Abstract
We previously characterized Tri1, a gene required for hydroxylation of the C-8 position during trichothecene mycotoxin biosynthesis in Fusarium sporotrichioides NRRL 3299. Sequence analysis of the region surrounding Tri1 revealed a gene, named Tri16, which could encode an acyltransferase. Unlike the wild-type parent strain NRRL 3299, which accumulates primarily T-2 toxin along with low levels of diacetoxyscirpenol (DAS) and neosolaniol (NEO) and trace amounts of 8-propionyl-neosolaniol (P-NEO) and 8-isobutyryl-neosolaniol (B-NEO), mutants containing a disruption of Tri16 were blocked in the production of the three C-8 esterified compounds T-2 toxin, P-NEO, and B-NEO and accumulated the C-8-hydroxylated compound NEO along with secondary levels of DAS. These data indicate that Tri16 encodes an acyltransferase that catalyzes the formation of ester side groups at C-8 during trichothecene biosynthesis. We also report the presence of a Tri16 ortholog in Gibberella pulicaris R-6380 that is likely linked to a presumably inactive ortholog for Tri1.
One of the major concerns regarding the presence of fungi in both agricultural settings and, more recently, indoor settings with compromised air quality is the ability of some species to produce trichothecene mycotoxins (18, 19, 33). The toxicity of these sesquiterpenoid fungal secondary metabolites to eukaryotic cells makes them important in human and animal health (19) and some cases of plant pathogenesis (5, 15).
As a group, trichothecenes are characterized by the presence of a 12,13-epoxy-trichothec-9-ene nucleus, a feature that is necessary for toxicity (13, 32). However, the addition of different combinations of side groups to this trichothecene core nucleus plays an important role in determining the level of toxicity of each trichothecene. Although there are some notable exceptions, such as the acetylation of the hydroxyl group at the C-3 position which reduces the toxicity of many trichothecenes (1), in general, the greater the number and the more elaborate the side groups, the greater the toxicity (35). One of the toxicologically most important examples involves 4,15-diacetoxyscirpenol (DAS), neosolaniol (NEO), and T-2 toxin. These three trichothecenes display increasing toxicity in mice, as demonstrated by 50% lethal doses of 23.0, 14.5, and 5.2 mg/kg, respectively (34). Significantly, they differ from one another only at the C-8 position by the presence of a hydrogen, a hydroxyl group, and an esterified hydroxyl group, isovaleroxy, respectively.
Fusarium sporotrichioides and F. sambucinum have been used as model organisms in studies directed toward the identification of the genes that encode the enzymes responsible for the hydroxylations and esterifications that occur during trichothecene biosynthesis. F. sporotrichioides NRRL 3299 makes a closely related series of type A trichothecenes in culture, with the predominant one being T-2 toxin. Minor components include DAS, NEO, 8-propionyl-neosolaniol (P-NEO), and 8-isobutyryl-neosolaniol (B-NEO) (8). All five of these trichothecenes, except DAS, result from a hydroxylation at position C-8 and, with the exception of NEO, a subsequent esterification at C-8-OH and therefore differ from one another only at the C-8 position. Some isolates of Gibberella pulicaris also possess the ability to synthesize NEO, 8-acetylneosolaniol (8-Ac-NEO), and/or T-2 toxin, in addition to DAS (3, 30). Despite a well-characterized biosynthetic pathway for type A trichothecenes (12) and the fact that most of the identified trichothecene (Tri) genes constitute a large gene cluster (10, 17), the gene required for the esterification reaction at C-8 has not been identified.
