Abstract
Burkholderia species are bacterial soil inhabitants that are capable of interacting with a variety of eukaryotes, in some cases occupying intracellular habitats. Pathogenic and nonpathogenic Burkholderia spp., including B. vietnamiensis, B. cepacia, and B. pseudomallei, were grown on germinating spores of the arbuscular mycorrhizal fungus Gigaspora decipiens. Spore lysis assays revealed that all Burkholderia spp. tested were able to colonize the interior of G. decipiens spores. Amplification of specific DNA sequences and transmission electron microscopy confirmed the intracellular presence of B. vietnamiensis. Twelve percent of all spores were invaded by B. vietnamiensis, with an average of 1.5 × 106 CFU recovered from individual infected spores. Of those spores inoculated with B. pseudomallei, 7% were invaded, with an average of 5.5 × 105 CFU recovered from individual infected spores. Scanning electron and fluorescence microscopy provided insights into the morphology of surfaces of spores and hyphae of G. decipiens and the attachment of bacteria. Burkholderia spp. colonized both hyphae and spores, attaching to surfaces in either an end-on or side-on fashion. Adherence of Burkholderia spp. to eukaryotic surfaces also involved the formation of numerous fibrillar structures.
Arbuscular mycorrhizal (AM) fungi are obligate symbionts of plants with the potential to enhance nutrient and water uptake by their hosts. Members of the genus Gigaspora are known to harbor intracellular bacterium-like organisms (7, 25, 33). In the case of Gigaspora margarita, the presence within its spores of a homogeneous population of endosymbionts belonging to the genus Burkholderia has been identified (7).
Burkholderia species constitute a diverse group of bacteria which are generally found in the rhizosphere, as are AM fungi. Initially, members included saprophytic plant-associated pathogens (2, 11, 31), although recently several species which are beneficial to plants have been recognized (3, 15, 34). In addition to causing disease in plants, several Burkholderia species pose serious threats to human and animal health. For example, B. cepacia and B. pseudomallei can cause fatal human disease. B. cepacia is a pathogen of compromised individuals, especially those with cystic fibrosis, in whom the species can cause a rapid and fatal necrotizing pneumonia known as cepacia syndrome. B. pseudomallei infection causes melioidosis, an infection that is endemic throughout the tropics, particularly in southeast Asia and northern Australia (24). For antibiotic-treated acute infections, these diseases are associated with mortality rates of 20 and 34% for B. cepacia (21) and B. pseudomallei (36), respectively. Persistence of these pathogens in humans is aided by their ability to survive within eukaryotic cells. The capacity to invade eukaryotic cells, as seen in Mycobacterium and Legionella spp., for example, can be regarded as a virulence determinant, allowing evasion of host defenses, tissue destruction, and systemic spread of the pathogen. B. cepacia, B. pseudomallei, and others within the genus (including nonpathogens) have been shown to survive within free-living amoebae (18, 23, 26). Organisms assigned to the B. cepacia complex can invade and survive within macrophages (27, 32), epithelial cells (12, 27), and human pneumocytes (22) in vitro.
Due to the ability of Burkholderia cells to interact with eukaryotic cells, it follows that these bacteria may serve as tools with which to decipher general mechanisms involved in prokaryote-eukaryote associations. This study investigated invasion of an AM fungus as a cellular model of Burkholderia infection.
MATERIALS AND METHODS
Bacterial suspensions.
The Burkholderia strains used for spore invasion studies were B. vietnamiensis strains WACC116 (clinical) and TVV75T (environmental), isolated from human blood and from rice paddies, respectively; B. cepacia ATCC 25416T (environmental) and PHLS Colindale 1125 (clinical, cblA positive); and B. pseudomallei 1026b (clinical) (13) and NCTC13177 (WACC56) (clinical) (19). Suspensions of rapidly motile bacteria grown to mid-lag phase in Trypticase soy broth (18) were washed twice in sterile 0.85% saline, harvested by centrifugation (5 min, 1,000 × g), and adjusted to 80% transmission at 450 nm (0.5 McFarland turbidity standard) in sterile distilled water, yielding approximately 108 CFU ml−1.
AM fungi.
Spores of Gigaspora decipiens Hall and Abbott were recovered from pot cultures of Trifolium subterraneum by wet sieving and gradient centrifugation (10). Spores were washed five times in sterile distilled water, surface disinfected with 4% (wt/vol) chloramine-T and 300 ppm of streptomycin (7) for 1 h, and then rinsed a further five times in sterile distilled water. Surface sterility was ascertained by plating out spores on 5% horse blood agar and by inoculation into brain heart infusion broth (Excel Laboratory Products) for a minimum of 48 h at 30°C. If any growth in the enrichment medium was recorded, the disinfection procedure was repeated. All washes of spores were carried out by very brief low-speed centrifugation (3 s, 800 × g) to collect spores, followed by aspiration of the supernatant.
