Abstract
A molecular screening approach was developed in order to amplify the genomic region that codes for the α- and β-subunits of the nitrile hydratase (NHase) enzyme in rhodococci. Specific PCR primers were designed for the NHase genes from a collection of nitrile-degrading actinomycetes, but amplification was successful only with strains identified as Rhodococcus erythropolis. A hydratase PCR product was also obtained from R. erythropolis DSM 43066T, which did not grow on nitriles. Southern hybridization of other members of the nitrile-degrading bacterial collection resulted in no positive signals other than those for the R. erythropolis strains used as positive controls. PCR-restriction fragment length polymorphism-single-strand conformational polymorphism (PRS) analysis of the hydratases in the R. erythropolis strains revealed unique patterns that mostly correlated with distinct geographical sites of origin. Representative NHases were sequenced, and they exhibited more than 92.4% similarity to previously described NHases. The phylogenetic analysis and deduced amino acid sequences suggested that the novel R. erythropolis enzymes belonged to the iron-type NHase family. Some different residues in the translated sequences were located near the residues involved in the stabilization of the NHase active site, suggesting that the substitutions could be responsible for the different enzyme activities and substrate specificities observed previously in this group of actinomycetes. A similar molecular screening analysis of the amidase gene was performed, and a correlation between the PRS patterns and the geographical origins identical to the correlation found for the NHase gene was obtained, suggesting that there was coevolution of the two enzymes in R. erythropolis. Our findings indicate that the NHase and amidase genes present in geographically distinct R. erythropolis strains are not globally mixed.
Nitrile hydratase (NHase) is a soluble bacterial metalloenzyme (41) that catalyzes the hydration of nitrile compounds to the corresponding amides, which may be converted by an amidase to the corresponding acids plus ammonia (40). NHase consists of two subunits (α and β), each with a molecular mass of approximately 23 kDa; the amino acid sequences of the subunits are not related, and the structural genes are normally adjacent in the same operon, although the α- and β-coding sequence order is variable (42). NHase is classified into two groups on the basis of the metal ion composing the catalytic center; an Fe-type NHase has a nonheme iron atom (74), and a Co-type NHase has a noncorrinoid cobalt atom (12) at the catalytic center. The Fe- and Co-type NHase enzymes differ in biotransformation activity and substrate specificity, although their amino acid sequences exhibit significant homology.
NHase enzymes have considerable practical importance as biocatalysts for the industrial production of acrylamide and nicotinamide (84) and also for environmental bioremediation, where they have been shown to be effective in the removal of nitriles from waste streams (83). However, despite this biotechnological relevance, information concerning the distribution of nitrile-hydrolyzing enzymes in microorganisms and in the environment is quite limited. Bacteria having NHase enzymes have been recovered from shallow marine sediment (45), deep-sea sediments (18, 31), geothermal habitats (65, 85), and various soils (21, 46, 47, 48, 81), including contaminated soils (6). Recently, we reported the widespread distribution of nitrile-hydrolyzing actinomycetes in marine sediments and terrestrial soils (9). A role for these enzymes in bacteria has recently been proposed in the metabolism of aldoximes, which are widely distributed intermediates in the biosynthesis of plant natural products (38).
The rapid increase in sequence information obtained from new cloned genes and from the increasing number of available genome sequences has enabled extensive alignment of homologous enzymes, thus allowing predictions of conserved regions that can serve as primers for molecular screening of both cultivated and uncultivated organisms. Molecular screening can be used to detect enzyme genes independent of the enzyme expression level by carefully choosing the enzyme nucleotide sequences from databases and using them to construct primers for recognizing and amplifying conserved regions directly from genomic DNA (20). In this paper we describe a molecular screening technique for amplifying the NHase and amidase enzyme genes from mycolic acid-containing actinomycetes recovered from geographically distinct locations; this technique was used to investigate the extent of conservation in this gene cluster.
The nitrile-transforming activities of Rhodococcus erythropolis strains recovered from widely separated geographic locations have been characterized previously, and all of the strains examined have been shown to contain a NHase-amidase hydrolyzing system (8). However, although these rhodococci belong to the same species, they exhibited highly variable transformation activities and substrate specificities with nitriles and amides, highlighting the possible diversity of the enzymes at the infraspecies level. Thus, the main objective of this work was to define the genetic diversity of the NHase and amidase genes in geographically disparate strains of R. erythropolis in order to obtain insight into the evolution and distribution of these enzymes. PCR-single-strand conformational polymorphism (PCR-SSCP) analysis (16) and PCR-restriction fragment length polymorphism-single-strand conformational polymorphism (PRS) analysis (11, 17) were used as rapid and reliable techniques to measure the diversity of the NHase and amidase genes amplified from R. erythropolis isolates. The NHase gene diversity was investigated further by sequencing.
MATERIALS AND METHODS
Chemicals.
Nitriles were purchased from Sigma-Aldrich Company Ltd. (Gillingham, United Kingdom) and were the highest purity available. Other chemicals were obtained from commercial sources and were molecular or reagent grade.
Organisms and growth conditions.
The nitrile-metabolizing strains of R. erythropolis were recovered from marine sediments or terrestrial soils as described previously (9, 19, 31) or were kindly provided by other laboratories (Table 1). Bacteria were grown on glucose yeast extract agar for 3 days at 30°C for DNA extraction (10) and in chelated mineral medium supplemented with 0.1% (vol/vol) acetonitrile or 0.1% (vol/vol) benzonitrile for 1 to 5 days at 30°C for growth experiments (31).
TABLE 1.
R. erythropolis strains used to verify the PRS pattern diversity of complete NHase genes (αβ-nhase) and amidase genes (amd), their growth on acetonitrile or benzonitrile, and their environmental sources and geographic origins
| R. erythropolis strain | PRS pattern code
|
Growth ona
|
Environmental source | Geographic originb | Reference(s) | Culture sourcec | ||
|---|---|---|---|---|---|---|---|---|
| αβ-nhase | amd | AN | BN | |||||
| N11T (= NCIB 9158T) | 1 | 1 | N | N | Soil | NAd | 24, 25 | M. Goodfellow |
| DSM 13002 | 2 | 2 | Y | N | Garden soil | Ludwigshafen, Germany | NA | C. Syldatk |
| 122-AN065 | 3 | 3 | Y | Y | Deep-sea sediment | Ryukyu Trench. northwest Pacific | 31 | UKC |
| 67-BEN001 | 4 | 4 | Y | Y | Deep-sea sediment | Japan Trench, northwest Pacific | 31 | UKC |
| ANT-AN007 | 4 | 4 | Y | Y | Lake sediment | Antarctica | 9 | UKC |
| IND-AN014 | 5 | 5 | Y | N | Mangrove mud | Indonesia | 9 | UKC |
| 870-AN019 | 6 | 6 | Y | Y | Deep-sea sediment | Suruga Bay, Japan | 9 | UKC |
| ARG-AN024 | 7 | 7 | Y | N | Subtropical rain forest soil | Argentina | 9 | UKC |
| ARG-AN025 | 4 | 4 | Y | ND | Subtropical rain forest soil | Argentina | 9 | UKC |
| ENG-AN033 | 8 | 8 | Y | N | Wet peat | United Kingdom | 9 | UKC |
| 871-AN042 | 9 | 9 | Y | ND | Deep-sea sediment | Suruga Bay, Japan | 9 | UKC |
| 871-AN053 | 10 | 10 | Y | Y | Deep-sea sediment | Suruga Bay, Japan | 9 | UKC |
| AJ192 | 11 | 11 | Y | Y | Contaminated river bank soil | Tyne, England | 5 | M. Goodfellow |
| AJ270 | 11 | 11 | Y | Y | Contaminated river bank soil | Tyne, England | 5, 6 | M. Goodfellow |
| N63 | 12 | 12 | N | N | NA | NA | 24, 25 | M. Goodfellow |
| N58 | 13 | 13 | Y | N | Soil | NA | 24, 25 | M. Goodfellow |
| N60 | —e | — | N | N | NA | NA | 24, 25 | M. Goodfellow |
| N53 (= NCIB 8147) | 14 | 14 | Y | N | Soil | England? | 4, 24, 25 | M. Goodfellow |
| N108 (= ATCC 4277T) | 1 | 1 | N | N | Soil | NA | 24, 25 | M. Goodfellow |
| N143 | 15 | 15 | N | N | NA | NA | 24 | M. Goodfellow |
| R285 (= NCIMB 9706) | 16 | 16 | Y | N | NA | NA | NA | M. Goodfellow |
| SRB1948-AO7 | 3 | 3 | Y | N | Deep-sea sediment | Suruga Bay, Japan | 19 | UKC |
| N186/21 | 17 | 17 | Y | N | Soil | Hungary? | 57 | R. De Mot |
| DSM 311 | 18 | 18 | Y | N | Soil | Japan? | NA | M. Barclay and C. J. Knowles |
| DSM 743 | 1 | 1 | N | N | Soil | NA | 24, 25 | R. De Mot |
| DSM 6344 | 19 | 19 | Y | N | Contaminated sludge | Switzerland | 71 | C. Syldatk |
| DSM 7337 | 20 | 20 | Y | N | Soil and water | The Netherlands | 28 | C. Syldatk |
| DSM 9675 | 21 | 21 | Y | N | Soil | Stuttgart, Germany | 47 | H.-J. Knackmuss |
| DSM 9685 | 22 | 22 | Y | N | Soil | Stuttgart, Germany | 47 | H.-J. Knackmuss |
| DSM 11397 | 22 | 22 | Y | N | Soil | Stuttgart, Germany | 46 | C. Syldatk |
| DSM 12789 | 23 | 23 | Y | N | River water and sediment | Elbe and Rhine, Germany | 39 | C. Syldatk |
| DSM 20665 | 24 | 24 | Y | N | Soil | United States? | 76 | C. Syldatk |
| DSM 43060 | 14 | 14 | Y | N | Soil | England? | 4 | C. Syldatk |
| DSM 43066T | 1 | 1 | N | N | Soil | NA | 24, 25 | C. Syldatk |
| DSM 43135 | 25 | 25 | Y | N | Soil | Scotland? | 82 | C. Syldatk |
| DSM 43188 | 26 | 26 | N | N | Chalky grassland soil | England? | 50 | C. Syldatk |
| DSM 43200 | 21 | 21 | Y | N | Soil | England? | 23 | C. Syldatk |
| DSM 43288 | 27 | 27 | Y | N | Sand | NA | NA | C. Syldatk |
| DSM 43296 | 14 | 14 | Y | N | NA | NA | NA | C. Syldatk |
| DSM 44235 | 28 | 28 | Y | N | River sediment | Rhine, The Netherlands | 72 | C. Syldatk |
| NCIMB 10422 | 29 | 29 | Y | N | Soil | Japan? | NA | M. Barclay and C. J. Knowles |
| NCIMB 11148T | 1 | 1 | N | N | Soil | NA | 24, 25 | M. Barclay and C. J. Knowles |
| GJ70 | 30 | 30 | Y | N | Contaminated sludge | Arnhem, The Netherlands | 35 | D. B Janssen |
| TB2 | 31 | 31 | Y | N | Contaminated site | United States | 68 | D. B. Janssen |
| DCL14 | — | — | N | N | Ditch sediment | Reeuwijk, The Netherlands | 78 | M. J. van der Werf |
| KA-2-5-1 | — | — | N | N | Soil | Japan | 44 | J. Konishi |
| N28f | 32 | — | N | N | NA | NA | NA | M. Goodfellow |
AN, acetonitrile; BN, benzonitrile; N, no growth; Y, growth; ND, not determined.
