Abstract
A locus encoding two repetitive proteins that have LPXTG cell wall anchoring signals from Lactobacillus fermentum BR11 has been identified by using an antiserum raised against whole L. fermentum BR11 cells. The first protein, Rlp, is similar to the Rib surface protein from Streptococcus agalactiae, while the other protein, Mlp, is similar to the mucus binding protein Mub from Lactobacillus reuteri. It was shown that multiple copies of mlp exist in the genome of L. fermentum BR11. Regions of Rlp, Mlp, and the previously characterized surface protein BspA were used to surface display or secrete heterologous peptides in L. fermentum. The peptides tested were 10 amino acids of the human cystic fibrosis transmembrane regulator protein and a six-histidine epitope (His6). The BspA promoter and secretion signal were used in combination with the Rlp cell wall sorting signal to express, export, and covalently anchor the heterologous peptides to the cell wall. Detection of the cell surface protein fusions revealed that Rlp was a significantly better surface display vector than BspA despite having lower cellular levels (0.7 mg per liter for the Rlp fusion compared with 4 mg per liter for the BspA fusion). The mlp promoter and encoded secretion signal were used to express and export large (328-kDa at 10 mg per liter) and small (27-kDa at 0.06 mg per liter) amino-terminal fragments of the Mlp protein fused to the His6 and CFTR peptides or His6 peptide, respectively. Therefore, these newly described proteins from L. fermentum BR11 have potential as protein production and targeting vectors.
Over the past several years investigation into the health-promoting, or probiotic, activity of certain strains of lactic acid bacteria in humans has been the subject of substantial research. Of particular interest are lactobacilli, which due to their nonpathogenic and mucosal surface-colonizing properties have been developed for recombinant protein delivery because they have the potential to express proteins at specific mucosal sites. For the most part, lactobacilli have been investigated as recombinant mucosal vaccines. There have been recent developments in the engineering of lactobacilli and other colonizing lactic acid bacteria so that they secrete or surface display single-chain Fv fragments with direct therapeutic effects (4, 16).
Lactobacillus secretion vectors have mainly utilized the secretion signals of proteins derived from bacteria other than lactobacilli, with examples including those of the M protein from Streptococcus pyogenes, Usp45 and PrtP from Lactococcus lactis, and protein A from Staphylococcus aureus (13, 31, 35). Lactobacillus secretion signals that have been used for extracellular heterologous protein production include the S-layer secretion signal from Lactobacillus brevis (33) and the α-amylase protein from Lactobacillus casei (16, 17). Protein surface display systems using lactobacilli can be divided into two classes, covalent and noncovalent anchoring systems. The covalent anchor system uses proteins that contain LPXTG cell wall sorting signals. The M protein has been used successfully (24, 27), while the PrtP proteins from L. casei and Lactobacillus delbrueckii have been used to anchor heterologous proteins on L. casei (16, 18) and Lactobacillus johnsonii, respectively (34). Noncovalent surface attachment systems utilize either the S-layer protein from L. brevis or Lactobacillus acidophilus (3, 36) or BspA from Lactobacillus fermentum (14, 43).
Our research is directed towards the development of therapeutic protein secretion and surface display expression systems based on native surface proteins of the guinea pig vaginal tract isolate L. fermentum BR11. L. fermentum BR11 is our model organism because we have shown that it is amenable to genetic manipulation (31, 42) and because the L. fermentum-Lactobacillus reuteri group are one of the bacterial groups found on a number of different mucosal surfaces in mammals, including the intestine. Thus far, we have identified and characterized BspA from L. fermentum BR11, an abundant basic surface protein which is anchored to the cell surface noncovalently (41). Closely related homologs of BspA include Cnb from L. reuteri NCIB11951 (29), a 29-kDa protein from L. fermentum RC-14 (11), and MapA from L. reuteri 104R (28). These proteins probably interact with teichoic acids via electrostatic interactions due to their strong cationic properties, which are shared with the anchoring domain of lactobacillus S-layer protein subunits (2, 41). BspA has been used as a fusion partner to express and surface display peptides from human immunodeficiency virus and Chlamydia as well as a large glucosyltransferase enzyme (14, 43).
Here we report the identification and characterization of two tandem genes of L. fermentum BR11 encoding proteins that have features similar to those of repetitive LPXTG covalently anchored surface proteins and have similarity to adhesins from other gram-positive bacteria. It is also shown that functional elements of these two proteins can facilitate efficient secretion and cell surface anchoring of heterologous peptides by L. fermentum and therefore may have potential for applications in other lactic acid bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
L. fermentum BR11 is a guinea pig vaginal tract isolate which was grown on solid MRS medium (Oxoid, Basingstoke, United Kingdom) anaerobically or in standing liquid culture tubes. Escherichia coli JM109, LE392, and recombinant derivatives were used in molecular cloning experiments. The E. coli-Streptococcus temperature-sensitive shuttle plasmid pJRS233 has previously been described (23). Ampicillin was used at a concentration of 100 or 200 μg per ml for E. coli, while erythromycin was used at concentrations of 750 μg per ml for E. coli and 10 μg per ml for L. fermentum. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a concentration of 1 mM for induction of proteins expressed from the E. coli clones. Plasmids pUC18, pBluescriptII, pGEM3zf, and pGEM-T Easy were used for routine cloning.
Genomic library constructions and screenings.
The L. fermentum BR11 pUC18 genomic library construction and screening with whole-cell antiserum have been described previously (41). The insert in pPNG602 was partially sequenced, while the insert in pPNG601 was sequenced fully. A λ library was constructed by using partially Sau3AI-digested L. fermentum BR11 DNA ligated into λ EMBL3 BamHI arms (Promega, Madison, Wis.). Packaging into phage particles and infection of E. coli LE392 were done according to the instructions of the manufacturer (Promega). The plaques were screened with a digoxigenin-11-dUTP-labeled PCR probe generated by using primers 5rev1 and 5for5 (Table 1) according to instructions of the manufacturer (Roche, Mannheim, Germany). DNA was purified from λ clones by a procedure described previously (32), and positive λ clones were confirmed by Southern blotting. λ8 was chosen for further study.
TABLE 1.