Qualitative spore invasion assay.
Invasion of G. decipiens spores by Burkholderia spp. was determined by using a variation of the gentamicin protection assay (20) modification of Burns et al. (12). Surface-disinfected spores were placed with flame-sterilized forceps on sterilized 0.8-μm-pore-size Millipore filter papers over 1.5% water agar. A 3-μl aliquot of bacterial suspension was added to each spore. Negative control spores were inoculated with 3 μl of sterile distilled water. Each treatment was set up in triplicate on separate petri dishes, which were then left at room temperature (24°C) for 21 days.
Following this coculture of the bacteria and fungal spores, the germination frequency of spores was recorded. Germinated spores were defined as those with a germ tube extended from the spore for a distance greater than the diameter of the spore (9). Statistical significance of germination outcomes was calculated by using Fisher's exact test (Prism GraphPad 2.01). G. decipiens spores were transferred with flame-sterilized forceps into sterile 1.5-ml microcentrifuge tubes. Spores were washed by centrifuging a minimum of three times in sterile distilled water and treated with 200 μl of antibiotic solution for 2 h at room temperature to clear bacteria present on the exterior of the spores. The most effective antibiotic solution was found to be 1 mg of ceftazidime per ml-500 μg of kanamycin per ml-0.5% Tween 20 in sterile distilled water. When green fluorescent protein (GFP)-marked B. vietnamiensis was used, this antibiotic mixture was modified to 5 mg of ceftazidime per ml-500 μg of gentamicin per ml-0.5% Tween 20 in sterile distilled water. Following antibiotic treatment, the spores were washed a further five times in sterile distilled water to remove residual antibiotics, spiral plated in 50-μl aliquots onto plate count agar (Excel Laboratory Products), and incubated at 37°C overnight. The antibiotic treatment and washes were repeated, often several times, until no bacteria were detected in the spore washes. Spores were then treated with the antibiotic solution and washed in water. The washes were plated out as described above immediately prior to lysis, since the time lapse necessary for incubating viable counts may have allowed residual bacteria to multiply. These final washes did not yield any viable organisms. Spores were bulked, and batches of five spores were lysed mechanically by being crushed beneath the tip of a flame-sterilized steel rod while immersed in the remaining final wash solution. Crushing of spores was audible and was accompanied by the extrusion of spore cytoplasmic contents from within the spore wall. This was confirmed by direct visual or microscopic examination. Bulked spore lysates were spiral plated in duplicate on plate count agar. A positive result for invasion of spores was recorded when bacterial colonies appeared on these spiral plates. This bacterial growth was always extensive, in excess of 105 CFU/ml.
Quantitative spore invasion assay.
Invasion of G. decipiens by B. pseudomallei 1026b or by B. vietnamiensis WACC116 was determined quantitatively by use of the procedure for the qualitative spore invasion assay. After washing of spores with antibiotics, individual spores were placed in sterile, 16-well microtiter trays with flame-sterilized forceps. Each spore was crushed individually as described above, and spore lysates were serially diluted and spiral plated onto plate count agar in duplicate while remaining separate. The numbers of intracellular bacteria calculated from the averages of the viable counts were expressed as CFU per spore. The identity of bacteria recovered from spore lysates was confirmed by carbohydrate utilization characteristics (API 50 CH microtube system) and by nucleic acid amplification with PCR and species-specific primers (4, 5).
DNA extraction and PCR.
To identify the presence of B. vietnamiensis within inoculated G. decipiens, spores were prepared and washed with antibiotics until culturable bacteria were no longer be detected, as described above. Final washes and lysates were collected from three batches of 20 spores and extracted for PCR with the Qiagen QIAamp viral RNA minikit according to the manufacturer's instructions. DNA preparations were diluted 20-fold to remove PCR inhibitors. PCRs were carried out in a final volume of 20 μl containing 8 μl of diluted DNA extract, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1× PE buffer II (10 mM Tris-HCl [pH 8.3], 50 mM KCl), a 0.2 μM concentration of each primer, 2.5 mM MgCl2, and 0.5 U of PE TaqGold (Applied Biosystems). The primers used for B. vietnamiensis were ViMaPS23-1 (forward) and CeVi-23-2 (reverse) (4, 5). Amplification was carried out with an Applied Biosystems GeneAmp PCR system 2700, with conditions as follows: hot start of 1 cycle of 94°C for 10 min, followed by 45 cycles at an annealing temperature of 50°C (94°C for 30 s, 50°C for 30 s, and 72°C for 45 s), and finally 72°C for 7 min and then holding at 4°C.