In a previous study, we isolated several new candidate trichothecene genes on the basis of an expression pattern that is dependent upon the presence of two trichothecene regulatory genes, Tri10 and Tri6 (24). One of these genes was Tri1, which encodes a cytochrome P450 monooxygenase required for oxygenation of position C-8 in the biosynthesis of T-2 toxin (23). Tri1 is not closely linked to the core trichothecene cluster. In the present study, we characterized Tri16, a gene that we identified while sequencing the DNA region adjacent to Tri1. The resulting data indicate that Tri16 encodes a C-8 acyltransferase required for the production of T-2 toxin and establish the presence of a second trichothecene gene cluster. Together, Tri16 and Tri1 are responsible for the addition of the C-8 substituents required for the production of the most toxic trichothecenes made by F. sporotrichioides.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Wild-type strains F. sporotrichioides NRRL 3299, G. pulicaris (F. sambucinum) R-6380, and Gibberella zeae (Fusarium graminearum) GZ3639 have been described previously (6, 9). The wild-type strains were maintained on V8 juice agar (200 ml of V8 juice, 3 g of CaCO3, and 20 g of agar per liter), while transformant strains were maintained on V8 juice agar plus 300 μg of hygromycin B (Calbiochem, La Jolla, Calif.) per ml. Standard light and temperature conditions were used for growth of strains on V8 juice agar slants and plates (23). Conidia for all strains were stored as frozen (−80°C) glycerol-water (15:85) stocks. All fungal liquid cultures were incubated at 28°C and shaken at 200 rpm on an orbital shaker after being inoculated as follows with mycelia or conidia harvested from cultures grown for 7 days on V8 juice agar plates. For DNA extraction, 100 ml of YEPD-2G medium (2% glucose, 0.3% yeast extract, 1% peptone) was inoculated with mycelia (from one plate) and incubated for 24 h; for RNA extraction and toxin analysis, 100 ml (RNA) or 25 ml (toxin) of YEPD-5G medium (5% glucose, 0.1% yeast extract, 0.1% peptone) was inoculated at a density of 104 conidia/ml and incubated for 23, 27, and 36 h (RNA) or 7 days (toxin); for toxin analysis involving coinoculation of strains and whole-cell feeding experiments, cultures were inoculated at a density of 5 × 104 conidia/ml. For whole-cell feeding studies, NEO (Sigma, St. Louis, Mo.), in acetone, was added to cultures 24 h following inoculation. The final concentration of NEO was 250 μM, and the final concentration of acetone was 1%. Storage of clones from the cDNA library has been described previously (24). All bacterial culture media were supplemented with ampicillin at 100 μg/ml.
Nucleic acid isolation.
Isolation of genomic DNA and RNA has been described previously (16, 29). Plasmid DNA was isolated with the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.). Restriction fragments were separated on 1.2% agarose (Boehringer Mannheim, Indianapolis, Ind.) gels and purified with the QIAquick gel extraction kit (Qiagen).
Disruption plasmid construction and fungal transformation.
pKO16, the Tri16 disruption plasmid, was constructed by cloning a fragment of the Tri16 sequence obtained from a cDNA clone into the previously described fungal cloning vector pUCH3 (22). Specifically, a 963-bp fragment, including 18 bp of the pBluescript SK (Stratagene, La Jolla, Calif.) vector sequence at the 5′ end, was cut from plasmid DNA of the Tri16 cDNA clone 14J4 from our cDNA library (24) with the restriction enzymes BamHI and SalI (New England Biolabs, Beverly, Mass.) and ligated to BamHI- and SalI-cut pUCH3. The resulting plasmid, pKO16, harbors a doubly truncated fragment of the Tri16 coding sequence adjacent to the hygromycin B resistance gene (hygB) in pUCH3. Protoplast formation and fungal transformation were conducted as previously described (26, 29).
PCR, Northern, and Southern analyses.
PCR analyses of Tri16 transformants were conducted with primers A-90 (5′-GCCCTTTCTAGATGCCTGAG-3′), A-91 (5′-GTTGGTTGCCTCCGTCTTGG-3′), and S-12 (5′-CAGGTGGGCCTTGACATGTG-3′). PCR conditions were 30 cycles of 30 s at 95°C, 60 s at 51°C, and 90 s at 72°C and 1 cycle of 10 min at 72°C. All reactions were performed with Taq DNA polymerase (Promega, Madison, Wis.) in accordance with the manufacturer's instructions. For Northern analyses, RNA electrophoresis was conducted with 1% agarose gels containing 1.1% formaldehyde and each lane was loaded with 5 μg of total RNA (27). For Southern analyses of Tri16 transformants, electrophoresis was performed in accordance with standard protocols (27) and 5 μg of DNA from each transformant strain (1 μg from NRRL 3299) was used in restriction digests. Genomic DNA from all strains was digested with BglII (New England Biolabs), which does not cut the pKO16 vector. For Southern analysis of the Tri16 and Tri1 genes in F. sporotrichioides and G. pulicaris, 5 μg of DNA was used for each restriction digest with ApaI, SmaI, or EcoRI (New England Biolabs, Boehringer Mannheim). These enzymes were chosen because sequence information indicated that Tri16 and Tri1 in F. sporotrichioides are on the same EcoRI and SmaI fragments but on different ApaI fragments. The Tri16 and Tri1 probes for all Southern and Northern analyses were the cDNA insert of library clone 14J4 excised with BamHI and SalI and the cDNA insert of library clone 1J3 cut with ApaI and SmaI, respectively. Individual radioactively labeled DNA probes were prepared using [α-32P]dCTP (Amersham, Piscataway, N.J.) with the Nick Translation System (GIBCO BRL, Rockville, Md.). All Northern and Southern blots were made on Hybond-N+ nylon membranes (Amersham) and hybridized, washed, and stripped under high stringency as specified by the Gene Images nonisotopic nucleic acid detection kit (United States Biochemical, Cleveland, Ohio). Autoradiographic images were produced on BIOMAX MR or X-Omat LS imaging film (Kodak, Rochester, N.Y.).