A question mark indicates that the origin of the sample is not certain.
Strains were isolated in our laboratory at University of Kent, Canterbury, United Kingdom (UKC), or were obtained from other sources.
NA, information not available or not found.
—, no gene amplification.
The 16S rRNA gene PRS profile is not related to the R. erythropolis profile.
Molecular screening.
The α- and β-subunits of the NHase gene from various Rhodococcus species deposited in the GenBank database were aligned, and primers were designed based on the conserved regions of the nucleotide alignment. The program Oligo Calculator (http://listeria.nwfsc.noaa.gov/protocols/oligoTMcalc.html) was used to calculate the theoretical melting temperatures and the G+C contents of the oligonucleotides designed and to select which primer set was the most appropriate primer set. Oligonucleotides were synthesized commercially (GENSET SA, Paris, France). The primers for the amidase gene were designed in a similar manner.
PCR amplification of the putative NHase and amidase genes.
The procedures used for extraction and purification of genomic DNA and the PCR conditions used for amplification of the putative NHase and amidase genes were those described previously for the 16S rRNA gene (10). The following combinations of primers were used to amplify the putative genes (Table 2): αNH1 and βNH2 for the complete NHase α- and β-subunit gene (αβ-nhase), αNH1 and αNH2 for the NHase α-subunit gene (α-nhase), βNH1 and βNH2 for the NHase β-subunit gene (β-nhase), and amd1 and amd2 for the complete amidase gene (amd).
TABLE 2.
Oligonucleotides designed for PCR amplification of the NHase α- and β-subunit genes and the amidase gene and for sequencing the NHase genea
| Primer | Sequence | Direction of reading | Positionb | Usec |
|---|---|---|---|---|
| NHase primers | ||||
| αNH1 | 5′-GTG AAC CAG ATG TCA GTA ACG ATC G | Forward | 1907 | Amp |
| αNH2 | 5′-CGC TCA GGC AGT CCT TGG TGA CG | Reverse | 2507 | Amp |
| βNH1 | 5′-GCA CAC CAT GGA TGG AGT ACA CG | Forward | 2559 | Amp |
| βNH2 | 5′-GCA GGC TCG AGG TAA CCC TCG | Reverse | 3197 | Amp |
| αNH4 | 5′-TCT ACG ACA CCA CCG CCG AAA CT | Forward | 2394 | Seq |
| βNH4 | 5′-CGA TGA CGT ACC TCT CGT AGT ACG | Reverse | 2800 | Seq |
| SV1 | 5′-GAC CAT GAT TTC CAG TGT CC | Reverse | 3308 | Seq |
| SV2 | 5′-GGC GAA GAA CCG ACG TAC C | Forward | 3088 | Seq |
| SV3 | 5′-GCC CGC ATA AGA AAA GGT GAA | Forward | 1891 | Amp |
| SV4 | 5′-TCA AAC GGT CTG GTC GGT ATA | Reverse | 1328 | Amp |
| Amidase primers | ||||
| amd1 | 5′-GTG AAG CCG ATC ACA TCA GGA GC | Forward | 245 | Amp |
| amd2 | 5′-CGG GTA CCA ATC CCT TAC CGT CG | Reverse | 2027 | Amp |
See Materials and Methods for details.
Position in the sequences of the Rhodococcus sp. strain N-774 NHase and amidase genes (30) except for the position of SV4, which is the position in the sequence of the Rhodococcus sp. strain N-774 NHase β-subunit downstream region Orf1188 (29). The DDBJ/EMBL/GenBank accession number for the NHase and amidase genes is X54074 and the accession number for Orf1188 is D30033.
Amp, amplification; Seq, sequencing.
PRS analysis.
The restriction enzymes used to digest the NHase and amidase genes were AccI and BcnI, respectively, which were selected by a method similar to that used by Brandão et al. (11) for the 16S rRNA gene. The PCR-restriction fragment length polymorphism reaction conditions were those used previously (11), except that the following digestion program was used for the enzyme genes: 16 h at 37°C, followed by 15 min at 65°C. The NHase gene PCR-restriction fragment length polymorphism fragments were electrophoresed in an SSCP gel (PRS) at 5 W (constant power) for about 10.5 kV · h, while the amidase gene fragments were electrophoresed at 3 W (constant power) for about 6.0 kV · h. The products obtained by PCR amplification of the separated α-nhase and β-nhase genes were not digested; consequently, a PCR-SSCP analysis was carried out for these PCR products (fragment sizes, approximately 600 bp). PCR-SSCP and PRS patterns were grouped by eye on the basis of similarity (17).
Sequencing and phylogenetic analysis of the NHase gene.
Amplified PCR products were purified from the reaction components by using the Wizard PCR Preps DNA purification system (Promega). The αβ-nhase sequences were determined by using a combination of sequencing procedures, as follows. (i) Amplified αβ-nhase products were ligated into the pGEM-T Easy vector (Promega) and transformed into competent Escherichia coli JM109 (Promega) by using the manufacturer's protocols; recombinant colonies were selected by blue-white screening, and the presence of the correct-size inserts was detected by PCR amplification directly from white colonies by using primers αNH1 and βNH2 (Table 2). Recombinant plasmids were extracted from bacterial clones representing each selected αβ-nhase PRS pattern by using the Wizard Plus Minipreps (Promega) system according to the manufacturer's instructions; sequencing was performed by using plasmid-based forward and reverse sequencing primers (T7 and SP6, respectively). (ii) Amplified αβ-nhase products were used as templates for sequencing of the NHase gene starting from the inner part of the coding region by using primers αNH4 and βNH4 (Table 2). All templates were commercially cycle sequenced (ABC, Imperial College School of Medicine, London, United Kingdom), and the fragments generated were assembled to form a complete gene sequence. All NHase gene sequences were aligned with DDBJ/EMBL/GenBank database sequences, and phylogenetic analyses were performed by using the least-squares, maximum-likelihood, maximum-parsimony, and neighbor-joining treeing algorithms, as described by Brandão et al. (10) for 16S rRNA genes, with a software package for inference of evolutionary trees, PAUP*, version 4.0 (75). Trees were displayed by using TreeView (63).
The complete N-terminal nucleotide sequence of the NHase β-subunit gene, which was not resolved with primer βNH2 (Table 2), and the sequence of a partial NHase activator gene (Orf1188) (29, 62) were determined for a selection of the R. erythropolis strains by using primers SV1 and SV2 (Table 2), respectively. PCR products obtained with primers SV3 and SV4 (Table 2), which amplified the complete αβ-NHase and NHase activator genes, were used as templates for sequencing with these primers.
Nucleotide sequence translation to amino acid sequence.
The deduced amino acid sequence was determined for each α-nhase and β-nhase sequence by using the sequence utilities from the web-based software of the Baylor College of Medicine (http://searchlauncher.bcm.tmc.edu).
Southern hybridization.
DNA (2.5 to 4 μg) was digested with BamHI for 12 h at 37°C, loaded into a 0.7% agarose gel, and electrophoresed for 4 h at 70 V. The electrophoresed DNA was transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) by using the Southern blotting protocols described by the membrane manufacturer. DNA fixation was performed with a Spectro Linker XL-1000 UV cross-linker (Spectronics Corporation) for 2 min at 254 nm. A Ready-To-Go DNA labeling bead (dCTP) kit (Amersham Pharmacia Biotech) was used to construct the α-32P-radiolabeled probe by labeling the PCR-amplified β-nhase gene from R. erythropolis 67-BEN001. Southern hybridization was performed by using the QuikHyb hybridization solution (Stratagene) and following the manufacturer's protocol for double-stranded probes. For low to medium stringency, the membrane was washed twice for 15 min at room temperature (∼21°C) with 2× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) containing 0.1% (wt/vol) sodium dodecyl sulfate (SDS) and three times for 10 min at room temperature with 1× SSC buffer containing 0.1% (wt/vol) SDS. A final high-stringency wash was performed twice for 10 min at 60°C with 0.1× SSC buffer containing 0.1% (wt/vol) SDS. After washing, the membrane was sealed in Saran Wrap and exposed to BioMax MS film (Kodak) in an autoradiography cassette for 17 h at −70°C. The film was developed with an automatic X-ray film processor (Amersham).
Nucleotide sequence accession numbers.
