gel permeation chromatography in the presence or absence of Tween 80 led us to the assumption that enterocin RJ-11 has a tendency to form aggregates (Fig. 3). The aggregated form (the first peak) also had antibacterial activity, and we assumed that it is composed of several or more monomeric peptides since it eluted in the void volume, in which peptides having molecular weights higher than 30,000 were expected to elute. The first peak corresponding to the aggregate form still remained when the concentration of Tween 80 was increased to 1.0% (data not shown), indicating that two aggregate forms might exist, an easily dissociated form and a tightly packed form. Other bacteriocins, including mutacin produced by Streptococcus mutans (13, 14) and lactacin F produced by Lactobacillus acidophilus (31), have also been reported to easily form aggregates. Addition of Tween 80 effectively dissociated the mutacin aggregate into its monomeric forms, and there was a simultaneous increase in activity. At present, we cannot postulate the actual form of enterocin RJ-11 produced in the original culture, since we could not rule out the possibility that the aggregation occurred artificially in the ammonium precipitation and/or dialysis step during the purification process.
The antibacterial activity of enterocin RJ-11 was highly resistant to heat, acid, and alkali treatments. It should be noted that enterocin RJ-11 is very stable at pH 12, a pH at which nisin rapidly loses activity (35). Enterocin RJ-11 also effectively inhibited a spore-forming Bacillus that often contaminates food production processes. These characteristics are promising for application of enterocin RJ-11 in the koji mold fermentation process in miso production and in other food production processes.
The purified enterocin RJ-11 was subjected to peptide sequencing, and a sequence comprising 44 amino acid residues was determined from the N terminus (Fig. 5). The calculated molecular weight of this sequence is 5,049, which is in good agreement with the molecular weight estimated by SDS-PAGE analysis, indicating that the sequence corresponds to almost the entire length of the enterocin RJ-11 polypeptide. The sequence was used as a query sequence to search for similar sequences deposited in the DDBJ database with BLAST. We found that the sequence exhibited a high level of similarity (75% identity) to the amino acid sequence of enterocin L50A (produced by E. faecium strain L50) (4, 5, 6) and enterocin I (produced by E. faecium strain 6T1a) (12), which are thought to be identical because they have the same amino acid sequence. Enterocin L50A exhibits antibacterial activity, but its activity is synergistically increased in the presence of enterocin L50B, which is produced by the same strain. The amino acid sequences of enterocins L50A and L50B are very similar (72% identity), and the genes encoding them (entL50A and entL50B) constitute an operon located in the megaplasmid harbored by E. faecium strain L50. Therefore, it is probable that enterocins L50A and L50B are produced concurrently and exert antibacterial activity by functioning together. However, we found only a single polypeptide with antibacterial activity in the culture fluid of E. faecalis RJ-11. Enterocin RJ-11 may be produced as a single peptide and may act alone, unlike enterocins L50A and L50B.
In most cases, the bacteriocins of LAB strains reported to date have a leader peptide or a signal sequence that is required for secretion of the precursor form (11, 36). Analysis of the gene structures of entL50A and entL50B revealed no additional sequence other than the primary sequence encoding enterocins secreted into the medium (5). The mechanism for secretion of enterocins L50A and L50B is unclear and remains to be elucidated. The amino acid sequence of enterocin RJ-11 is very similar to those of enterocins L50A and I, but there are differences in the N-terminal and C-terminal regions, in addition to replacement of five amino acid residues internally (Fig. 5). The characteristics of enterocin RJ-11 are also similar to those of enterocin L50A, but the antibacterial spectra of the two proteins are different. For example, enterocin RJ-11 is active against Leuconostoc mesenteroides, whereas enterocin I is not (12). Since this difference is likely to be caused by differences in the amino acid sequences, enterocin RJ-11 can be considered a novel bacteriocin. The bacteriocins produced by LAB strains are classified into four classes (classes I to IV) based on their structures and characteristics (23, 32). However, enterocins L50A and I have not been placed in any class because of their unique structures and rare appearance. This is the first report of bacteriocin production by E. faecalis, with the bacteriocin being similar to enterocins L50A and I originating from E. faecium. Isolation of a gene encoding enterocin RJ-11 is now under way so that the structure of this enterocin can be compared with the structures of enterocins L50AB and I. This should give us insight into the mechanism of secretion of enterocin RJ-11 and the possible involvement of other bacteriocin-related genes located in an operon.