DNA sequencing and sequence homology searches.
Inserts isolated from plasmid DNA preparations of cDNA clones were sequenced by using either the ABI Prism Dye Terminator cycle sequencing core kit or the BigDye Terminator cycle sequencing core kit (Perkin-Elmer, Boston, Mass.). All reactions were run on a 373 or 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) by the Gene Technologies Laboratory at Texas A&M University. Nucleotide sequences and their putative corresponding amino acid sequences were compared to the National Center for Biotechnology Information nonredundant database (GenBank+RefSeq Nucleotides+EMBL+DDBJ+PDB sequences) by using the blastn and blastp algorithms (4).
Toxin analyses.
Ethyl acetate extractions of whole-culture samples were performed as previously described (29). Trichothecenes in the ethyl acetate extracts were detected and identified by gas chromatography-mass spectrometry with a Hewlett-Packard 5890 gas chromatograph directly interfaced to a Hewlett-Packard 5970B mass spectrometer via a heated capillary interface. The gas chromatograph was configured with a splitless injection port and a DB-5ms fused-silica capillary column coated with a 0.25-μm film of stationary phase (30 m by 0.25 mm [internal diameter]; J&W Scientific, Folsom, Calif.). Sample volumes of 2 μl were introduced to a heated injection port. The injection and transfer line temperatures were 300°C. The oven temperature program consisted of an initial temperature of 175°C that was held for 2.0 min, ramped to 275°C at 23°C/min, held at 275°C for 2 min, ramped to 290°C at 30°C/min, and held at 290°C for 16 min. Helium was the carrier gas at a constant flow rate of 0.75 ml/min. A mass-selective detector was used in selective ion monitoring mode with an ionization energy of 70 eV and an ion source temperature of 180°C. T-2 toxin and NEO were purchased from Sigma and used without further purification. T-2 toxin was below the detection limit (<1 ppm) in the NEO standard.
Nucleotide sequence accession number.
The GenBank accession number for the Tri16 nucleotide and predicted protein sequences is AY187275.
RESULTS
Sequence of Tri16.
The largest putative open reading frame with a characteristic translational start site for Tri16 is 1,479 bp, which would encode a protein of 493 amino acids. Comparison of the genomic DNA sequence with cDNA sequence data generated by the F. sporotrichioides cDNA Sequencing Project (contig 799 and contig 873) and within our laboratory (clone 14J4) indicates the absence of any introns. Tri16's direction of transcription is the same as that of Tri1. The predicted protein sequence of TRI16 has the greatest identity (23 to 24%) with TRI101 (trichothecene 3-O-acetyltransferase) from various Fusarium species and with AYT1 from Saccharomyces cerevisiae (22%), which was designated an acetyltransferase on the basis of its ability to acetylate the C-3 position of isotrichodermol (2). Like those of TRI101 and AYT1, the TRI16 protein sequence contains a motif (HXXXDG) that is conserved among members of the BAHD superfamily of plant acyltransferase genes (28). However, the HXXXDG motif in TRI16 more closely resembles that of ATF1 (alcohol-O-acetyltransferase) from S. cerevisiae and TRI3 (trichothecene 15-O-acetyltransferase) from F. sporotrichioides and G. zeae because it does not contain the two-amino-acid insertion (HXXXDXXG) that is found in TRI101 and AYT1. A second motif in the carboxy terminus of BAHD acyltransferases, DFGXGKP, may be present in TRI16, but it would represent a unique case since there is an insertion of 11 amino acids (DFGXXXXXXXXXXXXGKP) rather than two amino acids (DFXXGXGKP) as found in TRI101 and AYT1. TRI3 and ATF1 do not possess this C-terminal motif (2).
Disruption of Tri16.