The DDBJ/EMBL/GenBank accession numbers for the reference NHase nucleotide sequences used in the phylogenetic analysis are as follows: R. erythropolis JCM 6823, D14454; Rhodococcus rhodochrous IFO 15564, E12518; Rhodococcus sp. strain N-771, AB016078; Rhodococcus sp. strain N-774, E02364; Rhodococcus sp. strain ACV2, Z48769; and Rhodococcus sp. strain AJ270, AJ278349. The R. erythropolis NHase gene sequences determined in this study have been deposited in the GenBank database under the following accession numbers: ANT-AN007, AY223827; IND-AN014, AY223828; ARG-AN024, AY223830; ARG-AN025, AY223831; ENG-AN033, AY223832; 870-AN019, AY223829; 871-AN042, AY223833; 871-AN053, AY223834; 122-AN065, AY223826; 67-BEN001, AY223825; DSM 13002, AY223836; and DSM 43066T, AY223835.
RESULTS
Molecular screening.
We have found previously that the NHase activities and substrate specificities of mycolate actinomycetes are highly variable, not only among nitrile-hydrolyzing environmental isolates belonging to different species but also among strains of R. erythropolis (8). For this reason we investigated whether the variable enzyme activities observed could be due to genomic variation of the NHase- and amidase-encoding genes.
The NHase nucleotide sequences of Rhodococcus species available in the DDBJ/EMBL/GenBank databases were aligned, but there was a low level of sequence homology between the genes encoding the iron-containing NHase enzymes of rhodococci and the genes encoding the high- and low-molecular-mass cobalt-containing NHase enzymes of R. rhodochrous J-1 (43) and M8 (79). Although we were interested in designing primers that amplified NHase genes from as many rhodococcal strains as possible, we biased the study for sequences of the Fe-type NHase enzymes since these enzymes exhibited higher levels of homology. Consequently, we used enzyme gene sequences of R. erythropolis JCM 6823 (22), R. rhodochrous IFO 15564 (DDBJ/EMBL/GenBank accession number E12518), and nontaxonomically defined Rhodococcus strains N-771 (62, 86), N-774 (33), AJ270 (DDBJ/EMBL/GenBank accession number AJ278349), and ACV2 (DDBJ/EMBL/GenBank accession number Z48769); the latter strain is a mutant of Brevibacterium sp. strain R312 (52, 53). Strains N-774 (and probably N-771) and R312 (and consequently ACV2) are considered very similar microorganisms and have been identified as R. erythropolis strains (15; C. M. Hjort et al., unpublished data), whereas strain AJ270 had a 16S rRNA gene PRS pattern identical to that of R. erythropolis (data not shown). Consequently, almost all of the NHase-encoding nucleotide sequences used to design the primers originated from strains of R. erythropolis; the only exception was the R. rhodochrous IFO 15564 sequence. The primers that were designed amplified the αβ-nhase gene (αNH1-βNH2 primer pair), the individual α-nhase (αNH1-αNH2 primer pair) and β-nhase (βNH1-βNH2 primer pair) genes, and the amd gene (amd1-amd2 primer pair) from all of the nitrile-metabolizing R. erythropolis strains, and the expected sizes were approximately 1,241, 602, 639, and 1,687 bp, respectively. The same primers also amplified PCR products that were the same size from the nitrile-metabolizing organism R. erythropolis DSM 13002. Most interestingly, these primers also amplified products from the reference strain R. erythropolis DSM 43066T, a microorganism not able to grow with acetonitrile or benzonitrile as the sole carbon source (Table 1); i.e., there was no indication that a PCR product could be obtained from a non-nitrile-metabolizing strain. However, the same oligonucleotide primers were not suitable for amplifying any nhase or amd gene from the environmental isolates that were not taxonomically related to R. erythropolis. Different PCR conditions were examined in an attempt to amplify these genes in the nitrile-metabolizing actinomycetes not related to R. erythropolis, but only nonspecific products were obtained. We pursued detection of the NHase gene in these strains by using a hybridization technique.
Southern hybridization.
Because the primers designed to amplify the αβ-nhase gene only worked for strains of R. erythropolis, Southern hybridization was performed in an attempt to determine the NHase gene sequences present in other environmental nitrile-metabolizing mycolate actinomycetes. The amplified β-nhase gene from R. erythropolis 67-BEN001 was used as a probe to examine the distribution of the NHase genes among 13 actinomycetes known to possess nitrile-transforming enzymes, including Rhodococcus opacus strains ANT-AN002 and 871-AN040, Rhodococcus ruber USA-AN012, Rhodococcus equi strains 871-AN029 and 871-AN030, R. rhodochrous ARG-BN062, and Gordonia namibiensis strains NAM-BN063AT and NAM-BN063B (9, 10), as well as R. rhodochrous strains LL100-21 (21) and NCIMB 11216 (27) and the positive control strains R. erythropolis 67-BEN001 (31), DSM 13002, and DSM 43066T. The expected positive hybridization signals at approximately 6-kb (22) were obtained only with the genomic DNA of R. erythropolis strains (7). Similar results were obtained both at normal stringency (as used by Duran et al.) and at high stringency.
Molecular profiling of enzyme genes.
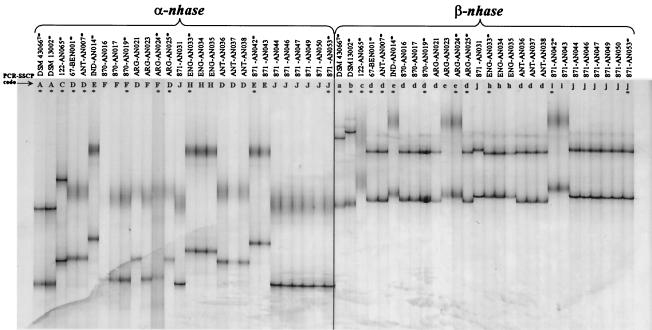
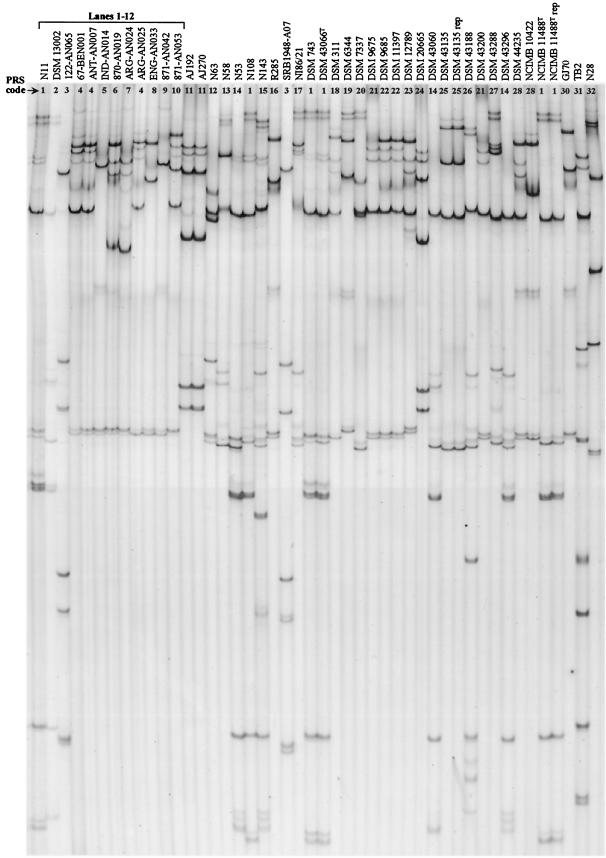
Only the strains that were identified as R. erythropolis were selected for further study of NHase and amidase gene diversity. The PCR-SSCP analysis of the independently amplified α-nhase and β-nhase genes of the R. erythropolis strains revealed a diversity of patterns (Fig. 1). Seven visually different patterns were produced as a result of PCR-SSCP analysis of the α-nhase genes (patterns A, C, D, E, F, H, and J) (Table 3), while the β-nhase gene analysis produced eight different patterns (patterns a, b, c, d, e, h, i, and j) (Table 3). The combinations of the α-nhase and β-nhase patterns resulted in different αβ-nhase patterns. For example, as shown in Table 3, strains DSM 43066T and DSM 13002 produced the same α-nhase pattern (pattern A). However, the corresponding β-nhase genes produced two different patterns (patterns a and b); therefore, the different combinations of the α-nhase and β-nhase genes produced two unique αβ-nhase PRS patterns for these two strains (patterns 1 and 2). When the complete αβ-nhase gene was used for PRS analysis of the same R. erythropolis strains, 10 different PRS patterns (patterns 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10) were obtained (Fig. 2, lanes 1 to 12, and Tables 1 and 3). Table 3 shows the relationships among the various α-nhase, β-nhase, and αβ-nhase patterns obtained for representative strains. Correlation of different α-nhase and β-nhase PCR-SSCP patterns for a strain always resulted in a unique αβ-nhase PRS pattern for the strain (Table 3). αβ-nhase patterns 1, 2, 3, 5, 6, 7, 8, 9, and 10 were unique for individual strains. Although strains 871-AN042 and 871-AN053 were recovered from the same deep-sea sediment (1,521 m, Suruga Bay) (9), they had different αβ-nhase PRS patterns (patterns 9 and 10, respectively) (Table 1 and Fig. 2). Similarly, strains ARG-AN024 and ARG-AN025, which were recovered from the same Argentinean soil, had αβ-nhase patterns 7 and 4, respectively (Table 1 and Fig. 2). The only isolates that produced the same αβ-nhase pattern (pattern 4) but were recovered from widely separated geographic locations were strains 67-BEN001, ANT-AN007, and ARG-AN025 (Table 1 and Fig. 2).
FIG. 1.
PCR-SSCP profiles of the NHase α-subunit gene (α-nhase) and β-subunit gene (β-nhase) present in R. erythropolis strains. A complete list of the recently recovered R. erythropolis strains used in the PCR-SSCP assay is available elsewhere (9). An asterisk indicates a representative strain to which a PCR-SSCP pattern code was assigned; PCR-SSCP pattern codes for representative strains are shown in Table 3.