Acknowledgments
We thank Goro Taguchi and Nobuaki Hayashida, Gene Research Center of Shinshu University, for helpful discussions. We are grateful to Kozue Oana and Masahiro Nogawa for their technical assistance and helpful discussions.
Footnotes
In tribute to the memory of Yoshikazu Togawa, chief research worker at Marukome Co., Ltd., who passed away in November 2002.
REFERENCES
- 1.Arai, Y., M. Uehara, Y. Sato, M. Kimira, A. Eboshida, H. Adlercreutz, and S. Watanabe. 2000. Comparison of isoflavones among dietary intake, plasma concentration and urinary excretion for accurate estimation of phytoestrogen intake. J. Epidemiol. 10:127-135. [DOI] [PubMed] [Google Scholar]
- 2.Arliss, R. M., and C. A. Biermann. 2002. Do soy isoflavones lower cholesterol, inhibit atherosclerosis, and play a role in cancer prevention? Holist. Nurs. Pract. 16:40-48. [DOI] [PubMed] [Google Scholar]
- 3.Chuyen, N. V., K. Ijichi, H. Umetsu, and K. Moteki. 1998. Antioxidative properties of products from amino acids or peptides in the reaction with glucose. Adv. Exp. Med. Biol. 434:201-212. [DOI] [PubMed] [Google Scholar]
- 4.Cintas, L. M., J. M. Rodriguez, M. F. Fernandez, K. Sletten, I. F. Nes, P. E. Hernandez, and H. Holo. 1995. Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cintas, L. M., P. Casaus, H. Holo, P. E. Hernandez, I. F. Nes, and L. S. Havarstein. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cintas, L. M., P. Casaus, C. Herranz, L. S. Havarstein, H. Holo, P. E. Hernandez, and I. F. Nes. 2000. Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182:6806-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholtz. 1996. Applications of the bacteriocin, nisin. Antonie Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 8.Devriese, L. A., B. Pot, and M. D. Collins. 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J. Appl. Bacteriol. 75:399-408. [DOI] [PubMed] [Google Scholar]
- 9.Ennahar, S., K. Sonomoto, and A. Ishizaki. 1999. Class IIa bacteriocins from lactic acid bacteria: antibacterial activity and food preservation. J. Biosci. Bioeng. 87:705-716. [DOI] [PubMed] [Google Scholar]
- 10.Ennahar, S., T. Zendo, K. Sonomoto, and A. Ishizaki. 1999. Investigation of bacteriocin production and purification from Nukadoko isolates displaying antimicrobial activity. Jpn. J. Lactic Acid Bacteria 10:29-37. [Google Scholar]
- 11.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 12.Floriano, B., J. L. Ruiz-Barba, and R. Jimenez-Diaz. 1998. Purification and genetic characterization of enterocin I from Enterococcus faecium 6T1a, a novel antilisterial plasmid-encoded bacteriocin which does not belong to the pediocin family of bacteriocins. Appl. Environ. Microbiol. 64:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukushima, H., J. Kelstrup, S. Fukushima, T. Umemoto, A. Kaibori, and H. Sagawa. 1985. Characterization and mode of action of a purified bacteriocin from the oral bacterium Streptococcus mutans RM-10. Arch. Oral Biol. 30:229-234. [DOI] [PubMed] [Google Scholar]
- 14.Hamada, S., H. Imanishi, and T. Ooshima. 1986. Isolation and mode of action of a cell-free bacteriocin (mutacin) from serotype g Streptococcus mutans MT3791. Zentralbl. Bakteriol. Mikrobiol. 261:287-298. [DOI] [PubMed] [Google Scholar]
- 15.Hurst, A. 1981. Nisin. Adv. Appl. Microbiol. 27:85-123. [Google Scholar]
- 16.Ishizaki, A., and T. Ohta. 1989. Batch culture kinetics of l-lactate fermentation employing Streptococcus sp. IO-1. J. Ferment. Bioeng. 67:46-51. [Google Scholar]