Unknown cultures obtained from uninoculated G. decipiens lysates were subjected to 16S rRNA amplification and dideoxy sequencing with an Applied Biosystems Prism 310 genetic analyzer.
To determine whether G. decipiens spores contained endobacteria related to Pandorea or Ralstonia, DNA extraction and PCR were performed as described by Bianciotto et al. (7). A Ralstonia paucula extract was used as a positive control.
Scanning electron microscopy.
Spores of G. decipiens were surface disinfected and then inoculated with either B. vietnamiensis, B. pseudomallei, or sterile distilled water as described above. Specimens were then fixed in 2.5% (vol/vol) glutaraldehyde in 0.05 M phosphate buffer (pH 7.0) for 24 h. Samples were washed twice in phosphate buffer (1 h total), dehydrated through a graded ethanol series over 7 days, transferred to 100% acetone, and then dried in a critical-point drying apparatus with liquid carbon dioxide. Spores were then sputter coated with carbon and gold and viewed with a scanning electron microscope (Philips XL30) at 20 to 25 kV. Regions of interest were further studied by using a field emission scanning electron microscope (JEOL JSM-6300F) at 3 kV. Control and inoculated spores were frozen in liquid nitrogen and observed with a Philips 505 scanning electron microscope fitted with a cryostage (Hexland) (−150°C, 6 kV) in order to reduce artifacts arising from sample fixation and dehydration.
Optical microscopy and transmission electron microscopy (TEM).
Negative control G. decipiens spores and spores cocultured with bacteria, as described above, were fixed and vacuum infiltrated (8 kPa for 5 min at room temperature) with a 2.5% (vol/vol) glutaraldehyde solution. While still in the glutaraldehyde solution, spore walls were punctured by using a laser microdissector (PALM Robot-Combisystem coupled to a Zeiss Axiovert 135) set at a cut focus of 67.49, a cut energy of 100.5%, and a pulse of 30/s to allow adequate resin infiltration. After 24 h in the glutaraldehyde solution, samples were rinsed twice in phosphate buffer (1 h total), postfixed in 2% osmium tetroxide in phosphate buffer (2 h), and rinsed a further two times in buffer prior to dehydration through graded ethanol and propylene oxide series. Specimens were then infiltrated with a Spurr's resin-propylene oxide mixture from 0 to 100% Spurr's resin over 5 days. Samples were then polymerized at 70°C for 18 h.
Semithin (0.5- to 1-μm-thick) and ultrathin (0.1-μm-thick) sections were cut on a Reichert Ultracut microtome with glass knives. Semithin sections were stained with 1% toluidine blue (pH 9) and examined with an optical microscope (Zeiss Axioskop 2). Ultrathin sections were placed onto copper grids, stained with 1% aqueous uranyl acetate and lead citrate, and inspected with a JEOL JEM-2000FX II electron microscope operating at 80 kV.
Construction of GFP-tagged Burkholderia strains.
B. vietnamiensis WACC116 and B. pseudomallei 1026b were used for all electroporation experiments. Plasmid PSMC21, constructed by Bloemberg and colleagues (8), was supplied by N. Banning, The University of Western Australia. Bacterial strains were grown to an optical density (640 nm) of between 0.9 and 1.1 in Luria broth at 37°C. These suspensions were washed twice in ice-cold sterile distilled water and twice in ice-cold sterile 10% glycerol solution and resuspended in a small volume (100 μl) of glycerol solution. Forty microliters of these cells was electroporated at 1.25 kV with approximately 85 μg of a PSMC21 plasmid preparation by using a Bio-Rad Gene Pulser II and 0.1- or 0.2-cm cuvettes. Electroporated cells were diluted in 1 ml of Luria broth, grown for 2 h at 37°C, and then spread onto Luria agar plates supplemented with 300 μg of kanamycin per ml. After 96 h at 37°C, an average of 11.2 recombinant colonies per 100 μl were recovered for B. vietnamiensis, i.e., 1.4 recombinants per ng of plasmid DNA.
Optical and confocal microscopy.
Hyphae of G. decipiens attached to germinating spores were inoculated with 10 μl of GFP-tagged B. vietnamiensis or B. pseudomallei suspension. Samples were examined immediately and after 7 days of coculture by using a differential interference contrast and fluorescence microscope (Olympus BX60F5) or a UV laser scanning confocal microscope (Bio-Rad MRC 1000/1024). All preparations were made up in sterile 0.85% saline in welled slides with coverslips of known thickness. Fluorescent samples were viewed by using the 488-nm blue line from the argon ion laser, a 522/35-nm band pass emission filter, and a Plan Apo 60x, NA1.2 water immersion objective lens (Nikon). Individual images in a stack were taken at 1.3-μm intervals.