Oligonucleotides used in this study (excluding those used for DNA sequencing)
| Oligonucleotide | Nucleotide sequence (5′ to 3′)a | Amplified product |
|---|---|---|
| 5rev1 | GTTCAATTGACGGATATG | mlp repeat region probe |
| 5for5 | CACCAGCACCAGTAATC | mlp repeat region probe |
| pUC-Bam | CTTGGATCCCTGCAGGTCGACTCTAG | 3′ end of mlp and downstream |
| Mlp-Paw-C | AAGGATCCCATGGAAGACCAAGAAGCT | 3′ end of mlp and downstream |
| Mlp-C-check | GCTATGAATCTGTTAAGCCAC | 3′ end of mlp and downstream |
| BspA-Xba | CGTTTCTAGAACTTGTTAGTAATGCCGG | bspA |
| BspA-LPX-Pst-His | AACTGCAGAGTGATGATGGTGATGATGTTCT CCTGTTGATGGTAATCCTCCTTCTGTAATATCCGCACC | bspA |
| Term-Xho | TAACTCGAGTTTTGCAGTTCATTCGTTAG | bspA terminator |
| Term-Hind | AAGGAAGCTTTGGTAATGGGGATTGCC | bspA terminator |
| BspA-Eco-stop | CCGAATTCTAACGAATGAACTGC | bspA |
| BspA-Eco-SS | AAGAATTCTAGCTTCGATGACAATGG | bspA 5′ end |
| BspA-SS-Pst-His | AACTGCAGAGTGATGATGGTGATGATGTACA TCATCAGATGCCGCATGAATACT | bspA 5′ end |
| Rlp′′-Pst-Xho | AGTCTGCAGCTCGAGCAGATGCAATTAAGAAT | rlp 3′ end |
| Rlp-Sal-Apa | TTTGGGCCCGTCGACTCCTCTTAATTCAATCGC | rlp 3′ end |
| MlpN-Xba | AAGTCTAGATAAGAAACGTAAGGCTG | mlp 5′ end |
| MlpN-Xho-His | TTACTCGAGTTAGTGATGATGGTGATGATGAACACTACCAAACGTGAC | mlp 5′ end |
| Mlprep-Xba | CATTCTAGATGACCCAACTGGCAA | mlp internal fragment |
| Mlprep-Pst-His | GTACTGCAGAGTGATGATGGTGATGATGATT ATCAATAGTTGCAGT | mlp internal fragment |
| CFTR-5′-Pst | AAACTGCAGGATCATATGATCCAGATAATAA GGAAGAACGTGGAGGAAGTTACGACCCGG ATAACAAGGAAGAACGTGGAGGA | CFTR, peptide-encoding region |
| CFTR-3′-Xho | AAACTCGAGAACGTTCTTCCTTATTATCTGGA TCATATGATCCTCCACGTTCTTCCTTGTTATCCGGGTCGTAACT | CFTR, peptide-encoding region |
Underlining indicates restriction enzyme cleavage sites.
Subcloning, DNA sequencing, and encoded-protein analysis of mlp and rlp.
A restriction endonuclease profile was determined for the λ8 DNA insert, and two SalI/EcoRI fragments of ∼5 and 10 kb were cloned into pBluescriptII to generate pPNG606 and pPNG607, respectively. The insert in pPNG606 was sequenced fully by primer walking. In the case of pPNG607, the presence of highly conserved repeats prevented complete first-pass primer walking. Therefore, subclones of HindIII and HindIII/XbaI fragments were constructed and the inserts were sequenced. The sequence of this region was confirmed by sequencing directly from pPNG607 with custom primers complementary to the second strand, and these sequence reads spanned the subcloned fragments to ensure correctness. The 3′ end of mlp was characterized by using the PCR-assisted gene walking (PAW) approach. DNA downstream of that cloned in pPNG607 was amplified by PCR with a primer which binds near the 3′ end of pPNG607 (Mlp-Paw-C) and a primer which binds to the pUC18 multiple cloning site (pUC-Bam). A mixed plasmid preparation from the L. fermentum BR11 pUC18 genomic library was used as the template, and the PCR was carried out with the Expand high-fidelity PCR system according to instructions of the manufacturer (Roche). A number of DNA fragments of different sizes were generated, as expected due to the different-sized inserts in the library. DNA of greater than 600 bp was purified from an agarose gel, cleaved with BamHI and XbaI, and then cloned into pUC18. A ∼1.6-kb fragment was cloned, partially sequenced, and found to contain the 3′ end of mlp and 350 bp of downstream sequence. Surprisingly, a single-base polymorphism was found when this sequence was compared with that in the 3′ end of the pPNG607 sequence, even though a proofreading DNA polymerase was used in the PCR. Therefore, a primer which is specific for a region 100 bp downstream of the mlp gene (Mlp-C-check) was used in combination with the Mlp-Paw-C primer to amplify the 3′ 1.2 kb of mlp and 100 bp downstream of mlp from genomic DNA as the template in order to determine if there is more than one mlp gene in L. fermentum BR11. This is further discussed in Results.
To determine the levels of similarity between Mlp, Rlp, and other proteins, the BlastX program was used (1). The signal sequence of Mlp was predicted by using the SignalP program (22). The protein repeats of Rlp and Mlp were analyzed with the Radar (10) and Blast2 (39) programs.
Construction of expression cassettes.
The first model peptide chosen for expression in this study was the His6 epitope. The second peptide corresponds to the 10 amino acids of the first extracellular domain of the human cystic fibrosis transmembrane conductance regulator (CFTR) protein (amino acids 108 to 117 of mature CFTR; peptide sequence, SYDPDNKEER). This peptide has previously been shown to inhibit internalization of Pseudomonas aeruginosa and Salmonella enterica serovar Typhi into lung and intestinal epithelial cells, respectively (25, 26). The expression constructs were generated following PCR amplification of DNA fragments and directional cloning with restriction endonuclease cleavage sites inserted onto the ends of oligonucleotides (Table 1). DNA encoding the His6 epitope was including in the oligonucleotides that contain His in their names. The bspA gene and terminator fragments used to generate the BspA-His6-CFTR construct were amplified by using oligonucleotides BspA-Xba with BspA-LPX-Pst-His and Term-Xho with Term-Hind, respectively, using pMFT1 as a template (41). DNA encoding three copies of the CFTR peptide was amplified by using overlapping oligonucleotides CFTR-5′-Pst and CFTR-3′-Xho. The bspA gene and DNA encoding the BspA secretion signal used to generate His6-CFTR-Rlp were amplified by using oligonucleotides BspA-Xba with BspA-Eco-stop and BspA-Eco-SS with BspA-SS-Pst-His, respectively. The rlp 3′ end and terminator were amplified by using oligonucleotides Rlp"-Pst-Xho and Rlp-Sal-Apa with pPNG606 as the template. The mlp DNA fragment to generate Mlp-His6 was amplified from pPNG606 by using oligonucleotides MlpN-Xba and MlpN-Xho-His. The mlp DNA fragment to generate Mlp-His6-CFTR was amplified from pPNG607 by using oligonucleotides Mlprep-Xba and Mlprep-Pst-His, respectively. All PCRs were performed with the Expand high-fidelity system (Roche), except for the CFTR peptide PCR, with which Platinum Taq (Invitrogen, Carlsbad, Calif.) was used.