TABLE 3.
Relationship between the PCR-SSCP patterns of the α-nhase and β-nhase genes and the PRS patterns of the complete NHase gene (αβ-nhase) obtained for representative R. erythropolis strains
| R. erythropolis strain | Pattern codea
|
||
|---|---|---|---|
| PCR-SSCP
|
PRS αβ-nhase | ||
| α-nhase | β-nhase | ||
| DSM 43066T | A | a | 1 |
| DSM 13002 | A | b | 2 |
| 122-AN065 | C | c | 3 |
| 67-BEN001 | D | d | 4 |
| ANT-AN007 | D | d | 4 |
| ARG-AN025 | D | d | 4 |
| IND-AN014 | E | e | 5 |
| 871-AN042 | E | i | 9 |
| 870-AN019 | F | d | 6 |
| ARG-AN024 | F | e | 7 |
| ENG-AN033 | H | h | 8 |
| 871-AN053 | J | j | 10 |
Boldface type indicates unique patterns.
FIG. 2.
PRS profiles of the complete NHase genes present in R. erythropolis strains isolated in our laboratory and obtained from other laboratories and culture collections. PCR products were digested with restriction enzyme AccI. The αβ-nhase PRS pattern codes assigned to the strains are shown in Table 1.
Molecular profiling of the amidase genes amplified from the same R. erythropolis strains that were used for the NHase gene diversity study resulted in 10 different PRS patterns (Table 1) (7). A correlation identical to that obtained for the αβ-nhase patterns was obtained for the amd PRS patterns of the R. erythropolis strains and the geographical sites from which the strains were recovered (Table 1).
Diversity of αβ-nhase and amd PRS patterns in R. erythropolis strains.
The fact that our collection of nitrile-metabolizing R. erythropolis strains produced distinct αβ-nhase and amd PRS patterns led us to question if such diversity existed in other, previously described R. erythropolis strains and if the primers could amplify genes from other strains of the same species (Table 1). Prior to this study we confirmed the taxonomic identities of all strains by PRS analysis of the 16S rRNA gene (data not shown), as described by Brandão et al. (11), and only the strains with a PRS pattern visually identical to the R. erythropolis DSM 43066T pattern were investigated further. All doubtful strains were reexamined for culture purity, DNA extraction, PCR amplification, and PRS analysis of the 16S rRNA gene. Occasionally, we came across a strain that did not produce the R. erythropolis 16S rRNA gene PRS pattern, which could have been due to misidentification of the microorganism or to contamination of the culture.
The PRS analysis of αβ-nhase of validated strains of R. erythropolis revealed considerable gene diversity (Fig. 2). Including the 10 αβ-nhase PRS patterns described above (Fig. 2, lanes 1 to 12), 31 αβ-nhase PRS patterns were found for the 45 isolates analyzed (Table 1). R. erythropolis strains N11T (= NCIB 9158T), N108T (= ATCC 4277T), DSM 743, and NCIMB 11148T are equivalent to DSM 43066T and, accordingly, produced the same αβ-nhase PRS pattern, pattern 1. Strains AJ192 and AJ270 produced the same PRS pattern (pattern 11), but these strains could have been subcultures of the same microorganism (5). Similarly, strains DSM 9675, DSM 9685, and DSM 11397, all isolated from soils from the same geographical region (46, 47), had very similar αβ-nhase PRS patterns (patterns 21 and 22); pattern 21 was also obtained for strain DSM 43200. The same αβ-nhase PRS profile (pattern 3) was obtained for deep-sea strains 122-AN065 and SRB1948-A07, although the organisms were recovered from geographically different sediments and depths. All other strains had unique αβ-nhase PRS patterns. Again, a correlation between the amd PRS patterns of the R. erythropolis strains and their geographic origins was observed, as found for the αβ-nhase patterns and the strains (Table 1) (7).
Although the primers that were designed detected unique αβ-nhase and amd genes in most of the strains of R. erythropolis, they could not amplify these genes from three strains (N60, DCL14, and KA2-5-1 [Table 1]) that were assumed to be R. erythropolis strains on the basis of 16S rRNA gene PRS analysis. Conversely, αβ-nhase was amplified from isolate N28 (Fig. 2, lane 32), a strain that did not generate an R. erythropolis 16S rRNA gene PRS pattern; however, the amd gene could not be amplified from this strain (Table 1).
NHase sequences.
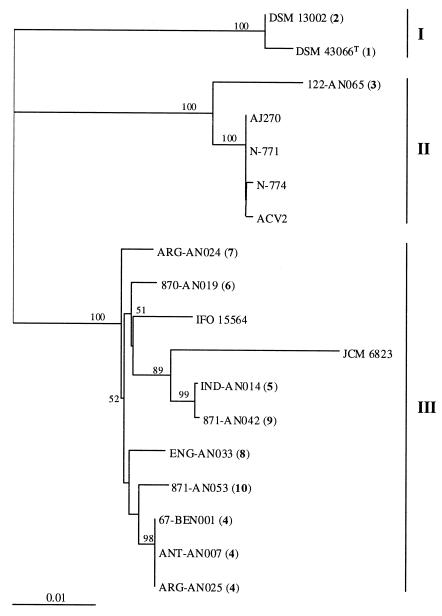
We sequenced the genes encoding the NHase α- and β-subunits from a selection of our R. erythropolis environmental strains based on their geographic origins and the unique PRS patterns that they represented; the strains studied were ANT-AN007, IND-AN014, ARG-AN024, ARG-AN025, ENG-AN033, 870-AN019, 871-AN042, 871-AN053, 122-AN065, 67-BEN001, DSM 13002, and DSM 43066T. The most closely related nucleotide sequences in the DDBJ/EMBL/GenBank databases were the sequences of R. rhodochrous IFO 15564, R. erythropolis JCM 6823, and Rhodococcus sp. strains N-771, N-774, ACV2, and AJ270 and the partial α-nhase sequences of Rhodococcus sp. strain R312 and R. erythropolis strains JCM 3191 and JCM 3132 (accession numbers M60264, AJ306169, and AJ306170, respectively). The α-nhase partial sequences were not included in the phylogenetic analysis. The levels of similarity between the αβ-nhase nucleotide sequences were between 92.4 and 100%. The phylogenetic relationships of the various αβ-nhase genes were represented in a neighbor-joining tree, which had three main clusters (clusters I, II, and III) (Fig. 3). The grouping integrity was supported by the 100% bootstrap value obtained for each cluster and by the results obtained from the application of all four treeing methods.
FIG. 3.
Neighbor-joining tree showing the phylogenetic relationships of the complete NHase genes (αβ-nhase) from R. erythropolis strains and other Rhodococcus species. The αβ-nhase PRS pattern code for each strain is indicated in parentheses (Fig. 2 and Table 1). The tree was constructed based on an examination of 1,282 NHase gene nucleotide positions. The values at the nodes indicate the levels of bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets. Scale bar = 0.01 substitution per nucleotide position.
Cluster I included the enzyme sequences of R. erythropolis strains DSM 13002 and DSM 43066T, while cluster II grouped the NHase sequence of the deep-sea strain R. erythropolis 122-AN065 with the sequences of four Rhodococcus sp. strains, N-771, N-774, AJ270, and ACV2. Cluster III grouped all the protein-coding sequences of the remaining nine environmental nitrile-metabolizing R. erythropolis strains, together with the sequences of R. erythropolis JCM 6823 and R. rhodochrous IFO 15564. Strain JCM 6823 is on a longer branch than the two most closely related strains, IND-AN014 and 871-AN042 (Fig. 3), due to its extra nucleotide substitutions. There was high congruence between the αβ-nhase PRS patterns obtained for the environmental isolates and the sequencing results, since each PRS pattern corresponded to a unique αβ-nhase sequence (Fig. 2 and 3 and Table 3). Although strains IND-AN014 and 871-AN042 (cluster III) had very similar αβ-nhase PRS patterns (patterns 5 and 9) (Fig. 2), their nucleotide sequences had one base difference (99.9% similarity) in the β-nhase region. As expected, this base shift was observed in their β-nhase PCR-SSCP patterns (patterns e and i) (Fig. 1 and Table 3) but not in their α-nhase patterns (pattern E) (Fig. 1 and Table 3).
The deduced amino acid sequences encoded by α-nhase and β-nhase were highly conserved in the R. erythropolis strains examined, with identity values ranging from 94.7 to 100% for the α-nhase product and from 93.9 to 100% for the β-nhase product. There were 15 positions in the α-nhase-encoded amino acid sequences studied with variable residues, and there were 19 positions in the β-nhase-encoded sequences with variable residues (Fig. 4). The substituted residues of the deduced α-nhase- and β-nhase-encoded amino acid sequences were not located at positions which are highly conserved in known Fe-type (22, 33, 49, 52, 61, 62, 86) and Co-type (43, 64) NHase and thiocyanate hydrolase (36) enzymes and which are thought to have direct effects on stabilization of the active cavity sites of the enzymes (56, 59). Nevertheless, some of these residues were located at neighboring positions or positions very close to other regions that were not directly related to the active site but were completely conserved in all the enzymes mentioned above, whose effects on the structure, activity, and substrate specificity of the final active enzyme are potentially significant.
FIG.4.