To determine whether unculturable endobacteria were present within uninoculated G. decipiens spores, batches of five surface-disinfected spores were crushed in 200-μl volumes of sterile distilled water and stained by using a Live/Dead BacLight bacterial viability kit (Molecular Probes, Eugene, Oreg.) according to the manufacturer's instructions. Preparations were examined under blue and green light with a fluorescence microscope (Olympus BX60F5).
RESULTS
Qualitative spore invasion assay.
All Burkholderia spp. tested were recovered from within G. decipiens spores following coculture (Table 1). The strains investigated were B. vietnamiensis WACC116 and TVV75T, B. cepacia ATCC25416T and PHLS Colindale 1125, and B. pseudomallei 1026b and NCTC13177. Similarly, the B. vietnamiensis strain marked with GFP was also found to invade G. decipiens spores. Recovered marked bacteria fluoresced green and grew on agar containing 300 μg of kanamycin per ml, confirming that these organisms were the same as those previously inoculated.
TABLE 1.
Recovery of Burkholderia spp. following repeated antibiotic washes and lysis of G. decipiens spores
| Strain | Bacterial growth after the following no. of antibiotic treatments and after spore lysisa:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Lysate | |
| WACC116 | + | + | + | − | − | − | + |
| TVV75T | + | + | − | − | + | ||
| ATCC 25416T | + | + | + | − | − | − | + |
| PHLS 1125 | + | + | + | − | − | − | + |
| NCTC 13177 | + | + | + | + | − | − | + |
| 1026b | + | + | − | − | + | ||
No further treatments were conducted after no bacterial growth was detected on two consecutive occasions. Each treatment consisted of incubation with antibiotics followed by five washes in sterile distilled water. Spores were lysed immediately after the final treatment.
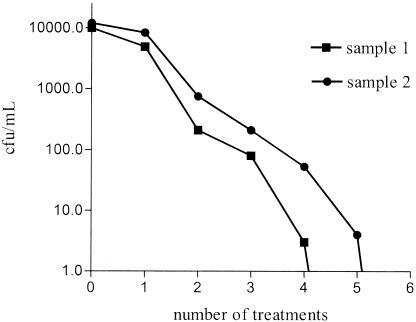
The combination of 1 mg of ceftazidime per ml, 500 μg of kanamycin per ml, and 0.5% Tween 20 in sterile distilled water was found to be effective for removing external bacteria (with the exception of the marked strain, which was kanamycin resistant). Figure 1 presents data showing the reduction in external numbers of B. vietnamiensis bacteria on G. decipiens spores.
FIG. 1.
Antibiotic clearance of B. vietnamiensis from G. decipiens spores. No bacteria were recovered following antibiotic treatments 5 and 6 for samples 1 and 2, respectively. Each treatment consisted of incubation with antibiotics followed by five washes in sterile distilled water.
Uninoculated spores rarely contained culturable organisms; however, two different colony types of gram-positive spore-forming bacteria were isolated from both the exteriors and the interiors of several spores on more than one occasion. Amplification and sequencing of 16S rRNA sequences revealed 99% sequence homology to Paenibacillus polymyxa (accession number AF181573) for one isolate and 96% sequence homology to various Bacillus and Paenibacillus spp. (accession numbers AF106597, PAZ251195, PAZ251192, PSP299573, UEU68623, and PSP131119) for the other isolate.
Germination of G. decipiens spores over water agar was poor (34%). An attempt was made to increase germination frequencies by altering exposure to light, culture conditions of bacterial inocula, depth in pot culture of isolated spores, and age of pot cultures and by treating spores with mild hypochlorite solution or by refrigeration of spores prior to use. No significant increase in spore germination frequencies was observed following variation of any of these factors, either in inoculated or in control spores. Overall, inoculation with B. vietnamiensis resulted in significant increases in spore germination (P = 0.0185), elevating the germination frequency to 41%. Inoculation with B. pseudomallei had little effect on spore germination, with an average of 31% of spores germinating.
Quantitative spore invasion assay.
Bacterial invasion of spores of G. decipiens was recorded in 12% of spores inoculated with B. vietnamiensis WACC116 and in 7% of spores inoculated with B. pseudomallei 1026b. Large numbers of bacteria were recovered from individual infected spores (Table 2). When B. vietnamiensis WACC116 or B. pseudomallei 1026b was inoculated onto spores, only the species inoculated could be recovered. Recovered bacteria were evaluated for carbohydrate utilization characteristics (API 50 CH microtube system) and subjected to nucleic acid amplification with PCR and species-specific primers (4, 5). Both of these tests indicated that only those species inoculated were recovered.
TABLE 2.