Transformation of L. fermentum BR11 and integration of plasmids.
Transformation of L. fermentum BR11 with plasmids was done by a procedure similar to that described previously (42). Following outgrowth in the presence of a subinhibitory concentration of erythromycin, the cells were plated onto MRS agar containing 10 μg of erythromycin per ml and grown at 37 instead of 30°C. After overnight growth, colonies were inoculated into MRS liquid medium containing 10 μg of erythromycin per ml and incubated at 40°C.
Cell fractionation, protein extraction, and Western blot analysis.
Cell extracts were prepared from early-stationary-phase cultures (optical density at 600 nm [OD600], ∼2.6), while supernatants were taken from exponentially growing cultures (OD600, ∼1.0). Two different whole-cell protein extraction methods were used. The first involved boiling cells in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (32) for 5 min. The other involved suspending the cells in 20 mM triethanolamine-HCl (pH 7.6) containing 0.1 mM EDTA and sonicating (3 30-s bursts at 10 W) with a 36810 series Torbeo ultrasonic cell disruptor (Cole-Parmer, Vernon Hills, Ill.). Extractions of L. fermentum strains with 5 M LiCl were done as described previously (42). Supernatants were filtered through 0.2-μm-pore-size membranes (Schleicher and Schuell, Dassel, Germany) and then precipitated with 5% (wt/vol) ice-cold trichloroacetic acid, washed with acetone, and resuspended in equal amounts of 50 mM NaOH and 2× SDS-PAGE loading buffer. Prior to loading of SDS-PAGE, all samples were boiled for 5 min. Proteins were transferred to nitrocellulose, blocked, and then probed with an anti-His5 monoclonal antibody (Qiagen, Hilden, Germany) at a 1:1,000 dilution. Following washes, the membrane was incubated with rabbit anti-mouse immunoglobulin-horseradish peroxidase conjugate (Dako, Glostrop, Denmark). The bound antibodies were detected by using a horseradish peroxidase chemiluminescence kit (Roche). To estimate levels of His6 proteins in extracts, various amounts of His6-labeled protein markers (Qiagen) were included alongside the samples. These markers have known quantities of His6-containing proteins in each band, allowing densitometry to be done on films with the TotalLab version 1.11 package (Phoretix, Newcastle upon Tyne, United Kingdom).
Cell surface display assay.
The assay of accessibility of the His6 epitope on whole cells was done similarly to that described previously (43). Exponentially grown cells were washed with phosphate-buffered saline (PBS) and resuspended to an OD600 of ∼14 in PBS. One milliliter of the cell suspension was blocked with 1% casein in PBS for 30 min and then reacted with anti-His5 antibody at a 1:1,000 dilution in PBS containing 1% casein for 30 min. Following two washes with PBS, the cells were suspended in rabbit anti-mouse immunoglobulin-alkaline phosphatase conjugate (Dako) at a 1:1,000 dilution in PBS containing 1% casein and incubated for 30 min. Following two further washes in PBS, the cells were suspended in 1 M Tris-HCl (pH 9). Several aliquots were taken and incubated with p-nitrophenyl phosphate substrate (Sigma, St. Louis, Mo.), and the absorbance at 405 nm was measured with an automated plate reader. In addition, 2 μl of each cell suspension was spotted onto nitrocellulose and then incubated with Western blue stabilized substrate for alkaline phosphatase (Promega).
Nucleotide sequence accession number.
The sequence of the rlp and mlp locus has been deposited in GenBank under accession number AY240028.
RESULTS
Cloning and characterization of a locus encoding two putative surface proteins from L. fermentum BR11.
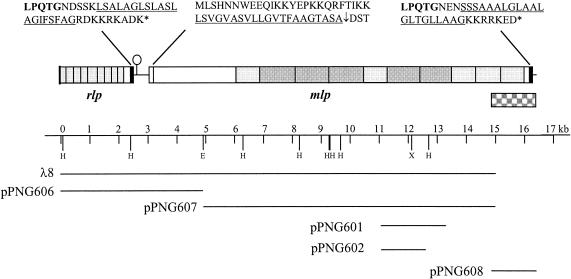
We have previously identified the gene encoding BspA by screening a pUC18 genomic library with antiserum raised against whole L. fermentum BR11 cells (41). Several more positive clones from this screen were identified and confirmed by Western blot analysis (data not shown). The plasmids from these clones were subjected to sequencing and were found to contain open reading frames (ORFs) encoding proteins similar to cell wall hydrolases, ClpC proteases (unpublished data), and the recently characterized mucus binding protein Mub from L. reuteri (30) (in pPNG601 and pPNG602). Two overlapping plasmids (pPNG601 and pPNG602) contained DNA similar to an internal region of mub, with the larger plasmid containing a 2.2-kb insert (Fig. 1). In order to clone a larger portion of this gene, termed mlp for Mub-like protein, a λ library was constructed and screened with a DNA probe containing part of the plasmid insert. Several positive plaques were isolated and were found to express large proteins reactive with whole-cell antiserum. These immunoreactive proteins were significantly larger than 200 kDa as determined by low-percent-polyacrylamide PAGE (data not shown). The 15-kb insert from one λ clone (λ8) was subcloned into two plasmids (pPNG606 and pPNG607) and fully sequenced (Fig. 1). The mlp gene was truncated at the 3′ end in λ8, so the sequence of the last 1.2 kb of the gene was obtained by using a PCR-based gene walking procedure (see Materials and Methods and “Identification of multiple mlp genes in L. fermentum BR11” below). The organization of open reading frames at this locus is shown in Fig. 1.