Alignment of deduced amino acid sequences of the α-subunit (A) and β-subunit (B) of R. erythropolis NHases. The residue numbers are in accordance with those of the Fe-type NHase from strain R312 (32). Note that because the methionine (M) residue at the N terminus is cotranslationally removed in the α-subunit of the strain N-771 enzyme, the residue numbers for this NHase α-subunit should be one less than the numbers for the other enzymes (77). The NHases of the following strains were used: N-774 (33), N-771 (62, 86), AJ270 (DDBJ/EMBL/GenBank accession number AJ278349), ACV2 (DDBJ/EMBL/GenBank accession number Z48769), R312 (49), 122-AN065 (122), ENG-AN033 (33), DSM 13002 (DSM), DSM 43066T (N11), ARG-AN024 (24), IND-AN014 (14), 871-AN042 (42), 871-AN053 (53), 870-AN019 (19), ANT-AN007 (7), ARG-AN025 (25), 67-BEN001 (67) (this study), R. rhodochrous IFO 15564 (IFO) (DDBJ/EMBL/GenBank accession number E12518), and JCM 6823 (JCM) (22). Different amino acids in a single sequence or in different sequences are indicated by a black background. The residues conserved in Fe- and Co-type NHases and thought to have direct effects in the active site are indicated by boldface type (32, 59). The gray background indicates the residues that form the iron active center of Fe-type NHase (32, 56). Asterisks indicate the conserved residues in Fe- and Co-type NHases and thiocyanate hydrolase (36, 59).
The partial nucleotide sequence of the NHase activator Orf1188 gene was determined for all of the R. erythropolis strains mentioned above except strains DSM 13002 and DSM 43066T. The most closely related nucleotide sequences in the databases were those of R. erythropolis JCM 6823 (accession number D14454) and Rhodococcus sp. strains N-771 (AB016078), N-774 (D30033), ACV2 (Z48769), and AJ270 (AJ278349 and AJ490527).
NHase cryptic genes.
Of the 43 strains of R. erythropolis from which there was NHase gene amplification, 8 strains were not able to grow on acetonitrile or benzonitrile (Table 1). This result suggested the presence of possible cryptic genes, which may be of two types (34): those which encode a known function that is not normally expressed and those which are expressed but whose function is unknown or whose protein product is simply inactive due, for example, to misfolding. Analysis of the NHase gene sequences showed that the non-nitrile-metabolizing strain R. erythropolis DSM 43066T contained a sequence that was different at only four nucleotides from the most similar NHase gene sequence present in the nitrile-metabolizing strain R. erythropolis DSM 13002 (99.7% identity) (data not shown). Three of the nucleotide substitutions were present in the β-nhase region, and this shift was also reflected by the different PCR-SSCP patterns obtained (cf. β-nhase patterns a and b in Fig. 1). Three of the four nucleotide base substitutions were responsible for the three different amino acid residues observed on the deduced NHase amino acid sequence of R. erythropolis DSM43066T; at one position (Arg184A, where the superscript A indicates the NHase α-subunit) in the α-subunit and at two positions (Ser71B and Ala118B, where the superscript B indicates the NHase β-subunit) in the β-subunit there were amino acid residues that were unique among all the R. erythropolis Fe-type NHase sequences analyzed (the original conserved residues were Gln184A, Pro71B, and Val118B, respectively) (Fig. 4).
DISCUSSION
NHse and amidase distribution and crypticity.
The previous observation that geographically distinct strains of R. erythropolis produced inducible or constitutive NHase and amidase enzymes with significantly variable activities and characteristically broad substrate specificities (8) highlighted the possible genetic diversity of the nitrile-transforming enzymes in a single species. The molecular screening approach used in this study allowed us to detect and describe the enzyme gene diversity directly from genomic DNA without resorting to more laborious and time-consuming protein purification and N-terminal sequencing methods.
Amplification of putative NHase and amidase genes gave PCR products of the expected sizes, but only for the environmental and laboratory strains of R. erythropolis. The fact that we used homologous NHase and amidase sequences mainly found in R. erythropolis strains or very closely species to design our oligonucleotide primers probably explains why the primers amplified only these genes from the same species. Further evidence has recently been provided by Precigou et al. (69), who used an approach similar to our approach for primer design. These authors constructed degenerate primers based on homologies found among Fe- and Co-type NHases to amplify NHase genes, but because the primers were too degenerate and amplified only nonspecific fragments, they decided to separate Fe- and Co-type NHase sequences in order to construct new primers. The redesigned Fe-type NHase primer set amplified only NHase genes from R. erythropolis strains JCM 3132 and JCM 3191 among the 16 actinomycete taxa that were studied (69), a result similar to our results. It should be noted that strains JCM 3132 and JCM 3139 are equivalent to strains DSM 43060 and DSM 43188, respectively, which were incorporated into our PRS profile of the NHase gene (patterns 14 and 26, respectively) (Table 1 and Fig. 2).
The fact that 14 of the 16 actinomycetes screened by Precigou et al. (69) for NHase genes did not produce expected PCR products, even with Co-type NHase primers, could have been because (i) the primers that were designed did not amplify the presumed NHases present in these bacteria or (ii) NHase genes were not present in these bacteria. Resolution of these questions is difficult, because the authors did not report whether they checked for growth on nitriles. Conversely, in the present study we tested our Fe-type NHase primers with known nitrile-metabolizing non-R. erythropolis strains; the lack of amplicon production suggested that primer specificity was responsible.
In our study, although strains N60, DCL14, and KA-2-5-1 had a 16S rRNA gene PRS pattern identical to that of R. erythropolis (results not shown), it was not possible to amplify the αβ-nhase or amd gene from their DNA (Table 1). As these genes were amplified from all other R. erythropolis strains tested, it cannot be assumed that this Fe-type NHase gene group is universally present in this species. The fact that R. erythropolis strains N60, DCL14, and KA-2-5-1 could not grow on acetonitrile or benzonitrile (Table 1) suggests that they do not possess nitrile-hydrolyzing enzymes. However, the data do not rule out the possible presence of cryptic Co-type NHase or nitrilase genes.
The primers which we designed to amplify the NHase and amidase genes did not work with all R. erythropolis strains; nevertheless, they could be useful for detecting these protein-coding genes directly in DNA extracted from environmental samples in order to help elucidate the geographic distribution of nitrile-metabolizing microorganisms, to understand the evolution of these enzymes in rhodococci, and to find novel NHase enzymes with unusual characteristics. This strategy has been tested by Precigou et al. (69) for rapid and specific identification of NHase-encoding genes in soils. These authors successfully detected Fe-type and Co-type NHases in all soil samples tested; some of these samples contained NHase sequences similar to the sequences of previously described enzymes, and others contained sequences never before reported for culturable nitrile-metabolizing microorganisms. Furthermore, these authors were able to express NHase genes recovered from soil in E. coli, although the activities were lower than that observed for R. erythropolis JCM 2892 NHase (22).
Duran et al. (22) examined the distribution of NHase genes in 31 species belonging to 17 genera of bacteria using the NHase gene from Rhodococcus sp. strain N-774 as a probe, and they reported positive Southern hybridization with only two strains of R. erythropolis that were analyzed. None of the other bacteria gave a positive result, suggesting that there was no NHase gene. However, because the latter strains were not tested for NHase activity, it is not possible to be categorical on this point. We used a hybridization method similar to that used by Duran et al. to analyze the presence of the NHase gene in actinomycetes known to have nitrile-transforming activities, but no hybridization occurred except for the hybridization that occurred with R. erythropolis strains used as positive controls (7). For this reason the possibility that there were NHase genes in the strains analyzed by Duran et al. (22) cannot be ruled out because the fact that no hybridization occurred does not mean that the bacteria did not harbor the NHase gene. Growth on or activity towards nitriles should be tested with pure cultures used in molecular screening of the NHase since the genes can be highly diverse in different bacterial genera and species, as shown by the nonspecific hybridization signal obtained with our probe for the non-R erythropolis nitrile-metabolizing isolates.
The fact that there was amplification of nhase and amd genes from strains N63, N143, DSM 43188, N11, N108, DSM743, NCIMB 11148T, and DSM 43066T, all of which were rhodococci that were unable to grow on the nitriles tested (Table 1), suggests that cryptic genes encoding NHase and amidase are present in these isolates. Cryptic genes are not usually expressed; i.e., they are phenotypically silent at the level of transcription or translation, but they do not have permanent loss of normal function (26). They can be periodically activated in a few members of a population by decryptification (reversion) under appropriate environmental circumstances to provide a selective advantage. Thus, cryptic genetic systems involving cycles of inactivation and reactivation represent a form of long-term gene regulation of rarely utilized functions operating at the level of bacterial populations (26). A silent gene that produces an inactive or relatively inactive product may be involved in the evolution of new enzymes or novel enzymatic specificities (34). If a certain metabolic demand is no longer required, a gene may acquire a mutation which either eliminates expression of the gene or eliminates activity of the gene product (2); in the latter case, a mutation which results in incorrect polypeptide folding is most likely. The accumulation of mutations free from the pressure of natural selective constraints may result in genes from which a new or improved enzymatic specificity may emerge (2).
NHase and amidase diversity in R. erythropolis.
We have shown previously that PRS analysis can be used as a presequencing step to verify the diversity of the 16S rRNA genes in Rhodococcus species (9, 11). We extended this approach to PRS analysis of the NHase and amidase genes and found a high degree of diversity among geographically dispersed strains of R. erythropolis. A selection of genes from the NHase PRS patterns were sequenced, and in most cases, the sequences were unique to individual R. erythropolis strains (Fig. 3), showing that these protein-encoding genes, although geographically well distributed, appear not to be globally mixed.
One should be cautious when referring to the occurrence of cosmopolitan microorganisms (ubiquitous geographic distribution) or endemic microorganisms (restricted to one geographic location) in the environment, and a set of postulates has been proposed, the fulfillment of which would be necessary to categorize an organism as cosmopolitan (73). Testing these postulates was not an aim of our investigation, and for this reason the data are not sufficient to infer that there are geovars (geographic varieties of microorganisms that are endemic to a specific area or host [73]) or candidate endemic species among our collection of geographically distinct R. erythropolis strains. Nevertheless, the fact that strains of R. erythropolis from different geographical areas possess biogeographically distinct NHase genes with high levels of sequence similarity (92 to 100%) suggests that there is an evolutionary trait that is dependent on habitat characteristics. This infraspecific variation of NHase genes in R. erythropolis demonstrates the intimate relationship between environmental or geographic factors and the speciation of microorganisms (14).