Invasion of G. decipiens spores by B. vietnamiensis WACC116 and B. pseudomallei 1026b
| Species | Mean (SEM) bacterial CFU per infected spore | Mean (SEM) % invasion frequency |
|---|---|---|
| B. vietnamiensis | 1.5 × 106 (1.3 × 106)a | 12.1 (1.4)b |
| B. pseudomallei | 5.5 × 105 (2.0 × 105)c | 7.3 (1.2)d |
Data derived from triplicate viable counts for each of 13 invaded spores.
Data from a total of 162 spores tested.
Data derived from triplicate viable counts for each of 15 invaded spores.
Data from a total of 199 spores tested.
PCR.
B. vietnamiensis primers consistently amplified DNA extracted from G. decipiens lysates of spores after coculture with B. vietnamiensis. No amplification products were obtained from either the interior or exterior of uninoculated spores. Although washes of cocultured G. decipiens spores were culture negative, on one occasion B. vietnamiensis was detected by PCR in washes of spores. In this instance, a 100-fold dilution resulted in these bands being no longer detectable, although amplification of sequences from spore lysates still occurred at this dilution. All banding patterns observed were indistinguishable from those produced by cell lysates of B. vietnamiensis.
No Pandorea or Ralstonia DNA was detected in extracts of 50 surface-disinfected G. decipiens spores. R. paucula controls yielded strong bands even when diluted 100-fold, while similar dilutions of spore extracts did not yield any amplification products.
SEM.
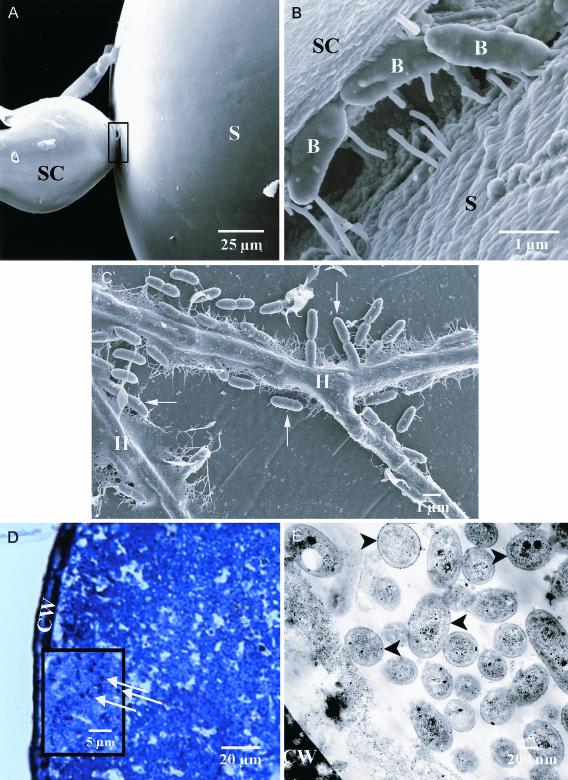
Scanning electron microscopy (SEM) was used to show colonization patterns of bacteria on spore surfaces and to provide insights into the means of bacterial entry into spores. Following 21 days of coculture with B. vietnamiensis (WACC116) or B. pseudomallei, G. decipiens had few bacteria present on external spore surfaces but many bacteria colonizing the hyphae. Spores were generally smooth, with little debris present (Fig. 2A). Surface morphology observations were not attributed to specimen preparation, as spores examined by SEM with a cryostage for sample preparation appeared similar to those analyzed by conventional SEM. In one case, B. pseudomallei was found to preferentially colonize the junction between the spore itself and the sporogenous cell (Fig. 2A and B). Field emission SEM analysis of this region revealed the presence of fibrillar structures often linking the prokaryotic and eukaryotic cells (Fig. 2B). Similar structures were consistently observed on G. decipiens hyphae following inoculation with Burkholderia spp. Figure 2C depicts G. decipiens hyphae, adherent B. pseudomallei, and associated fibrillar material. This image also illustrates the presence of both end-on and side-on attachment of B. pseudomallei to G. decipiens. No bacteria were seen on uninoculated spore controls.
FIG. 2.
(A) SEM of a G. decipiens spore (S) with attached sporogenous cell (SC) inoculated with B. pseudomallei. (B) Field emission SEM of the junction between the spore and sporogenous cell shown in panel A. B, bacterium. (C) Field emission SEM of G. decipiens hyphae, adherent B. pseudomallei (arrows), and associated fibrillar material. H, hypha. (D) Optical semithin section of B. vietnamiensis-inoculated G. decipiens stained with toluidine blue. Bacteria (arrows) are present throughout the cytoplasm. CW, cell wall. (E) TEM of G. decipiens cytoplasm containing bacteria (arrowheads).
Optical microscopy and TEM of spores.