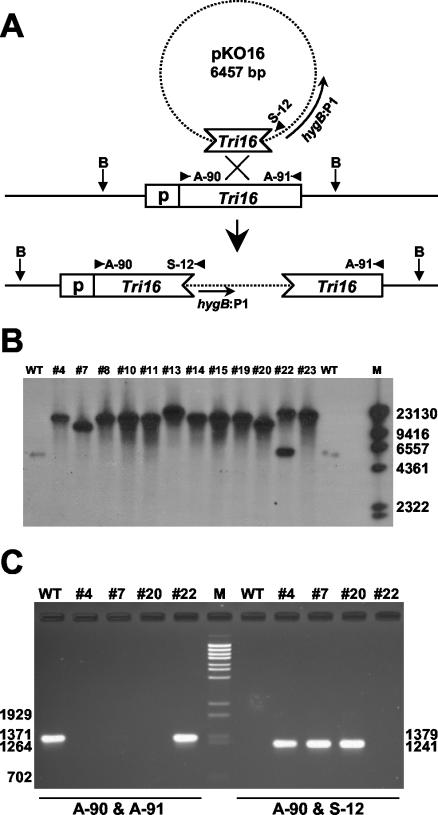
Tri16 mutants were generated by transforming F. sporotrichioides NRRL 3299 with pKO16. This vector contains a doubly truncated fragment of the Tri16 coding region. Homologous integration of a single copy of this construct results in two nonfunctional copies of the Tri16 gene, one truncated at the amino terminus (by 88 amino acids) and the other truncated at the carboxy terminus (by 91 amino acids) (Fig. 1A). Transformation of the wild-type strain yielded 28 hygromycin-resistant transformants. Southern analysis revealed that at least 11 of these transformants no longer contained a wild-type Tri16 gene. Two of these 11, Tx #7 and Tx #20, resulted from the homologous integration of a single copy of pKO16; the remainder resulted from the homologous integration of multiple tandem copies, e.g., Tx #4 (Fig. 1B). In contrast, transformant Tx #22 contained a wild-type-sized Tri16 BglII fragment, as well as a second, larger Tri16-hybridizing BglII fragment indicative of the ectopic integration of the transforming vector. These integration events were confirmed in a PCR with primer A-90 and vector-specific primer S-12 (Fig. 1A) that would yield a 1,241-bp product only if homologous integration occurred. Transformants Tx #4, Tx #7, and Tx #20 all produced this product (Fig. 1C), while neither the wild-type strain nor transformant Tx #22 did. We also conducted a PCR with primers A-90 and A-91 (Fig. 1A) and found that the wild-type strain and Tx #22 yielded the expected 1,379-bp PCR product for the wild-type Tri16 gene sequence but that Tx #4, Tx #7, and Tx #20 did not (Fig. 1C). Thus, Tx #4, Tx #7, and Tx #20 are transformants resulting from homologous integration of pKO16 while the transformed DNA in Tx #22 appears to have been inserted into the genome ectopically.
FIG. 1.
Disruption of Tri16 in F. sporotrichioides NRRL 3299. (A) Expected result for insertion of the Tri16 gene disruption construct pKO16 into the host genome via a single homologous integration event. B, BglII restriction sites. (B) Southern blot analysis of selected transformant strains. Wild-type (WT) and transformant genomic DNAs were cut with BglII, and blots were probed with a doubly truncated fragment of the Tri16 gene. M, lambda DNA cut with HindIII. All transformants except Tx #22 lack the wild-type Tri16 BglII fragment. Instead, transformants Tx #7 and Tx #20 contain a single fragment consistent with the expected combined size (approximately 13 kb) of the wild-type Tri16 BglII fragment and one homologously integrated copy of pKO16. The remaining transformants contain a larger fragment indicative of the presence of two (expected size, approximately 20 kb) or more tandem, homologously integrated copies of pKO16. Band sizes (bases) of the marker are shown at the right. (C) PCR analysis of wild-type and selected transformant strains. PCRs were performed with primers A-90 and A-91 and primers A-90 and S-12 (positions are shown in panel A). PCR products were resolved on an agarose gel and stained with ethidium bromide. M, lambda DNA cut with BstEII. Band sizes (bases) of the marker are shown at the left, and band sizes of the PCR products are shown at the right.
Northern analyses.
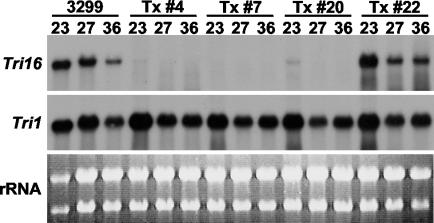
When RNA blots were hybridized with a probe for the Tri16 gene (Fig. 2), Tri16 transcripts accumulated at all three of the time points examined (23, 27, and 36 h) for the wild-type strain (NRRL 3299) and ectopic strain (Tx #22) while the three Tri16 disruption transformants (Tx #4, Tx #7, and Tx #20) accumulated little, if any, of this transcript. Tri1 transcript accumulation was similar in all of the strains and at all three time points. These results confirm the loss of Tri16 gene expression in the ΔTri16 transformants.
FIG. 2.
Northern analysis of Tri16 and Tri1 transcript accumulation by F. sporotrichioides strains grown in liquid culture. Total RNA was isolated for each strain from mycelia collected from cultures grown for 23, 27, and 36 h in YEPD-5G medium. Strains are (left to right) the wild type (NRRL 3299), ΔTri16 transformants (Tx #4, Tx #7, and Tx #20), and a transformant (Tx #22) resulting from ectopic integration of pKO16. rRNA bands from the agarose gel stained with ethidium bromide are shown.