Aldoximes are widely distributed as intermediates in the biosynthesis of plant cyanogenic glucosides (80), and the presence in bacteria of aldoxime dehydratases together with nitrile-metabolizing enzymes has been reported previously (1, 37). The fact that NHase genes were found to be present in geographically separated R. erythropolis strains may signify an important catabolic function, such as a function involved in the metabolism of aldoximes (38).
It was interesting that without exception, the NHase and amidase genes of each R. erythropolis strain had complementary PRS patterns (Table 1) related to the site from which the organisms were recovered. This finding suggests that these genes, which have adjacent locations in many of the NHase-amidase systems that have been genetically analyzed (42), have undergone coevolution. Comparison of amidase phylogeny with NHase phylogeny revealed similar evolution of these two enzymes (3, 15, 69). The physical proximity and coevolution of NHase and amidase genes point to the emergence of this gene cluster for a specific metabolic function, namely, the transformation of nitriles, via amides, into acids.
The NHase genes of R. erythropolis strains 67-BEN001, ANT-AN007, and ARG-AN025 were the only genes that had identical sequences, even though the organisms were recovered from geographically different regions and dramatically different habitats. The activities of strains 67-BEN001 and ANT-AN007 towards nitriles and amides have been shown to be slightly different (8, 31). Therefore, even though geographically distinct environmental strains may have identical NHase gene sequences, it does not follow that their activity profiles are identical. Similar findings were obtained for Rhodococcus strains N-771, N-774, and AJ270, isolated from different soil samples, which although possessing identical NHase gene and amino acid sequences, had distinct nitrile-transforming activities (6, 81).
Amino acid sequences of NHase.
The deduced amino acid sequences of the NHases of R. erythropolis were very similar; however, although the substituted residues were located not in the NHase active site cavity (32, 59) but in nearby regions, they could be responsible for the observed variability of activities (8). Thus, Rhodococcus strain ACV2 (originating from R. erythropolis R312 [53]) had a single substitution (Met40B → Val40B) located at a position that forms the entrance channel from the bulk solvent to the active site (59); the mutant NHase activity differed from the wild-type enzyme activity in its pI, optimum pH, and rates of hydrolysis of cyanovaleramide and cyanovaleric acid, which were 30- and 15-fold greater (52). It has been proposed that modification of residues that form the entrance channel of an active site may enable design of novel NHase enzymes having wider substrate specificities (59). Nevertheless, the possibility should also be considered that the diversity observed among the various NHase sequences may not be related to the variability detected in the NHase activities, which were measured by using whole cells (8), but rather may be dependent on infraspecies diversity of other kind.
Translation of the NHase nucleotide sequence of R. erythropolis DSM 43066T revealed positions in both the α- and β-subunits containing amino acid residues that were unique among all the R. erythropolis sequences analyzed (Fig. 4). These amino acid changes could play an important role in the lack of expression of the NHase enzyme and consequently could affect the growth of R. erythropolis DSM 43066T on nitrile compounds. Particularly interesting was the Pro71B substitution in the β-subunit, since this is the neighbor position of two residues (Tyr72B and Tyr73B) of the highly conserved amino acids that surround the active site structure in the NHase family, whose stabilized conformation is achieved by hydration water molecules (59). The interference of this substitution with the extensive network of hydrogen bonds that surround the active site could be responsible for the nonfunctionality of this enzyme. By generating a point mutation at arginine residue 56 in the NHase β-subunit (Arg56B) of Rhodococcus sp. strain N-771, Piersma et al. (66) obtained mutant enzymes with limited stability and no activity or low activity compared to that of the wild-type enzyme. This residue, which is conserved in most known NHases (22, 33, 43, 49, 52, 61, 62, 64, 86; DDBJ/EMBL/GenBank accession numbers E12518 and AJ278349) and also in the homologous thiocyanate hydrolase (36), is considered to be essential for the catalytic activity of NHase, as it is involved not only in substrate binding but also in positioning the iron catalytic center so that it is suitable for hydration of nitriles by formation of hydrogen bonds between its guanidium group and modified cysteine residues (66). Another possibility to explain the lack of activity of R. erythropolis DSM 43066T is impaired interactions between the α- and β-subunits; the substitution is next to Tyr72B, which is one of the amino acids engaged in subunit interactions for the formation of a stable αβ-NHase structure (59). The lack of effective interactions between subunits could jeopardize the enzyme stability and hence the activity of the enzyme. However, substitutions near amino acids involved in subunit interactions or substitutions of such amino acids could also be responsible for altered activity and/or substrate specificity of the enzyme, as is the case for Gly22A, Ser19B, Ser158B, and Leu200B in R. erythropolis JCM 6823 and Ile148A in R. rhodochrous IFO 15564, which are unique among the Fe-type NHases (Fig. 4), or for Ile156A/Val156A, Asp93B/Glu93B, Ser154B/Ala154B, and Ser158B/Thr158B, which are different in different groups of R. erythropolis Fe-type NHase sequences (Fig. 4) (59). The highly conserved β-nhase amino acid Leu208B found in most NHase and thiocyanate hydrolase enzymes was replaced by a phenylalanine (Phe208B) in R. erythropolis strain 870-AN019 NHase (Fig. 4B), suggesting that this change could be responsible for the different catalytic activities. Currently, this strain is being considered as a candidate for biotransformation of a particular nitrile into an industrially important sterically hindered amide (P. F. B. Brandão and C. Syldatk, unpublished data).
The Fe-type NHases, particularly that of Rhodococcus sp. strain N-771, have been more thoroughly characterized than the Co-type NHases because of their unique photoreactivity (54). In vivo, the enzymatic activity of the N-771 NHase during aerobic incubation is subject to dark inactivation, but it is recovered almost completely upon light irradiation (photoreactivation). However, the photoreactivated enzyme cannot be inactivated by darkness in vitro (55). Similar results have been obtained for other Fe-type NHases, including those of Rhodococcus sp. strain N-774 (58) and Rhodococcus sp. strain R312 (13), which have amino acid sequences identical to that of Rhodococcus sp. strain N-771 (Fig. 4) (33, 49, 62). The NHase activities of our strains were not tested under dark and light conditions for a comparison of the effects on the NHase enzyme activity; however, the high levels of amino acid sequence identity (>92%) among all the environmental R. erythropolis NHases sequenced and the NHases from Rhodococcus sp. strains N-771, N-774, and R312 (Fig. 4A and B) suggest that the former enzymes could also be subject to photoreactivity.
Investigation of microorganisms at the infraspecific level is crucial in the context of biotechnological discoveries because many sought-after properties are known to be strain specific as opposed to species specific (14). The considerable molecular diversity which we found among the NHases and amidases of geographically distinct deep-sea and terrestrial R. erythropolis strains demonstrates clearly the extent of infraspecies variability. The amino acid variation found for NHases could make these enzymes prime candidates for artificial enzyme evolution development (51, 60, 67) and attempts to obtain novel activities. Raillard et al. (70) used directed evolution for two highly homologous triazine hydrolases and found that the shuffled library contained enzymes that not only exhibited up to 150-fold-greater transformation rates than either parental enzyme exhibited but also hydrolyzed five of eight triazines that were not substrates for either starting enzyme. Modification of the physical or catalytic properties of nitrile-transforming enzymes could produce novel biocatalysts of major industrial interest, for example, for the stereoselective transformation of α-aminonitriles and/or α-hydroxynitriles into l- or d-amino acids.
Acknowledgments
P.F.B.B. acknowledges Fundação para a Ciência e a Tecnologia, Portugal, for a postgraduate research scholarship (PRAXIS XXI/BD/11463/97).
We thank Stefan Verseck and coworkers from Degussa AG, Hanau-Wolfgang, Germany, for their help in part of the sequencing experiments. We also thank C. J. Knowles, M. Barclay, R. De Mot, M. Goodfellow, D. B. Janssen, H.-J. Knackmuss, J. Konishi, C. Syldatk, and M. J. van der Werf for kindly providing some of the R. erythropolis strains used in this study.
REFERENCES
- 1.Asano, Y., and Y. Kato. 1998. Z-phenylacetaldoxime degradation by a novel aldoxime dehydratase from Bacillus sp. strain OxB-1. FEMS Microbiol. Lett. 158:185-190. [Google Scholar]
- 2.Beacham, I. R. 1987. Silent genes in prokaryotes. FEMS Microbiol. Rev. 46:409-417. [Google Scholar]
- 3.Bigey, F. 1995. Ph.D. thesis. Ecole Nationale Superieure Agronomique de Montpellier, Montpellier, France.
- 4.Bisset, K. A., and F. Moore. 1950. Jensenia, a new genus of the Actinomycetales. J. Gen. Microbiol. 4:280. [DOI] [PubMed] [Google Scholar]
- 5.Blakey, A. J. 1994. Isolation of nitrile utilising microorganisms and physiological and biochemical investigation of Rhodococcus nov. sp. AJ270 and investigation of its nitrile hydratase. Ph.D. thesis. University of Sunderland, Sunderland, United Kingdom.
- 6.Blakey, A. J., J. Colby, E. Williams, and C. O'Reilly. 1995. Regio-specific and stereo-specific nitrile hydrolysis by the nitrile hydratase from Rhodococcus AJ270. FEMS Microbiol. Lett. 129:57-61. [Google Scholar]
- 7.Brandão, P. F. B. 2001. Diversity and biotransformation activities of nitrile metabolising actinomycetes from deep-sea sediments and terrestrial soils. Ph.D. thesis. University of Kent at Canterbury, Canterbury, United Kingdom.