FIG. 1.
The 16,401-bp DNA region, showing the arrangement of mlp and rlp, and the λ and plasmid clones used to determine the sequence. The letters above the boxes indicate the cell wall sorting (black boxes) and secretion signals of the encoded proteins. The underlined sequences indicate hydrophobic domains. The shaded boxes indicate the encoded amino acid repeat regions, with light and dark boxes indicating the different-sized repeats in Mlp. The lollipop indicates a potential stem-loop structure. The checkered box indicates the DNA region which was amplified by PCR to determine the 3′ end sequence of mlp and to identify multiple mlp genes. The oligonucleotides Mlp-Paw-C and Mlp-C-check (Table 1) were used to amplify this region.
Mlp is predicted to have a 46-amino-acid secretion signal, a 957-amino-acid N-terminal nonrepetitive sequence, an internal repetitive region consisting of five imperfect repeats of 275 to 280 amino acids each and five imperfect repeats of 371 to 407 amino acids each, and a typical C-terminal gram-positive bacterial cell wall sorting signal containing the sequence LPQTG (Fig. 1). Mlp has similarity to several hypothetical proteins from the unpublished Lactobacillus gasseri genome which have potential LPXTG cell wall sorting signals (Lgas0953 and Lgas0817) and to some without LPXTG cell wall sorting signals (Lgas0292 and Lgas0954) (Table 2). Mlp also has strong similarity to the human and bovine hr44 proteins and similarity to the L. lactis YwfG protein (Table 2).
TABLE 2.
Proteins similar to Mlp and Rlp
| Protein (no. amino acids) | Similar proteins | % Identity/no. of amino acids | Function of similar protein if known | Reference |
|---|---|---|---|---|
| Mlp (4,427) | Lactobacillus gasseri | |||
| Lgas0953 | 28/3,825 | |||
| Lgas0817 | 23/1,737 | |||
| Lgas0292 | 26/602 | |||
| Lgas0954 | 21/874 | |||
| Lactobacillus reuteri Mub | 21/2,038 | Mucus binding | 30 | |
| Homo sapiens hr44 | 58/224 | 6 | ||
| Lactococcus lactis YwfG | 22/702 | 5 | ||
| Rlp (831) | Streptococcus agalactiae Rib | 38/747 | 44 | |
| Streptococcus pyogenes R28 | 38/736 | Adhesion to epithelial cells | 37 | |
| Enterococcus faecalis Esp | 33/756 | Biofilm formation | 40 | |
| Streptococcus agalactiae α | 32/768 | 20 | ||
| Lactobacillus gasseri | ||||
| Lgas1358 | 29/802 | |||
| Lgas0230 | 29/745 | |||
| Lgas0701 | 23/246 | |||
| Staphylococcus aureus Bap | 22/528 | Biofilm formation | 7 |
Upstream of mlp is an ORF which potentially encodes another cell wall-anchored surface protein (Fig. 1). The protein product has similarity to the surface protein Rib from Streptococcus agalactiae (Table 2) and therefore has been termed Rlp, for Rib-like protein. The partial sequence obtained shows that Rlp contains at least nine imperfect amino acid repeats of between 69 and 100 amino acids (average of 80 amino acids) and a typical C-terminal gram-positive bacterial cell wall sorting signal containing the sequence LPQTG (Fig. 1). Rlp also has similarity to the R28 putative adhesin from S. pyogenes, the Esp surface protein from Enterococcus faecalis, the α protein from S. agalactiae, several hypothetical proteins from the unpublished L. gasseri genome containing LPXTG type cell wall sorting signals (Lgas1358, Lgas0230, and Lgas0701), and biofilm-associated surface protein Bap from S. aureus (Table 2). Therefore, this chromosomal locus in L. fermentum BR11 appears to encode at least two repetitive surface proteins that have similarity to adhesion, pathogenicity, or colonization determinants.
Identification of multiple mlp genes in L. fermentum BR11.
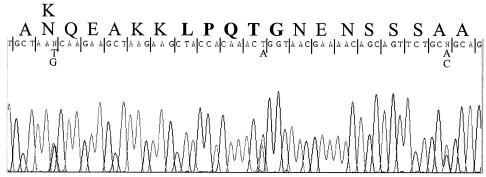
PCR was carried out with chromosomal DNA as a template and primers which flank the 3′ end of mlp (Fig. 1). Partial sequencing of the amplified 1.3-kb DNA fragment revealed the presence of a number of nucleotide polymorphisms. An example of part of the sequence trace obtained with the downstream primer (Mlp-C-check) is shown in Fig. 2. To ensure that this was not due to misincorporations occurring during the PCR, another PCR was carried out and the product was partially sequenced. The same polymorphisms were also clearly present in this sequence trace. Polymorphisms also are present in a nonrepetitive region (around the LPXTG sorting signal); therefore, they are not due to Mlp-Paw-C oligonucleotide mispriming in different repeats. This result demonstrates that L. fermentum BR11 contains multiple mlp genes and that they are very conserved, at least at the 3′ end. The amplified DNA from the PCR was cloned into pGEM-T, and partial sequence analysis of six plasmids revealed four different sequence types. At least two mlp sequence types have a 100% identical overlapping sequence (94 bp) with the 3′ end of pPNG607, which therefore makes it impossible to ascertain which is the correct 3′ sequence of the mlp gene cloned in pPNG607. Of the 900 bp sequenced from these two sequence types, only two nucleotides are different and there is no change in the amino acid sequence. Due to the high similarity of these two sequence types, the full sequence of one of the inserts (in pPNG608) was arbitrarily assigned as the 3′ end of mlp.
FIG. 2.
Reverse complement of a DNA sequencing trace of the 1.3-kb mlp 3′ PCR fragment (checkered box in Fig. 1) with the Mlp-C-check oligonucleotide. Above the DNA sequence is the encoded amino acid sequence of the Mlp protein, with the LPQTG motif in boldface. Double peaks indicate polymorphic nucleotides, which are included under the assigned sequence. The first polymorphism from the left shows that the encoded amino acid is different in the two Mlp proteins.