Sectioning of G. decipiens spores inoculated with B. vietnamiensis (WACC116) was undertaken to confirm the presence of intrasporular bacteria as indicated by in vitro invasion assays. Although the majority of spores sectioned contained no objects resembling bacteria, large numbers of bacterium-like objects were observed in some specimens that had been inoculated with B. vietnamiensis (Fig. 2D). These were located on the interior of spores, adjacent to intact spore walls, and throughout the spore cytoplasm. TEM analysis revealed that these objects were bacteria (Fig. 2E), measuring approximately 1.0 μm in length and 0.3 to 0.4 μm in width, which is typical of B. vietnamiensis (16). Bacterial cell membranes were well defined, and electron-dense bodies were apparent within the bacilli. G. decipiens spores that had not been inoculated with bacteria were also sectioned. No objects resembling bacteria were observed in any of these negative control spores.
Optical and confocal microscopy.
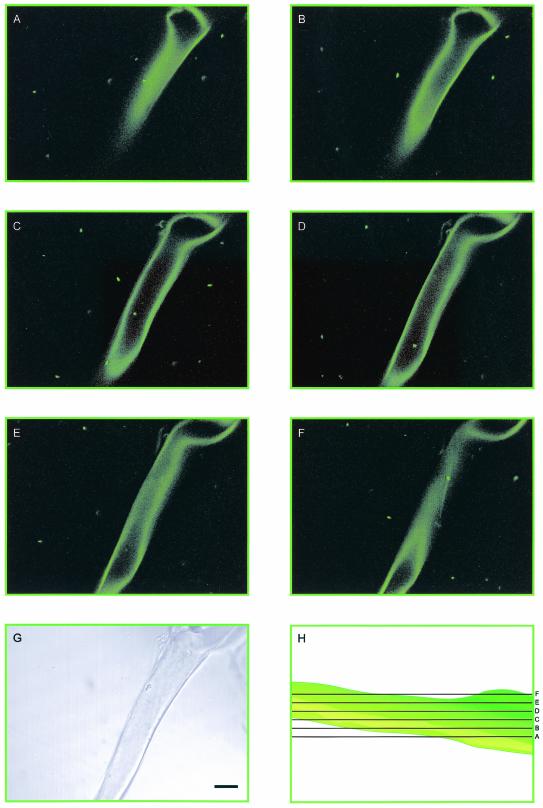
In contrast to the spores themselves, hyphal surfaces of G. decipiens were readily colonized by B. pseudomallei and B. vietnamiensis. Differential interference contrast microscopy of hyphae exposed to B. pseudomallei and B. vietnamiensis revealed attachment of live bacteria to hyphae. Confocal laser microscopy allowed discrimination between fluorescent bacilli attached to external surfaces of G. decipiens hyphae and those located within the hyphae, as hyphal walls autofluoresce. An example of GFP-expressing B. pseudomallei internalized within a hypha is shown in Fig. 3. The confocal images are representatives taken from a stack of 13 slices through a G. decipiens hypha.
FIG. 3.
(A to F) Series of confocal laser scanning micrographs of GFP-labeled B. pseudomallei and a G. decipiens hypha. Motile, fluorescent bacteria are apparent in the aqueous medium, attached to the external surfaces of the hypha, and internalized within the hypha (C and D). (G) Nonconfocal transmission image. (H) Diagrammatic representation of the optical sectioning of the hypha. Bar, 30 μm. Images were falsely colored green using Adobe Photoshop 7.0.
Morphological observations of uninoculated G. decipiens spores by using fluorescent dyes indicated that endobacteria may be present in some G. decipiens spores. Of four preparations of five spores each, one contained both red (dead) and green (live) fluorescent objects. All of these objects were coccoid and ranged from approximately 1 to 10 μm in diameter. More green than red fluorescent objects were apparent, indicating that the majority were alive.
DISCUSSION
In this study of cellular interactions between rhizospheric bacterial and fungal species, we demonstrated that Burkholderia spp. have the capacity to enter the spores of the AM fungus G. decipiens. This extends the known interactions of Burkholderia spp. to encompass all four kingdoms of eukaryotes, namely, Plantae, Animalia, Protista, and Fungi.
Lysis assays of G. decipiens spores indicate that variable and often large numbers of bacteria can be recovered from a proportion (7 to 12%) of spores. The variable nature of this observation may simply reflect variation in spore size. Carbohydrate utilization characteristics, nucleic acid amplification, and use of a GFP-marked strain revealed that the bacteria released from spores upon lysis were the same as those previously introduced by deliberate inoculation in vitro. Microscopic observations and amplification of PCR products from whole spore extracts confirmed the presence of intracellular bacteria. In the case of TEM, spores that contained bacteria displayed amorphous cytoplasm that lacked defined organelles. This appearance is consistent with cell death but does not resolve whether spore death occurred before or after bacterial invasion. No actively dividing bacteria have been observed within spores, consistent with other studies of eukaryotic Burkholderia associations (12, 32), which report an absence of significant intracellular bacterial replication.