- 8.Brandão, P. F. B., and A. T. Bull. 2003. Nitrile hydrolysing activities of deep-sea and terrestrial mycolate actinomycetes. Antonie Leeuwenhoek 84:89-98. [DOI] [PubMed]
- 9.Brandão, P. F. B., J. P. Clapp, and A. T. Bull. 2002. Discrimination and taxonomy of geographically diverse strains of nitrile-metabolising actinomycetes using chemometric and molecular sequencing techniques. Environ. Microbiol. 4:262-276. [DOI] [PubMed] [Google Scholar]
- 10.Brandão, P. F. B., L. Maldonado, A. C. Ward, A. T. Bull, and M. Goodfellow. 2001. Gordonia namibiensis sp. nov., a novel nitrile metabolising actinomycete recovered from an African sand. Syst. Appl. Microbiol. 24:510-515. [DOI] [PubMed] [Google Scholar]
- 11.Brandão, P. F. B., M. Torimura, R. Kurane, and A. T. Bull. 2002. Dereplication for biotechnology screening: PyMS analysis and PCR-RFLP-SSCP (PRS) profiling of 16S rRNA genes of marine and terrestrial actinomycetes. Appl. Microbiol. Biotechnol. 58:77-83. [DOI] [PubMed] [Google Scholar]
- 12.Brennan, B. A., G. Alms, M. J. Nelson, L. T. Durney, and R. C. Scarrow. 1996. Nitrile hydratase from Rhodococcus rhodochrous J1 contains a non-corrin cobalt ion with two sulfur ligands. J. Am. Chem. Soc. 118:9194-9195. [Google Scholar]
- 13.Brennan, B. A., J. G. Cummings, D. B. Chase, I. M. Turner, Jr., and M. J. Nelson. 1996. Resonance Raman spectroscopy of nitrile hydratase, a novel iron-sulfur enzyme. Biochemistry 35:10068-10077. [DOI] [PubMed] [Google Scholar]
- 14.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chebrou, H., F. Bigey, A. Arnaud, and P. Galzy. 1996. Study of the amidase signature group. Biochim. Biophys. Acta 1298:285-293. [DOI] [PubMed] [Google Scholar]
- 16.Clapp, J. P. 1999. The identification of root-associated fungi by polymerase chain reaction-single-strand conformational polymorphism (PCR-SSCP), p. 1-18. In A. D. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Clapp, J. P., I. Mansur, J. C. Dodd, and P. Jeffries. 2001. Ribotyping of rhizobia nodulating Acacia mangium and Paraserianthes falcataria from different geographical areas in Indonesia using PCR-RFLP-SSCP (PRS) and sequencing. Environ. Microbiol. 3:273-280. [DOI] [PubMed] [Google Scholar]
- 18.Colquhoun, J. A., S. C. Heald, L. Li, J. Tamaoka, C. Kato, K. Horikoshi, and A. T. Bull. 1998. Taxonomy and biotransformation activities of some deep-sea actinomycetes. Extremophiles 2:269-277. [DOI] [PubMed] [Google Scholar]
- 19.Colquhoun, J. A., J. Mexson, M. Goodfellow, A. C. Ward, K. Horikoshi, and A. T. Bull. 1998. Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie Leeuwenhoek 74:27-40. [DOI] [PubMed] [Google Scholar]
- 20.Dalbøge, H., and L. Lange. 1998. Using molecular techniques to identify new microbial biocatalysts. Trends Biotechnol. 16:265-272. [DOI] [PubMed] [Google Scholar]
- 21.DiGeronimo, M. J., and A. D. Antoine. 1976. Metabolism of acetonitrile and propionitrile by Nocardia rhodochrous LL100-21. Appl. Environ. Microbiol. 31:900-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duran, R., M. Nishiyama, S. Horinouchi, and T. Beppu. 1993. Characterization of nitrile hydratase genes cloned by DNA screening from Rhodococcus erythropolis. Biosci. Biotechnol. Biochem. 57:1323-1328. [DOI] [PubMed] [Google Scholar]
- 23.Fredricks, K. M. 1967. Products of the oxidation of n-decane by Pseudomonas aeruginosa and Mycobacterium rhodochrous. Antonie Leeuwenhoek 33:41-48. [DOI] [PubMed] [Google Scholar]
- 24.Goodfellow, M. 1971. Numerical taxonomy of some nocardioform bacteria. J. Gen. Microbiol. 69:33-80. [DOI] [PubMed] [Google Scholar]
- 25.Goodfellow, M., and G. Alderson. 1977. The actinomycete genus Rhodococcus: a home for the ‘rhodochrous’ complex. J. Gen. Microbiol. 100:99-122. [DOI] [PubMed] [Google Scholar]
- 26.Hall, B. G., S. Yokoyama, and D. H. Calhoun. 1983. Role of cryptic genes in microbial evolution. Mol. Biol. Evol. 1:109-124. [DOI] [PubMed] [Google Scholar]
- 27.Harper, D. B. 1977. Microbial metabolism of aromatic nitriles, enzymology of C-N cleavage by Nocardia sp. (Rhodococcus group) NCIB 11216. Biochem. J. 165:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmans, S., M. W. Jansen, M. J. van der Werf, and J. A. de Bont. 1991. Bacterial metabolism of 3-chloroacrylic acid. J. Gen. Microbiol. 137:2025-2032. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto, Y., M. Nishiyama, S. Horinouchi, and T. Beppu. 1994. Nitrile hydratase gene from Rhodococcus sp. N-744: requirement for its downstream region for efficient expression. Biosci. Biotechnol. Biochem. 58:1859-1865. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto, Y., M. Nishiyama, O. Ikehata, S. Horinouchi, and T. Beppu. 1991. Cloning and characterisation of an amidase gene from Rhodococcus species N-744 and its expression in Escherichia coli. Biochim. Biophys. Acta 1088:225-233. [DOI] [PubMed] [Google Scholar]
- 31.Heald, S. C., P. F. B. Brandão, R. Hardicre, and A. T. Bull. 2001. Physiology, biochemistry and taxonomy of deep-sea nitrile metabolising Rhodococcus strains. Antonie Leeuwenhoek 80:169-183. [DOI] [PubMed] [Google Scholar]
- 32.Huang, W., J. Jia, J. Cummings, M. Nelson, G. Schneider, and Y. Lindqvist. 1997. Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold. Structure 5:691-699. [DOI] [PubMed] [Google Scholar]
- 33.Ikehata, O., M. Nishiyama, S. Horinouchi, and T. Beppu. 1989. Primary structure of nitrile hydratase deduced from the nucleotide sequence of a Rhodococcus species and its expression in Escherichia coli. Eur. J. Biochem. 181:563-570. [DOI] [PubMed] [Google Scholar]
- 34.Innes, D., I. R. Beacham, C. A. Beven, M. Douglas, M. W. Laird, J. C. Joly, and D. M. Burns. 2001. The cryptic ushA gene (ushAc) in natural isolates of Salmonella enterica (serotype Typhimurium) has been inactivated by a single missense mutation. Microbiology 147:1887-1896. [DOI] [PubMed] [Google Scholar]
- 35.Janssen, D. B., D. Jager, and B. Witholt. 1987. Degradation of n-haloalkanes and α,ω-dihaloalkane by wild type and mutants of Acinetobacter sp. strain GJ70. Appl. Environ. Microbiol. 53:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama, Y., Y. Matsushita, M. Kaneko, M. Kondo, T. Mizuno, and H. Nyunoya. 1998. Cloning of genes coding for the three subunits of thiocyanate hydrolase of Thiobacillus thioparus THI 115 and their evolutionary relationships to nitrile hydratase. J. Bacteriol. 180:2583-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato, Y., R. Ooi, and Y. Asano. 1998. Isolation and characterisation of a bacterium possessing a novel aldoxime-dehydration activity and nitrile-degrading enzymes. Arch. Microbiol. 170:85-90. [DOI] [PubMed] [Google Scholar]
- 38.Kato, Y., R. Ooi, and Y. Asano. 2000. Distribution of aldoxime dehydratase in microorganisms. Appl. Environ. Microbiol. 66:2290-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsivela, E., D. Bonse, A. Kruger, C. Strompl, A. Livingston, and R. M. Wittich. 1999. An extractive membrane biofilm reactor for degradation of 1,3-dichloropropene in industrial waste water. Appl. Microbiol. Biotechnol. 52:853-862. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi, M., and S. Shimizu. 1998. Metalloenzyme nitrile hydratase: structure, regulation, and application to biotechnology. Nat. Biotechnol. 16:733-736. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi, M., and S. Shimizu. 2000. Nitrile hydrolases. Curr. Opin. Chem. Biol. 4:95-102. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi, M., T. Nagasawa, and H. Yamada. 1992. Enzymatic synthesis of acrylamide: a success story not yet over. Trends Biotechnol. 10:402-408. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi, M., M. Nishiyama, T. Nagasawa, S. Horinouchi, T. Beppu, and H. Yamada. 1991. Cloning, nucleotide sequence and expression in Escherichia coli of two cobalt-containing nitrile hydratase genes from Rhodococcus rhodochrous J1. Biochim. Biophys. Acta 1129:23-33. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi, M., T. Onaka, Y. Ishii, J. Konishi, M. Takaki, H. Okada, Y. Ohta, K. Koizumi, and M. Suzuki. 2000. Desulfurization of alkylated forms of both dibenzothiophene and benzothiophene by a single bacterial strain. FEMS Microbiol. Lett. 187:123-126. [DOI] [PubMed] [Google Scholar]
- 45.Langdahl, B. R., P. Bisp, and K. Ingvorsen. 1996. Nitrile hydrolysis by Rhodococcus erythropolis BL1, an acetonitrile-tolerant strain isolated from a marine sediment. Microbiology 142:145-154. [DOI] [PubMed] [Google Scholar]
- 46.Layh, N., B. Hirrlinger, A. Stolz, and H.-J. Knackmuss. 1997. Enrichment strategies for nitrile-hydrolysing bacteria. Appl. Microbiol. Biotechnol. 47:668-674. [Google Scholar]
- 47.Layh, N., A. Stolz, J. Bohme, F. Effenberger, and H.-J. Knackmuss. 1994. Enantioselective hydrolysis of racemic naproxen nitrile and naproxen amide to S-naproxen by new bacterial isolates. J. Biotechnol. 33:175-182. [DOI] [PubMed] [Google Scholar]
- 48.Martínková, L., J. Hrůzová, F. Machek, L. Seichert, J. Panoš, and P. J.ůzlová. 1992. Isolation of acetonitrile-utilising bacteria. Folia Microbiol. 37:372-376. [Google Scholar]
- 49.Mayaux, J. F., E. Cerebelaud, F. Soubrier, D. Faucher, and D. Petre. 1990. Purification, cloning, and primary structure of an enantiomer-selective amidase from Brevibacterium sp. strain R312: structural evidence for genetic coupling with nitrile hydratase. J. Bacteriol. 172:6764-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metcalfe, G., and M. E. Brown. 1957. Nitrogen fixation by new species of Nocardia. J. Gen. Microbiol. 17:567-572. [DOI] [PubMed] [Google Scholar]
- 51.Minshull, J., and W. P. Stemmer. 1999. Protein evolution by molecular breeding. Curr. Opin. Chem. Biol. 3:284-290. [DOI] [PubMed] [Google Scholar]
- 52.Moreau, J. L., S. Azza, A. Arnaud, and P. Galzy. 1993. Purification and characterisation of nitrile hydratase of a mutant strain of Brevibacterium sp. J. Basic Microbiol. 33:323-329. [Google Scholar]
- 53.Moreau, J. L., N. Bernet, A. Arnaud, and P. Galzy. 1993. Isolation of Brevibacterium sp. R312 mutant potentially useful for the enzymatic production of adipic acid. Can. J. Microbiol. 39:524-528. [Google Scholar]
- 54.Nagamune, T., H. Kurata, M. Hirata, J. Honda, A. Hirata, and I. Endo. 1990. Photosensitive phenomena of nitrile hydratase of Rhodococcus sp. N-771. Photochem. Photobiol. 51:87-90. [Google Scholar]
- 55.Nagamune, T., H. Kurata, M. Hirata, J. Honda, H. Koike, M. Ikeuchi, Y. Inoue, A. Hirata, and I. Endo. 1990. Purification of inactivated photoresponsive nitrile hydratase. Biochem. Biophys. Res. Commun. 168:437-442. [DOI] [PubMed] [Google Scholar]
- 56.Nagashima, S., M. Nakasako, N. Dohmae, M. Tsujimura, K. Takio, M. Odaka, M. Yohda, N. Kamiya, and I. Endo. 1998. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. Nat. Struct. Biol. 5:347-351. [DOI] [PubMed] [Google Scholar]
- 57.Nagy, I., J. Nagy, J. Matyas, and M. Kecskés. 1987. Decomposition of EPTC by soil microbes in two soils, p. 525-530. In Proceedings of the British Crop Protection Conference—Weeds, vol. 1. Lavenham Press Ltd., Lavehnam, United Kingdom.