Cell surface display of the CFTR and His6 peptides by using BspA and Rlp.
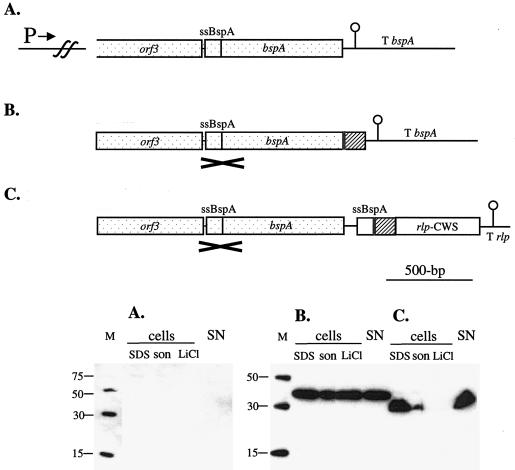
The cell wall sorting signal of Rlp was compared with BspA in its ability to display heterologous peptides on the surface of L. fermentum BR11. The human CFTR peptide was used, since it has potential as a therapeutic agent because it has been shown to reduce uptake of S. enterica serovar Typhi into human intestinal epithelial cells (26). The His6 peptide was also used to facilitate detection of the fusion proteins. To enable direct comparison, both constructs were integrated into the chromosome downstream of the bspA promoter (Fig. 3). The BspA fusion protein construct was generated similarly to that previously described (43). The Rlp fusion protein construct was placed downstream of a full-length and unaltered bspA gene which served as the homologous DNA for the chromosomal integration process. The last 161 amino acids of the Rlp protein were used as the cell wall anchor signal, and the BspA secretion signal was used to direct export of this Rlp fusion protein.
FIG. 3.
Gene constructs used to integrate into the L. fermentum BR11 bspA locus (top panels) and Western blot detection of fusion proteins in cell extracts and in the supernatant with anti-His5 antibody (bottom panels) for the native bspA genomic locus (A), the BspA-His6-CFTR fusion protein (B), and the His6-CFTR-Rlp fusion protein (C). In the diagrams in the top panels, the bspA upstream promoter (P→), the bspA terminator (T bspA), the rlp terminator (T rlp), and DNA encoding the BspA secretion signal (ssBspA), His6 (grey box), three copies of the CFTR peptide (hatched box), and the cell wall sorting signal of Rlp (rlp-CWS) are indicated. The DNA region which is the site of single-crossover homologous recombination into the bspA locus is stippled and marked with a cross. In the bottom panels, the Western blots contained His6 molecular mass markers (with sizes in kilodaltons) (lanes M); cell extracts prepared by boiling in 2× loading dye (lanes SDS), by sonication (lanes son), and with 5 M LiCl (lanes LiCl); and precipitated supernatant fractions (lanes SN). The amounts of cells or medium loaded in each lane are equivalent to 500 μl (lanes SDS), 50 μl (lanes son), 160 μl (lanes LiCl), and 625 μl (lanes SN) of culture.
Western blot analysis with an anti-His5 antibody showed that both fusion proteins were associated with the cells and also in the supernatant (Fig. 3). The predicted molecular mass of the Rlp fusion protein is 19 kDa; however, the band was approximately 28 kDa. The BspA fusion protein is 32 kDa, but the band was approximately 38 kDa. Fourteen percent of the BspA fusion protein was found in the supernatant, while 34% of the Rlp fusion protein was found in the supernatant (Table 3). The amount of protein associated with cells was estimated at 4 mg per liter for the BspA fusion protein and at 0.7 mg per liter for the Rlp fusion protein. Taking the sizes of the fusion proteins into account, cells expressing the BspA fusion contained approximately three times more His6 peptide than cells expressing the Rlp fusion protein. The percentage of BspA fusion protein released from cells by using 5 M LiCl was 65%, compared with only 6% for the Rlp fusion protein (Table 3).
TABLE 3.
Levels and distribution of His6-containing fusion proteins in L. fermentum strains
| Vector type | Fusion protein (size in kDa) | Protein (mg/liter of culture)
|
Surface display efficiencyf (mean ± SD) | ||||
|---|---|---|---|---|---|---|---|
| Total (cell extract + supernatant)a | SDSb | Sonc | LiCld | SNe | |||
| Surface display | BspA-His6-CFTR (32) | 4.7 | 0.78 | 4.0 | 2.6 | 0.68 | 2.03 ± 0.01 |
| His6-CFTR-Rlp (19) | 1.1 | 0.47 | 0.71 | 0.04 | 0.37 | 10.01 ± 0.03 | |
| Secretion | Mlp-His6 (27) | 2.2 | 0.46 | 2.1 | 0 | 0.06 | ND |
| Mlp-His6-CFTR (328) | 12.0 | 1.7 | 1.2 | 0 | 10.3 | ND | |
Calculated by using the cell extract method which yielded the greatest amount of protein (either SDS or Son).
SDS, proteins extracted from cells by boiling in 2× SDS loading buffer.
Son, proteins extracted from cells by sonication.
LiCl, proteins extracted from cells by 5 M LiCl.
SN, proteins from supernatant.
Whole-cell ELISA results presented as the fold increase in OD405 signal per OD600 of cells ± over that obtained with wild-type L. fermentum BR11. The OD405 signal per OD600 of cells for L. fermentum BR11 was 0.0226 ± 0.0006. gND, not determined.
To test the ability of Rlp to display the heterologous peptides on the cell surface, a whole-cell enzyme-linked immunosorbent assay was used (43). The cells expressing the BspA fusion protein bound significantly more anti-His5 antibody than wild-type cells, while the cells expressing the Rlp fusion protein bound significantly more anti-His5 antibody than those expressing the BspA fusion protein (Table 3). A similar result was also seen when 2 μl of the antibody-reacted cells was spotted onto nitrocellulose and incubated with substrate (data not shown). Therefore despite having ∼3-fold-fewer cell-associated molecules, the Rlp cell wall anchor fusion protein is much more efficiently recognized by antibodies than the BspA fusion protein on whole cells.
Secretion of the CFTR and His6 peptides by using Mlp.