The bacteria observed within G. decipiens spores with TEM were not antibody labeled and therefore could not be conclusively identified as those previously inoculated. Furthermore, G. decipiens spores may contain resident populations of coccoid endobacteria. The granular bodies within the intracellular bacteria are likely to be cytoplasmic accumulations of polyhydroxyalkanoates which are known to occur in Burkholderia spp. (17, 29). Lysates of inoculated spores consistently yielded PCR products identical to those of the inoculated bacteria. No bacterial DNA product was ever amplified from either the interior or the exterior of surface-disinfected, uninoculated spores.
The frequency with which bacterial invasion occurred was more constant than the number of bacteria observed per spore. This may have reflected the characteristics of the spore population, such as age, dormancy, or viability. It is interesting that the increase in germination frequencies attributed to inoculation with B. vietnamiensis (14%) matched the proportion of spores invaded (12%). Bianciotto et al. (7) proposed that Burkholderia spp. within G. margarita migrated from spores to hyphae to allow for vertical transmission of the symbiotic prokaryotes. In the case of G. decipiens and Burkholderia spp., although there is no evidence of any symbiotic relationship, bacterial access to fungal hyphae may allow for movement into the spores and subsequent colonization of this intracellular habitat. Individual spores of G. decipiens are able to germinate on more than one occasion, resulting in old germ tubes and associated damaged or open-ended hyphae, which may act as entry points for bacterial access to spores. As Burkholderia spp. are generally free-living, this may facilitate continuous but low-level bacterial uptake from the environment by mycorrhizal fungi.
The germination frequency of spores (G. decipiens) was significantly enhanced by incubation with B. vietnamiensis but not by incubation with B. pseudomallei. It has been shown that bacteria can have either positive or negative effects on germination of spores of AM fungi (1, 35, 37). In several cases positive effects on spore germination and mycorrhiza formation in plants have been attributed to volatile compounds produced by bacteria (30, 35). Bacteria are also thought to play a more indirect role by detoxifying the fungal culture medium (14). This is believed to reduce the inhibition of hyphal growth that results from an accumulation of fungal biosynthetic metabolites in the medium.
We have conclusively demonstrated, using a variety of techniques, that exogenous bacteria can be recovered from within spores of an AM fungus following a period of coculture in vitro. This observation has possible implications for the dissemination of disease-causing bacteria. In the case of Legionella spp., intra-amoebic survival and escape are regarded as a potential route of infection (6). In a similar fashion, inhalation of soilborne particles containing spores of AM fungi could theoretically result in disease, if harmful bacteria occupy those spores. Implications regarding the role of AM fungi in agriculture also arise. In the symbiosis described between G. margarita and a Burkholderia endosymbiont, nitrogen fixation (nif) genes possessed by the bacterial partner were proposed to play a role in nitrogen acquisition by the fungus (28). As bacterial uptake by at least one species of AM fungal spores (G. decipiens) occurs, possible beneficial effects on fungal growth and mycorrhiza formation should be further investigated.
This study highlights the wide range of potential cellular hosts for Burkholderia spp. in the natural environment. Our observations emphasize the need for a more comprehensive ecological appraisal of Burkholderia spp. and their interactions with eukaryotes in the rhizosphere.
Acknowledgments
We thank Paul Rigby from the Biomedical Confocal Microscopy Research Centre supported by the Lotteries Commission of Western Australia and Kathryn Heel from the Lotteries Laser Microdissection Facility for their technical assistance. We also thank Donald Woods for kindly supplying B. pseudomallei strain 1026b and Lee Ingram for his assistance with the digital images.
REFERENCES
- 1.Ali, N., and R. Jackson. 1989. Stimulation of germination of spores of some ectomycorrhizal fungi by other microorganisms. Mycol. Res. 93:182-186. [Google Scholar]
- 2.Azegami, K., K. Nishiyama, Y. Watanabe, I. Kadota, A. Ohuchi, and C. Fukazawa. 1987. Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int. J. Syst. Bacteriol. 37:531-539. [Google Scholar]
- 3.Baldani, V., J. Baldani, and J. Döbereiner. 2000. Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol. Fertil. Soils 30:485-489. [Google Scholar]
- 4.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1998. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J. Clin. Microbiol. 36:2748-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianciotto, V., C. Bandi, D. Minerdi, M. Sironi, H. Tichy, and P. Bonfante. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloemberg, G., G. Otoole, B. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brundrett, M., and S. Juniper. 1995. Non-destructive assessment of spore germination of vam fungi and production of pot cultures from single spores. Soil Biol. Biochem. 27:85-91. [Google Scholar]
- 10.Brundrett, M. C., N. Bougher, B. Dell, T. Grove, and N. Malajczuk. 1996. Working with Glomalean fungi, p. 155-158. In P. Lynch (ed.), Working with mycorrhizas in forestry and agriculture. Pirie Printers, Canberra, Australia.