- 58.Nakajima, Y., T. Doi, Y. Satoh, A. Fujiwara, and I. Watanabe. 1987. Photoactivation of nitrile hydratase in Corynebacterium sp. N-774. Chem. Lett. 9:1767-1770. [Google Scholar]
- 59.Nakasako, M., M. Odaka, M. Yohda, N. Dohmae, K. Takio, N. Kamiya, and I. Endo. 1999. Tertiary and quaternary structures of photoreactive Fe-type nitrile hydratase from Rhodococcus sp. N-771: roles of hydration water molecules in stabilizing the structures and the structural origin of the substrate specificity of the enzyme. Biochemistry 38:9887-9898. [DOI] [PubMed] [Google Scholar]
- 60.Ness, J. E., S. B. Del Cardayre, J. Minshull, and W. P. Stemmer. 2000. Molecular breeding: the natural approach to protein design. Adv. Protein Chem. 55:261-292. [DOI] [PubMed] [Google Scholar]
- 61.Nishiyama, M., S. Horinouchi, M. Kobayashi, T. Nagasawa, H. Yamada, and T. Beppu. 1991. Cloning and characterization of genes responsible for metabolism of nitrile compounds from Pseudomonas chlororaphis B23. J. Bacteriol. 173:2465-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nojiri, M., M. Yohda, M. Odaka, Y. Matsushita, M. Tsujimura, T. Yoshida, N. Dohmae, K. Takio, and I. Endo. 1999. Functional expression of nitrile hydratase in Escherichia coli: requirement of a nitrile hydratase activator and post-translational modification of a ligand cysteine. J. Biochem. 125:696-704. [DOI] [PubMed] [Google Scholar]
- 63.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 64.Payne, M. S., S. Wu, R. D. Fallon, G. Tudor, B. Stieglitz, M. Ivan, J. Turner, and M. J. Nelson. 1997. A stereoselective cobalt-containing nitrile hydratase. Biochemistry 36:5447-5454. [DOI] [PubMed] [Google Scholar]
- 65.Pereira, R. A., D. Graham, F. A. Rainey, and D. A. Cowan. 1998. A novel thermostable nitrile hydratase. Extremophiles 2:347-357. [DOI] [PubMed] [Google Scholar]
- 66.Piersma, S. R., M. Nojiri, M. Tsujimura, T. Noguchi, M. Odaka, M. Yohda, Y. Inoue, and I. Endo. 2000. Arginine 56 mutation in the beta subunit of nitrile hydratase: importance of hydrogen bonding to the non-heme iron center. J. Inorg. Biochem. 80:283-288. [DOI] [PubMed] [Google Scholar]
- 67.Pikkemaat, M. G., and D. B. Janssen. 2002. Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox. Nucleic Acids Res. 30:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poelarends, G. J., M. Zandstra, T. Bosma, L. A. Kulakov, M. J. Larkin, J. R. Marchesi, A. J. Weightman, and D. B. Janssen. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Precigou, S., P. Goulas, and R. Duran. 2001. Rapid and specific identification of nitrile hydratase (NHase)-encoding genes in soil samples by polymerase chain reaction. FEMS Microbiol. Lett. 204:155-161. [DOI] [PubMed] [Google Scholar]
- 70.Raillard, S., A. Krebber, Y. Chen, J. E. Ness, E. Bermudez, R. Trinidad, R. Fullem, C. Davis, M. Welch, J. Seffernick, L. P. Wackett, W. P. Stemmer, and J. Minshull. 2001. Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chem. Biol. 8:891-898. [DOI] [PubMed] [Google Scholar]
- 71.Scholtz, R., A. Schmuckle, A. M. Cook, and T. Leisinger. 1987. Degradation of eighteen 1-monohaloalkanes by Arthrobacter sp. strain HA1. J. Gen. Microbiol. 133:267-274. [Google Scholar]
- 72.Schraa, G., B. M. Bethe, A. R. W. van Neerven, W. J. J. van den Tweel, E. van der Wende, and A. J. B. Zehnder. 1987. Degradation of 1,2-dimethylbenzene by Corynebacterium strain C125. Antonie Leeuwenhoek 53:159-170. [DOI] [PubMed] [Google Scholar]
- 73.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 74.Sugiura, Y., J. Kuwahara, T. Nagasawa, and H. Yamada. 1987. Nitrile hydratase: the first non-heme iron enzyme with a typical low-spin Fe(III)-active center. J. Am. Chem. Soc. 109:5848-5850. [Google Scholar]
- 75.Swofford, D. L. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, Mass.
- 76.Tate, R. L., and J. C. Ensign. 1974. A new species of Arthrobacter which degrades picolinic acid. Can. J. Microbiol. 20:691-694. [DOI] [PubMed] [Google Scholar]
- 77.Tsujimura, M., N. Dohmae, M. Odaka, M. Chijimatsu, K. Takio, M. Yohda, M. Hoshino, S. Nagashima, and I. Endo. 1997. Structure of the photoreactive iron center of the nitrile hydratase from Rhodococcus sp. N-771. Evidence of a novel post-translational modification in the cysteine ligand. J. Biol. Chem. 272:29454-29459. [DOI] [PubMed] [Google Scholar]
- 78.van der Werf, M. J., and J. A. M. Bont. 1998. Screening for microorganisms converting limonene into carvone. Stud. Org. Chem. 53:231-234. [Google Scholar]
- 79.Veiko, V. P., A. S. Yanenko, M. G. Alekseeva, A. A. Sintin, L. B. Gulko, K. I. Ratmanova, I. V. Ovcharova, L. B. Astaurova, I. N. Poljakova, V. N. Paukov, S. P. Voronin, and V. G. Debabov. 1995. Cloning, nucleotide sequence of nitrile hydratase gene from Rhodococcus rhodochrous M8. Biotekhnologiya. 5:3-5. [Google Scholar]
- 80.Vetter, J. 2000. Plant cyanogenic glycosides. Toxicon 38:11-36. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe, I., Y. Satoh, and K. Enomoto. 1987. Screening, isolation and taxonomical properties of microorganisms having acrylonitrile-hydrating activity. Agric. Biol. Chem. 51:3193-3199. [Google Scholar]
- 82.Webley, D. M., and P. C. de Kock. 1952. The metabolism of some saturated aliphatic hydrocarbons, alcohols and fatty acids by Proactinomyces opacus Jensen (Nocardia opaca Waksman and Henrici). Biochem. J. 51:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wyatt, J. M., and C. J. Knowles. 1995. The development of a novel strategy for the microbial treatment of acrylonitrile effluents. Biodegradation 6:93-107. [DOI] [PubMed] [Google Scholar]
- 84.Yamada, H., and M. Kobayashi. 1996. Nitrile hydratase and its application to industrial production of acrylamide. Biosci. Biotechnol. Biochem. 60:1391-1400. [DOI] [PubMed] [Google Scholar]
- 85.Yamaki, T., T. Oikawa, K. Ito, and T. Nakamura. 1997. Cloning and sequencing of a nitrile hydratase gene from Pseudonocardia thermophila JCM 3095. J. Ferment. Bioeng. 83:474-477. [Google Scholar]
- 86.Yohda, M., J. Honda, T. Nagamune, I. Endo, T. Yoshida, and K. Miura. 1994. Molecular cloning and nucleotide sequence of the gene coding photosensitive nitrile hydratase. Ann. N. Y. Acad. Sci. 721:158-159. [DOI] [PubMed] [Google Scholar]