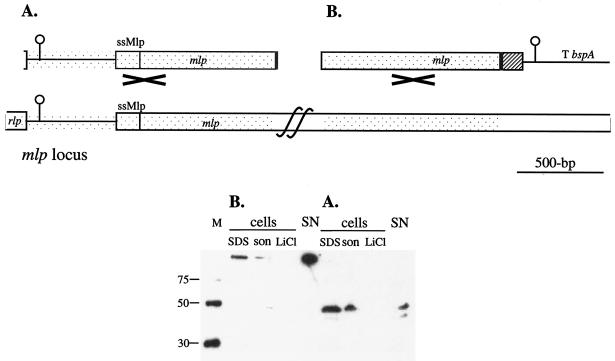
To test Mlp as a secretion vector in L. fermentum BR11, two different constructs were made and integrated into the mlp gene such that they are under the control of the mlp promoter. The first fusion protein consisted of the Mlp putative secretion signal and the next 261 amino acids of the nonrepetitive amino-terminal region, as well as a His6 peptide at the very carboxyl terminus (Fig. 4). The second fusion protein consisted of the Mlp putative secretion signal, 3,014 amino acids of Mlp including the nonrepetitive region and nearly six repeats, the His6 peptide, and the CFTR peptide at the carboxyl terminus (Fig. 4). Western blot analysis revealed that both fusion proteins were secreted and also associated with the cells (Fig. 4). The small Mlp-His6 fusion protein produced two bands in the SDS and supernatant extracts but only a single band in the sonication extracts (Fig. 4A). The bands resolved at approximately 40 to 45-kDa which is in contrast to its calculated size of 27-kDa (Fig. 4A). The large Mlp-His6-CFTR fusion protein, which has a predicted molecular mass of 328 kDa, resolves as a single band which just enters the separating gel (Fig. 4B). For the cells expressing the small Mlp-His6 fusion protein, only 3% of the His6-containing protein was found in the supernatant, while cells expressing the large Mlp-His6-CFTR fusion protein had 86% of the His6-containing protein in the supernatant (Table 3). The level of the small Mlp-His6 fusion protein detected in the supernatant was only 0.06 mg per liter, while 10.3 mg of the large Mlp-His6-CFTR fusion protein per liter was detected in the supernatant (Table 3). When the sizes of these two proteins are taken into account, there was approximately 14 times more large Mlp-His6-CFTR fusion protein than small Mlp-His6 fusion protein detected in the supernatant. This result shows that secretion of a very large heterologous fusion protein by L. fermentum BR11 can be very efficient.
FIG. 4.
Gene constructs used to integrate into the L. fermentum BR11 mlp locus (top panels) and Western blot detection of fusion proteins in cell extracts and in the supernatant with anti-His5 antibody (bottom panels) for the Mlp-His6 fusion protein (A) and the Mlp-His6-CFTR fusion protein (B). In the diagrams in the top panels, the bspA terminator (T bspA) and DNA encoding the Mlp secretion signal (ssMlp), His6 (grey box), and three copies of the CFTR peptide (hatched box) are indicated. The DNA region which is the site of single-crossover homologous recombination into the mlp locus is stippled and marked with a cross. Due to the large size of mlp, the full length of the gene could not be shown. In the bottom panels, the Western blots contained His6 molecular mass markers (with sizes in kilodaltons) (lanes M); cell extracts prepared by boiling in 2× loading dye (lanes SDS), by sonication (lanes son), and with 5 M LiCl (lanes LiCl); and precipitated supernatant fractions (lanes SN). The amounts of cells or medium loaded in each lane are the equivalent to 500 μl (lanes SDS), 50 μl (lanes son), 160 μl (lanes LiCl), and 625 μl (lanes SN) of culture.
DISCUSSION
This paper describes the identification and characterization of two tandem genes from L. fermentum BR11 which encode proteins similar to surface proteins from other gram-positive bacteria and the construction of protein surface display and secretion systems by using these two proteins. Although Mlp and Rlp have not definitively been shown to be surface localized, they do contain features very typical of cell wall covalently anchored surface proteins, including amino acid repeat regions, LPXTG cell wall anchoring signals, and, in the case of Mlp, a putative secretion signal (21). The functions of Mlp and Rlp are as yet unknown; however, due to their similarity to putative adhesin proteins such as Mub and R28, respectively, they may function as colonization determinants. Like the case of Mlp in L. fermentum BR11, a number of other gram-positive bacteria have multiple copies of cell wall-anchored surface proteins, including the CshA and CshB as well as the SspA and SspB proteins of Streptococcus gordonii (8, 19).
Mlp is one of the largest bacterial surface proteins thus far characterized; it is predicted to have 4,427 amino acids and a molecular mass of 473 kDa. Mlp is more similar to Lgas0953 from L. gasseri than to Mub from the much more closely related L. reuteri. It is possible that multiple Mub homologs which are more closely related to Mlp exist in L. reuteri. Although L. fermentum BR11 contains multiple genes with similarity to mub, we have been unable to demonstrate binding of L. fermentum BR11 cells to pig gastric mucin (M1778; Sigma) by using a method similar to that used previously to demonstrate L. reuteri and Mub binding (30). This is probably not due to L. fermentum BR11 mlp being silent, since part of this gene was initially cloned from a pUC18 genomic library by using an antiserum raised against whole cells. It is possible that Mlp may bind to a different receptor than Mub, since there is reasonable divergence between the two protein sequences. Mlp contains larger repeats than Mub (275 to 407 amino acids long in Mlp and 183 to 197 amino acids long in Mub), and Mlp contains an approximately 60-amino-acid proline-rich region (around 30% proline) in every repeat which is not present in Mub. This proline-rich region is present in at least some repeats of the Lgas0953 protein and is also in the human and bovine hr44 proteins.