- 11.Burkholder, W. H. 1950. Sour skin: a bacterial rot of onion bulbs. Phytopathology 40:15-23. [Google Scholar]
- 12.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duponnois, R., and J. Garbaye. 1990. Some mechanisms involved in growth stimulation of ectomycorrhizal fungi by bacteria. Can. J. Bot. 68:2148-2152. [Google Scholar]
- 15.Estrada-De Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillis, M., T. Vanvan, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 17.Gomez, J., M. Rodrigues, R. Alli, B. Torres, C. Bueno Netto, M. Oliveira, and L. da Silva. 1996. Evaluation of soil gram-negative bacteria yielding polyhydroxyalkanoic acids from carbohydrates and propionic acid. Appl. Microbiol. Biotechnol. 45:785-791. [Google Scholar]
- 18.Inglis, T. J., P. Rigby, T. A. Robertson, N. S. Dutton, M. Henderson, and B. J. Chang. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect. Immun. 68:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inglis, T. J., Garrow, S. C., Adams, C., Henderson, M., Mayo, M., and B. J. Currie. 1999. Acute melioidosis outbreak in Western Australia. Epidemiol. Infect. 123:437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isberg, R. R., and S. Falkow. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262-264. [DOI] [PubMed] [Google Scholar]
- 21.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 22.Keig, P. M., E. Ingham, and K. G. Kerr. 2001. Invasion of human type II pneumocytes by Burkholderia cepacia. Microb. Pathog. 30:167-170. [DOI] [PubMed] [Google Scholar]
- 23.Landers, P., K. G. Kerr, T. J. Rowbotham, J. L. Tipper, P. M. Keig, E. Ingham, and M. Denton. 2000. Survival and growth of Burkholderia cepacia within the free-living amoeba Acanthamoeba polyphaga. Eur. J. Clin. Microbiol. Infect. Dis. 19:121-123. [DOI] [PubMed] [Google Scholar]
- 24.Leelarasamee, A., and S. Bovornkitti. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11:413-425. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald, R. M., R. Muriel, M. R. Chandler, and B. Mosse. 1982. The occurrence of bacterium-like organelles in the vesicular arbuscular mycorrhizal fungi. New Phytol. 90:659-663. [Google Scholar]
- 26.Marolda, C., B. Hauroder, M. John, R. Michel, and M. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 27.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minerdi, D., R. Fani, R. Gallo, A. Boarino, and P. Bonfante. 2001. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 67:725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitomo, H., T. Takahashi, H. Ito, and T. Saito. 1999. Biosynthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) produced by Burkholderia cepacia D1. Int. J. Biol. Macromol. 24:311-318. [DOI] [PubMed] [Google Scholar]
- 30.Mugnier, J., and B. Mosse. 1987. Spore germination and viability of a vesicular arbuscular mycorrhizal fungus Glomus-mosseae. Trans. Br. Mycol. Soc. 88:411-413. [Google Scholar]
- 31.Palleroni, N. 1984. Gram-negative aerobic rods and cocci, p. 175-178. In N. R. Kreig and J. G. Holt (ed.), Bergey's manual of systemic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 32.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 33.Scannerini, S., and P. Bonfante-Fasolo. 1991. Bacteria and bacteria-like objects in endomycorrhizal fungi (Glomaceae), p. 273-287. In L. Margulis and R. Fester (ed.), Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press, Cambridge, Mass. [PubMed]
- 34.TrÂn Van, V., O. Berge, S. Ke, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 35.Tylka, G. L., R. S. Hussey, and R. W. Roncadori. 1991. Axenic germination of vesicular-arbuscular mycorrhizal fungi effects of selected Streptomyces-spp. Phytopathology 81:754-759. [Google Scholar]
- 36.White, N. J., D. A. B. Dance, W. Chaowagul, Y. Wattanagoon, V. Wuthiekanun, and N. Pitakwatchara. 1989. Halving of mortality of severe melioidosis by ceftazidime. Lancet ii:697-701. [DOI] [PubMed] [Google Scholar]
- 37.Will, M. E., and D. M. Sylvia. 1990. Interaction of rhizosphere bacteria fertilizer and vesicular-arbuscular mycorrhizal fungi with sea oats. Appl. Environ. Microbiol. 56:2073-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]