Toxin analyses.
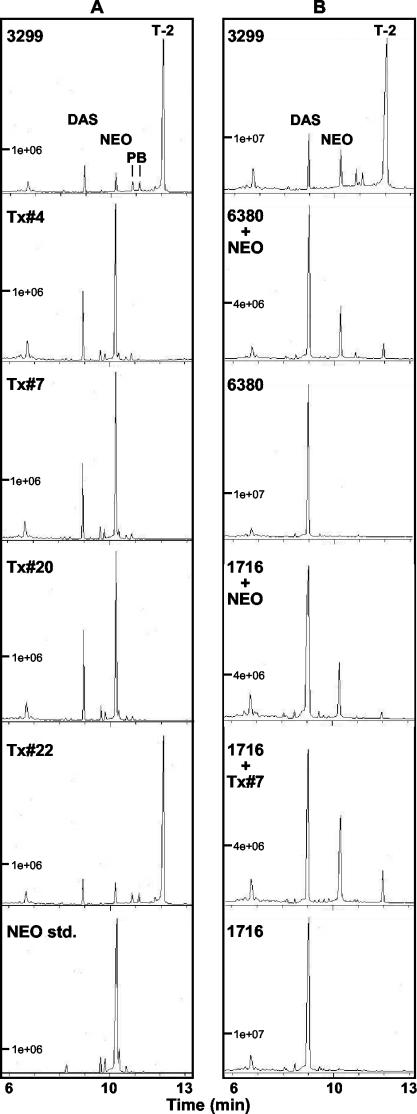
To determine the effect of Tri16 gene disruption on T-2 toxin production, trichothecene profiles were examined for the wild-type and four transformant strains (Fig. 3A). The wild-type strain, NRRL 3299, produced T-2 toxin, DAS, NEO, P-NEO, and B-NEO as expected. While the ectopic transformant, Tx #22, retained a wild-type toxin profile, all three Tri16 disruption transformants, Tx #4, Tx #7, and Tx #20, produced no detectable T-2 toxin. Instead, they accumulated primarily NEO and DAS. In addition, no detectable amounts of P-NEO or B-NEO were produced by these three transformants. Minor peaks with retention times similar to those of P-NEO and B-NEO were present; however, analysis of the mass spectra of the compounds in these peaks failed to identify a base peak of m/z 382 that is common to both P-NEO and B-NEO. Thus, the disruption of Tri16 blocks C-8 esterification during trichothecene biosynthesis.
FIG. 3.
Chromatograms of ethyl acetate extracts of 7-day Fusarium cultures. (A) Analysis of the Tri16 transformants. P, P-NEO; B, B-NEO. (B) Precursor feeding and coinoculation experiments. std., standard.
Detection of Tri16 in G. pulicaris.
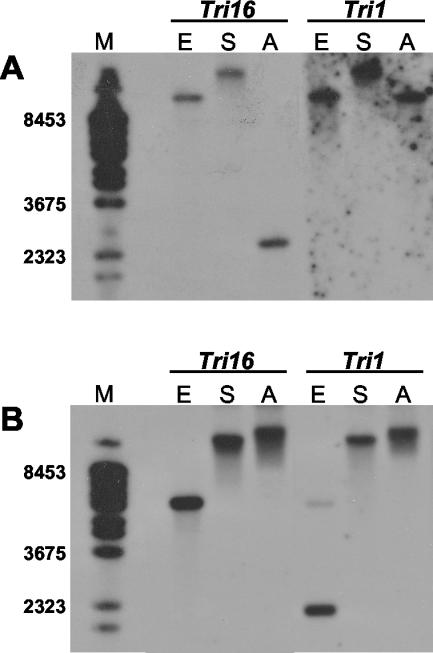
We previously identified a putative, presumably inactive, Tri1 ortholog in G. pulicaris (F. sambucinum) R-6380 (23). In this study, we detected an ortholog of Tri16 in the same strain (Fig. 4B). The Southern hybridization pattern for Tri1 and Tri16 in F. sporotrichioides NRRL 3299 (Fig. 4A) is consistent with the restriction sites predicted from sequence data that placed Tri1 and Tri16 on the same EcoRI and SmaI fragments but on different ApaI fragments. In G. pulicaris R-6380 (Fig. 4B), the Tri16 ortholog lies on restriction fragments (for all three enzymes) of the same size as Tri1. Thus, as in F. sporotrichioides, Tri16 and Tri1 appear to be physically linked in G. pulicaris.
FIG. 4.
Southern blots of F. sporotrichioides NRRL 3299 (A) and G. pulicaris (F. sambucinum) R-6380 (B) probed with the Tri16 and Tri1 genes. E, EcoRI; S, SmaI; A, ApaI; M, lambda DNA digested with BstEII. Sizes (bases) of selected marker bands are shown at the left.