The discovery of Rlp represents the first report of a Rib family protein present in a nonpathogenic organism. Rlp contains nonidentical repeats, unlike Rib and α from S. agalactiae and R28 from S. pyogenes, which contain identical repeats. The repeats of Rib, α, and R28, however, are not identical between proteins (37). The repeats from the streptococcal proteins are 79 to 82 amino acids long, while L. fermentum BR11 Rlp repeats range from 69 to 100 amino acids in length, with the majority (six of nine) ranging from 73 to 79 amino acids in length. The size of Rlp is not known; however, the sizes of the putative Rlp homologs from L. gasseri suggest that it may be another very large surface protein (Lgas1358, Lgas0203, and Lgas0701 are 2,549, 2,456, and 268 amino acids long, respectively). Attempts to clone the 5′ end of rlp have been unsuccessful. Similarly, the cloning of rib in high-copy-number plasmids in E. coli was found to be unstable (44). Rlp has similarity to the R28 protein, which has been implicated in promoting adhesion of S. pyogenes to epithelial cells (37), and also to Esp from E. faecalis and Bap from S. aureus, both of which have been shown to be involved in biofilm formation (7, 40). It is therefore probable that Rlp may function as an adhesin and/or may be involved in biofilm development.
The carboxyl-terminal 161-amino-acid region of Rlp containing a cell wall anchoring signal has been shown to be a significantly better peptide surface display vector than the previously developed noncovalently anchored BspA system. This was despite there being threefold more cell-associated BspA fusion molecules than Rlp fusion molecules. This may be because either (i) a major portion of BspA is located not on the outside of the cell wall but instead between the cytoplasmic membrane and the peptidoglycan or (ii) a significant amount of the BspA fusion is washed away from the cells during the wash steps of the whole-cell enzyme-linked immunosorbent assay. Interestingly, a very closely related BspA homolog from L. fermentum RC-14 can be removed from cells by suspension in PBS for 2 h, suggesting that BspA is easily washed from cells even by unconcentrated salt solutions (11). Both the Rlp and BspA fusion proteins were detected in concentrated supernatants. This may be due to cell wall turnover, cell lysis, or limiting sortase enzyme in the case of the Rlp fusion protein (9). A number of LPXTG-containing proteins from their native host have been detected in supernatants. These include M6 from S. pyogenes and Mub from L. reuteri (9, 30). Recently, a closely related homolog of BspA (MapA from L. fermentum 104R; accession number AJ293860) has been detected in supernatant fractions as well as cell surface protein extracts (28). The absence of multiple bands in the protein extracts of cells expressing the BspA and Rlp fusion proteins suggests that the BspA secretion signal is very efficient in driving secretion of heterologous proteins.
Large and small Mlp fragments were tested as potential secretion fusion partners. The significant difference in secretion efficiency between the small and large Mlp fusion proteins is probably due to stability of the His6-containing proteins in the supernatant in the presence of cell surface and secreted proteases. Indeed, no His6-containing large Mlp-His6-CFTR fusion protein could be detected from overnight stationary-phase culture supernatants, indicating degradation of at least the His6 peptide. Mlp fusion protein associated with the cell appears to be more stable than that in the supernatant, since the large Mlp-His6-CFTR protein could be detected in overnight stationary-phase culture cell extracts (data not shown). Similarly, when L. fermentum BR11 was used to express and secrete the amino terminus of protein A, most (80%) of the protein was found associated with cells and not in the supernatant (31). The small Mlp-His6 fusion protein migrated as a 45-kDa protein in SDS-PAGE, a molecular mass which is much greater than the predicted 27 kDa. This may be due to glycosylation of this protein. Like for Mub, the mature 144 amino acids of Mlp are rich in serine (23%), which may be potential O-linked glycosylation sites. Recently, the LPXTG-containing Fap1 fimbrial adhesin of Streptococcus parasanguis was confirmed to be a glycoprotein containing sugar residues that are O linked to serine amino acids (38).
Levels of heterologous protein produced by lactobacilli are reported to be 2 to 4 mg per liter with M6 as a cell surface fusion protein (9), 3 mg per liter with BspA as a fusion protein (43), and 10 mg per liter with M6 or protein A as a secretion fusion protein (13, 31). The number of cell-associated BspA fusion protein molecules detected in this study is similar to that reported previously, while the number of cell-associated Rlp fusion protein molecules is ∼3-fold lower. This suggests that the bspA promoter is strong and able to drive expression of high levels of protein from a single chromosomally integrated gene, compared with other expression systems which are plasmid encoded. The number of cell-associated and secreted Mlp fusion protein molecules is two- to fourfold lower than the number of cell-associated and secreted BspA fusion protein molecules. This suggests that the Mlp promoter is also highly active. Recently, L. brevis and L. acidophilus S-layers have been investigated for the cell surface display of heterologous peptides by modification of the chromosomal S-layer genes (3, 36). Although S-layers are expressed at very high levels, a major limitation of S-layer-based expression systems is that many fusion partners disrupt the ability of the S-layer subunit to polymerize into the S-layer lattice. In contrast, LPXTG and BspA protein surface display systems have been proven to be versatile and to be capable of accommodating very large fusion partners (12, 14).
A number of therapeutic proteins which interfere with attachment of pathogenic organisms to host surfaces have been characterized (15). It has been shown that a peptide corresponding to the pathogen receptor region of the human CFTR protein, which was inserted into the BspA, Rlp, and Mlp fusion proteins, binds to lipopolysaccharides of S. enterica serovar Typhi and P. aeruginosa and can reduce their internalization by human intestinal or airway epithelial cells (25, 26). Further work is needed to examine the ability of CFTR peptide-expressing L. fermentum strains and purified CFTR fusion proteins to bind to lipopolysaccharide and prevent pathogen internalization.