- 17.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamei, H., T. Koide, Y. Hashimoto, T. Kojima, T. Umeda, and M. Hasegawa. 1997. Tumor cell growth-inhibiting effect of melanoidins extracted from miso and soy sauce. Cancer Biother. Radiopharm. 12:405-409. [DOI] [PubMed] [Google Scholar]
- 19.Kato, T., K. Maeda, H. Kasuya, and T. Matsuda. 1999. Complete growth inhibition of Bacillus subtilis by nisin-producing lactococci in fermented soybeans. Biosci. Biotechnol. Biochem. 63:642-646. [DOI] [PubMed] [Google Scholar]
- 20.Kato, T., L. Inuzuka, M. Kondo, and T. Matsuda. 2001. Growth of nisin-producing lactococci in cooked rice supplemented with soybean extract and its application to inhibition of Bacillus subtilis in rice miso. Biosci. Biotechnol. Biochem. 65:330-338. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, P. B., J. A. Duke, H. Brielmann, J. Boik, and J. E. Hoyt. 1997. A comparative survey of leguminous plants as sources of the isoflavones, genistein and daidzein: implications for human nutrition and health. J. Altern. Complement. Med. 3:7-12. [DOI] [PubMed] [Google Scholar]
- 22.Klaenhammer, T. R. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337-349. [DOI] [PubMed] [Google Scholar]
- 23.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 24.Konings, W. N., J. Kok, O. P. Kuipers, and B. Poolman. 2000. Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, B., J. E. Suarez, and A. Rodriguez. 1996. Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142:2393-2398. [DOI] [PubMed] [Google Scholar]
- 26.Matsusaki, H., N. Endo, K. Sonomoto, and A. Ishizaki. 1996. Development of purification method and identification of a peptide antibiotic produced by Lactococcus lactis IO-1. Food Sci. Technol. 2:157-162. [Google Scholar]
- 27.Matsusaki, H., N. Endo, K. Sonomoto, and A. Ishizaki. 1996. Lantibiotic nisin Z fermentative production by Lactococcus lactis IO-1: relationship between production of the lantibiotic and lactate and cell growth. Appl. Microbiol. Biotechnol. 45:36-40. [DOI] [PubMed] [Google Scholar]
- 28.Mayr-Harting, A., A. J. Hedges, and R. C. W. Berkeley. 1972. Methods for studying bacteriocins. Methods Microbiol. 7:315-422. [Google Scholar]
- 29.Messina, M. J., V. Persky, K. D. Setchell, and S. Barnes. 1994. Soy intake and cancer risk: a review of the in vitro and in vivo data. Nutr. Cancer 21:113-131. [DOI] [PubMed] [Google Scholar]
- 30.Montville, T. J., and Y. Chen. 1998. Mechanistic action of pediocin and nisin: recent progress and unresolved questions. Appl. Microbiol. Biotechnol. 50:511-519. [DOI] [PubMed] [Google Scholar]
- 31.Muriana, P. M., and T. R. Klaenhammer. 1991. Purification and partial characterization of lactacin F, a bacteriocin produced by Lactobacillus acidophilus 11088. Appl. Environ. Microbiol. 57:114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 33.Parente, E., and A. Ricciardi. 1999. Production, recovery and purification of bacteriocins from lactic acid bacteria. Appl. Microbiol. Biotechnol. 52:628-638. [DOI] [PubMed] [Google Scholar]
- 34.Pitcher, D. G., N. A. Saunders, and R. G. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 35.Rollema, H. S., O. P. Kuipers, P. Both, W. M. de Vos, and R. J. Siezen. 1995. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 61:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 37.Schagger, H. J., and G. Von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of protein in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 38.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, S., T. Sobue, M. Kobayashi, S. Sasaki, and S. Tsugane. 2003. Soy, isoflavones, and breast cancer risk in Japan. J. Natl. Cancer Inst. 95:906-913. [DOI] [PubMed] [Google Scholar]