C-8 acylation in G. pulicaris.
To test whether the Tri16 ortholog in G. pulicaris R-6380 could be functional, we assessed the ability of this strain to convert NEO to T-2 toxin by growing it with exogenously added NEO for 7 days in YEPD-5G liquid medium. We found that R-6380 could convert NEO to T-2 toxin at a very low rate (Fig. 3B), as could MB1716, an F. sporotrichioides NRRL 3299-derived tri1 mutant that retains C-8 acylation activity (7). Both of these results are consistent with the previously observed poor conversion of NEO to T-2 toxin by the F. sporotrichioides NRRL 3299-derived ΔTri5 transformant 8-3-9 (25). These data indicate that R-6380, like MB1716, can esterify C-8 although it cannot hydroxylate this carbon. Since MB1716 converted the exogenously supplied NEO to T-2 toxin inefficiently, this strain was coinoculated with a Tri16 mutant (Tx #7) into YEPD-5G medium to test for production of T-2 toxin under these conditions. This mixed culture was capable of producing T-2 toxin, but it still accumulated primarily DAS and NEO (Fig. 3B).
DISCUSSION
We previously reported the discovery of a putative acyltransferase gene 2.5 kb downstream of Tri1 that has a Tri10- and Tri6-dependent expression pattern similar to that of other Tri genes (23). In the present study, we demonstrated that this gene is necessary for esterification of the C-8 position during T-2 toxin biosynthesis and have thus designated it Tri16. Disruption of the Tri16 coding region in F. sporotrichioides prevents the production of compounds esterified at C-8 and results in accumulation of the less toxic metabolite NEO. The predicted TRI16 protein sequence possesses motifs that are found in a group of plant acyltransferase enzymes and thus appears to be a C-8 acyltransferase.
The most obvious phenotype of the Tri16 disruption mutants is that they accumulate NEO and do not produce T-2 toxin. This result is consistent with the inability to add the isovaleryl moiety to the C-8 hydroxyl group. A previous study (8) showed that a leucine auxotroph of NRRL 3299 produced less T-2 toxin and more P-NEO and B-NEO than the wild-type parent when leucine was limited during growth in YEPD-5G medium. A wild-type toxin profile was restored by the addition of sufficient levels of leucine to the medium. Since leucine is a precursor to the C-8 isovaleryl moiety (8), the overproduction of P-NEO and B-NEO was attributed to increased competitive esterification of propionyl and isobutyryl groups at C-8 due to the decreased availability of isovalerate. If the addition of the propionyl and butyryl moieties were catalyzed by an enzyme other than TRI16, then P-NEO and B-NEO levels should increase in the ΔTri16 mutants. They did not. Instead, they declined such that P-NEO and B-NEO were no longer detectable. The failure of ΔTri16 mutants to accumulate P-NEO and B-NEO indicates that TRI16 also is responsible for the esterification reactions that result in the production of P-NEO and B-NEO.
In the proposed metabolic grid for trichothecene biosynthesis (23), the placement of Tri16 would fall between Tri1 and Tri8, which encodes a C-3 esterase (20). 3-Acetylneosolaniol is thought to be the preferred substrate for TRI16. However, the production of 4-deoxy-T-2 toxin and HT-2 toxin by Tri13 (C-4 hydroxylase) deletion mutants and Tri7 (C-4 acetylase) deletion mutants (10, 11), respectively, suggests that 8-hydroxycalonectrin and 3,15-diacetyl-T-2 tetraol also may serve as substrates for TRI16. Therefore, in addition to being able to catalyze the addition of multiple acyl (isovaleryl, propionyl, and butyryl) groups to C-8-OH, TRI16 appears to accommodate multiple trichothecene substrates.
Tri16 is the fourth cloned acyltransferase gene for trichothecene biosynthesis, but among the protein products of those genes, TRI16 is significantly similar only to TRI101. Aside from one conserved motif in TRI3, TRI3 and TRI7 are not significantly similar to TRI16 or TRI101 and have previously been reported not to be similar to known fungal acetyltransferases (10, 21, 22).
Tri1 and Tri16 are adjacent to each other but not to the core trichothecene gene cluster (20). On the basis of the toxicological differences between DAS and T-2 toxin, this organizational pattern has potential implications for the evolution of the production of more toxic trichothecenes and for the evolution of gene clusters for fungal secondary metabolism. Tri1 and Tri16 encode enzymes for the two consecutive steps that extend the trichothecene pathway beyond DAS to the most toxic type A trichothecene, T-2 toxin. A similar pattern exists for another fungal secondary metabolite. The last three steps in the cephalosporin biosynthetic pathway in Acremonium chrysogenum are catalyzed by two adjacent genes that are not located in the cluster of genes required for the first portion of the pathway (14). Adjacent genes also encode enzymes that catalyze consecutive biosynthetic steps in fungal secondary metabolic pathways for the gibberellins (31) and the aflatoxins (36), but they are not organized in a separate second gene cluster. Certainly, the comparative analysis of Tri1 and Tri16 relative to the core trichothecene gene cluster in other Fusarium species will be most interesting.