Acknowledgments
This work was supported by the Australian Dairy Research and Development Corporation (grant QUT10723), a Queensland University of Technology Encouragement Award, and Queensland University of Technology Faculty of Science and School of Life Sciences.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antikainen, J., L. Anton, J. Sillanpaa, and T. K. Korhonen. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46:381-394. [DOI] [PubMed] [Google Scholar]
- 3.Avall-Jaaskelainen, S., K. Kyla-Nikkila, M. Kahala, T. Miikkulainen-Lahti, and A. Palva. 2002. Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl. Environ. Microbiol. 68:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beninati, C., M. R. Oggioni, M. Boccanera, M. R. Spinosa, T. Maggi, S. Conti, W. Magliani, F. De Bernardis, G. Teti, A. Cassone, G. Pozzi, and L. Polonelli. 2000. Therapy of mucosal candidiasis by expression of an anti-idiotype in human commensal bacteria. Nat. Biotechnol. 18:1060-1064. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, G., N. M. McKechnie, and W. Gurr. 1995. Molecular and immunological characterization of hr44, a human ocular component immunologically cross-reactive with antigen Ov39 of Onchocerca volvulus. J. Exp. Med. 182:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 9.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J.-C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heger, A., and L. Holm. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41:224-237. [DOI] [PubMed] [Google Scholar]
- 11.Heinemann, C., J. E. van Hylckama Vlieg, D. B. Janssen, H. J. Busscher, H. C. van der Mei, and G. Reid. 2000. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol. Lett. 190:177-180. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, A. R., C. Gilbert, J. M. Wells, and H. F. Jenkinson. 1998. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hols, P., P. Slos, P. Dutot, J. Reymund, P. Chabot, B. Delplace, J. Delcour, and A. Mercenier. 1997. Efficient secretion of the model antigen M6-gp41E in Lactobacillus plantarum NCIMB 8826. Microbiology 143:2733-2741. [DOI] [PubMed] [Google Scholar]
- 14.Hung, J., C. Rathsam, N. A. Jacques, and P. M. Giffard. 2002. Expression of a streptococcal glucosyltransferase as a fusion to a solute-binding protein in Lactobacillus fermentum BR11. FEMS Microbiol Lett. 211:71-75. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, C. G., D. Medaglini, J. S. Younson, and G. Pozzi. 2001. Biotechnological approaches to fight pathogens at mucosal sites. Biotechnol. Genet. Eng. Rev. 18:329-347. [DOI] [PubMed] [Google Scholar]
- 16.Kruger, C., Y. Hu, Q. Pan, H. Marcotte, A. Hultberg, D. Delwar, P. J. van Dalen, P. H. Pouwels, R. J. Leer, C. G. Kelly, C. van Dollenweerd, J. K. Ma, and L. Hammarstrom. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat. Biotechnol. 20:702-706. [DOI] [PubMed] [Google Scholar]
- 17.Maassen, C. B. M., J. D. Laman, M. J. Heijne den Bak-Glashouwer, F. J. Tielen, J. C. P. A. van Hotlen-Neelen, L. Hoogteijling, C. Antonissen, R. J. Leer, P. H. Pouwels, W. J. A. Boersma, and D. M. Shaw. 1999. Instruments for oral disease-intervention strategies: recombinant Lactobacillus casei expressing tetanus toxin fragment C for vaccination or myelin proteins for oral tolerance induction in multiple sclerosis. Vaccine 17:2117-2128. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, B., J. Sillanpaa, E. Smit, T. K. Korhonen, and P. H. Pouwels. 2000. Expression of cbsA encoding the collagen-binding S-protein of Lactobacillus crispatus JCM5810 in Lactobacillus casei ATCC 393T. J. Bacteriol. 182:6857-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNab, R., H. F. Jenkinson, D. M. Loach, and G. W. Tannock. 1994. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 14:743-754. [DOI] [PubMed] [Google Scholar]
- 20.Michel, J. L., L. C. Madoff, K. Olson, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89:10060-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 24.Piard, J. C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pier, G. B., M. Grout, and T. S. Zaidi. 1997. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc. Natl. Acad. Sci. USA 94:12088-12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pier, G. B., M. Grout, T. Zaidi, G. Meluleni, S. S. Mueschenborn, G. Banting, R. Ratcliff, M. J. Evans, and W. H. Colledge. 1998. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393:79-82. [DOI] [PubMed] [Google Scholar]
- 27.Reveneau, N., M. C. Geoffroy, C. Locht, P. Chagnaud, and A. Mercenier. 2002. Comparison of the immune responses induced by local immunizations with recombinant Lactobacillus plantarum producing tetanus toxin fragment C in different cellular locations. Vaccine 20:1769-1777. [DOI] [PubMed] [Google Scholar]
- 28.Rojas, M., F. Ascencio, and P. L. Conway. 2002. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roos, S., Aleljung, P., N. Robert, B. Lee, T. Wadstrom, M. Lindberg, and H. Jonsson. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol. Lett. 144:33-38. [DOI] [PubMed] [Google Scholar]
- 30.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 31.Rush, C., L. Hafner, and P. Timms. 1997. Protein A as a fusion partner for the expression of heterologous proteins in Lactobacillus. Appl. Microbiol. Biotechnol. 47:537-542. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Savijoki, K., M. Kahala, and A. Palva. 1997. High level heterologous protein production in Lactococcus and Lactobacillus using a new secretion system based on the Lactobacillus brevis S-layer signals. Gene 186:255-262. [DOI] [PubMed] [Google Scholar]
- 34.Scheppler, L., M. Vogel, A. W. Zuercher, M. Zuercher, J. E. Germond, S. M. Miescher, and B. M. Stadler. 2002. Recombinant Lactobacillus johnsonii as a mucosal vaccine delivery vehicle Vaccine. 20:2913-2920. [DOI] [PubMed]
- 35.Slos, P., P. Dutot, J. Reymund, P. Kleinpeter, D. Prozzi, M. P. Kieny, J. Delcour, A. Mercenier, and P. Hols. 1998. Production of cholera toxin B subunit in Lactobacillus. FEMS Microbiol. Lett. 169:29-36. [DOI] [PubMed] [Google Scholar]
- 36.Smit, E., D. Jager, B. Martinez, F. J. Tielen, and P. H. Pouwels. 2002. Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324:953-964. [DOI] [PubMed] [Google Scholar]
- 37.Stalhammar-Carlemalm, M., T. Areschoug, C. Larsson, and G. Lindahl. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33:208-219. [DOI] [PubMed] [Google Scholar]
- 38.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 39.Tatusova, T. A., and T. L. Madden. 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 40.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner, M. S., P. Timms, L. M. Hafner, and P. M. Giffard. 1997. Identification and characterization of a basic cell-surface-located protein from Lactobacillus fermentum BR11. J. Bacteriol. 179:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner, M. S., T. Woodberry, L. M. Hafner, and P. M. Giffard. 1999. The bspA locus of Lactobacillus fermentum BR11 encodes an l-cystine uptake system. J. Bacteriol. 181:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, M. S., and P. M. Giffard. 1999. Expression of Chlamydia psittaci- and human immunodeficiency virus-derived antigens on the cell surface of Lactobacillus fermentum BR11 as fusions to BspA. Infect. Immun. 67:5486-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wastfelt, M., M. Stalhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]