As determined by Southern hybridization, G. pulicaris R-6380 contains a putative Tri16 ortholog that is physically linked to a potential ortholog of Tri1. This DAS-accumulating strain, like the tri1 DAS-accumulating mutant of F. sporotrichioides, can convert NEO to T-2 toxin at a low rate and therefore can add an isovalerate moiety to C-8-OH. Although some naturally occurring isolates of G. pulicaris produce T-2 toxin (3, 30), to our knowledge, this is the first report that R-6380 can produce T-2 toxin under laboratory conditions. The ability of R-6380 to produce T-2 toxin from NEO suggests that it lacks only the ability to hydroxylate the C-8 position of trichothecenes. Disruption of Tri16 in R-6380 will be necessary to prove that the TRI16 homolog catalyzes the C-8 esterification. Some strains of G. pulicaris produce 8-Ac-NEO as their primary trichothecene metabolite (7). Whether TRI16 or another enzyme in Fusarium can add the acetyl side group to C-8 also remains to be determined. In addition, both genetic and molecular studies can be used to reveal the location of Tri16 relative to the other Tri genes in G. pulicaris.
We previously reported the failure to detect a Tri1 ortholog in G. zeae (F. graminearum) GZ3639, a type B trichothecene producer, by Southern analysis (23). Similarly, we were not able to detect an ortholog of Tri16 in GZ3639. However, examination of the initial release of the draft sequence of the F. graminearum PH-1 genome (Fusarium graminearum Sequencing Project, Whitehead Institute/MIT Center for Genome Research [http://www-genome.wi.mit.edu]) revealed possible orthologs for both of these genes. As in F. sporotrichioides NRRL 3299, the putative orthologs are adjacent to each other and are not associated with the core trichothecene gene cluster, but their directions of transcription diverge rather than converge. While the Tri16-like sequence appears to be inundated with stop codons and therefore nonfunctional, the Tri1 ortholog could be functional. Analysis of the function of the Tri1 ortholog in F. graminearum would be critical to answering the question of whether this species uses this gene or a different one to hydroxylate the C-8 position in the biosynthesis of type B trichothecenes.
The characterization of Tri16 completes the assignment of genes for the last eight steps in the trichothecene biosynthetic pathway in F. sporotrichioides NRRL 3299 and completes the identification of genes for the esterifications required at C-5, C-4, C-15, and C-8 (10, 21, 22; this study) in the synthesis of T-2 toxin. In contrast to TRI101, the activity of TRI16, like that of TRI7, increases trichothecene toxicity.
Acknowledgments
This work was supported in part by USDA/CSREES NRICGP grant 9503702.
REFERENCES
- 1.Alexander, N. J., S. P. McCormick, and S. L. Ziegenhorn. 1999. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat. Toxins 7:265-269. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, N. J., S. P. McCormick, and T. M. Hohn. 2002. The identification of the Saccharomyces cerevisiae gene AYT1 (ORF-YLL063c) encoding an acetyltransferase. Yeast 19:1425-1430. [DOI] [PubMed] [Google Scholar]
- 3.Altomare, C., A. Logrieco, A. Bottalico, G. Mule, A. Moretti, and A. Evidente. 1995. Production of type A trichothecenes and enniatin B by Fusarium sambucinum Fuckel sensu lato. Mycopathologia 129:177-181. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, G. H., A. E. Desjardins, and R. D. Plattner. 2002. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153:91-98. [DOI] [PubMed] [Google Scholar]
- 6.Beremand, M. N. 1987. Isolation and characterization of mutants blocked in T-2 toxin biosynthesis. Appl. Environ. Microbiol. 53:1855-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beremand, M. N., and S. P. McCormick. 1992. Biosynthesis and regulation of trichothecene production by Fusarium species, p. 359-384. In D. Bhatnagar, E. B. Lillehoj, and D. K. Arora (ed.), Handbook of applied microbiology. Marcel Dekker, Inc., New York, N.Y.
- 8.Beremand, M. N., F. Van Middlesworth, S. Taylor, R. D. Plattner, and D. Weisleder. 1988. Leucine auxotrophy specifically alters the pattern of trichothecene production in a T-2 toxin-producing strain of Fusarium sporotrichioides. Appl. Environ. Microbiol. 54:2759-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowden, R. L., and J. F. Leslie. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182-188. [DOI] [PubMed] [Google Scholar]
- 10.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 36:224-233. [DOI] [PubMed] [Google Scholar]
- 12.Desjardins, A. E., T. M. Hohn, and S. P. McCormick. 1993. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol. Rev. 57:595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grove, J. F., and P. H. Mortimer. 1969. The cytotoxicity of some transformation products of diacetoxyscirpenol. Biochem. Pharmacol. 18:1473-1478. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez, S., F. Fierro, J. Casqueiro, and J. F. Martin. 1999. Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 75:81-94. [DOI] [PubMed] [Google Scholar]
- 15.Harris, L. J., A. E. Desjardins, R. D. Plattner, P. Nicholson, G. Butler, J. C. Young, G. Weston, R. H. Proctor, and T. M. Hohn. 1999. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 83:954-960. [DOI] [PubMed] [Google Scholar]
- 16.Hohn, T. M., and A. E. Desjardins. 1992. Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol. Plant-Microbe Interact. 5:249-256. [DOI] [PubMed] [Google Scholar]
- 17.Hohn, T. M., A. E. Desjardins, S. P. McCormick, and R. H. Proctor. 1995. Biosynthesis of trichothecenes, genetic and molecular aspects, p. 239-248. In M. Eklund, J. L. Richard, and K. Mise (ed.), Molecular approaches to food safety: issues involving toxic microorganisms. Alaken, Inc., Ft. Collins, Colo.
- 18.Jarvis, B. B., W. G. Sorenson, E. L. Hintikka, M. Nikulin, Y. Zhou, J. Jiang, S. Wang, S. Hinkley, R. A. Etzel, and D. Dearborn. 1998. Study of toxin production by isolates of Stachybotrys chartarum and Memnoniella echinata isolated during a study of pulmonary hemosiderosis in infants. Appl. Environ. Microbiol. 64:3620-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identity and mycotoxicology. The Pennsylvania State University Press, University Park, Pa.
- 20.McCormick, S. P., and N. J. Alexander. 2002. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 68:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick, S. P., N. J. Alexander, S. E. Trapp, and T. M. Hohn. 1999. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 65:5252-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick, S. P., T. M. Hohn, and A. E. Desjardins. 1996. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meek, I. B., A. W. Peplow, C. Ake, Jr., T. D. Phillips, and M. N. Beremand. 2003. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl. Environ. Microbiol. 69:1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peplow, A. W., A. G. Tag, G. F. Garifullina, and M. N. Beremand. 2003. Identification of new genes positively regulated by Tri10 and a regulatory network for trichothecene mycotoxin production. Appl. Environ. Microbiol. 69:2731-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salch, Y. P., and M. N. Beremand. 1993. Gibberella pulicaris transformants: state of transforming DNA during asexual and sexual growth. Curr. Genet. 23:343-350. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.St-Pierre, B., and V. De Luca. 2000. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism, p. 285-315. In J. T. Romeo, R. Ibrahim, L. Varin, and V. De Luca (ed.), Evolution of metabolic pathways. Pergamon, Amsterdam, The Netherlands.
- 29.Tag, A. G., G. F. Garifullina, A. W. Peplow, C. Ake, Jr., T. D. Phillips, T. M. Hohn, and M. N. Beremand. 2001. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67:5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thrane, U., and U. Hansen. 1995. Chemical and physiological characterization of taxa in the Fusarium sambucinum complex. Mycopathologia 129:183-190. [DOI] [PubMed] [Google Scholar]
- 31.Tudzynski, B. P. Hedden, E. Carrera, and P. Gaskin. 2001. The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthesis pathway. Appl. Environ. Microbiol. 67:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueno, Y. 1977. Mode of action of trichothecenes. Ann. Nutr. Aliment. 31:885-900. [DOI] [PubMed] [Google Scholar]
- 33.Ueno, Y., M. Sawano, and K. Ishii. 1975. Production of trichothecene mycotoxins by Fusarium species in shake culture. Appl. Microbiol. 30:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueno, Y., N. Sato, K. Ishii, K. Sakai, H. Tsunoda, and M. Enomoto. 1973. Biological and chemical detection of trichothecene mycotoxins of Fusarium species. Appl. Microbiol. 25:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei, C. M., and C. S. McLaughlin. 1974. Structure-function relationship in the 12,13-epoxytrichothecenes: novel inhibitors of protein synthesis. Biochem. Biophys. Res. Commun. 57:838-844. [DOI] [PubMed] [Google Scholar]
- 36.Yu., J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, B. Montalbano, J. M. Dyer, D. Bhatnagar, and T. E. Cleveland. 1998. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 64:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]