Abstract
The luminous bacterium Vibrio fischeri colonizes a specialized light-emitting organ within its squid host, Euprymna scolopes. Newly hatched juvenile squid must acquire their symbiont from ambient seawater, where the bacteria are present at low concentrations. To understand the population dynamics of V. fischeri during colonization more fully, we used mini-Tn7 transposons to mark bacteria with antibiotic resistance so that the growth of their progeny could be monitored. When grown in culture, there was no detectable metabolic burden on V. fischeri cells carrying the transposon, which inserts in single copy in a specific intergenic region of the V. fischeri genome. Strains marked with mini-Tn7 also appeared to be equivalent to the wild type in their ability to infect and multiply within the host during coinoculation experiments. Studies of the early stages of colonization suggested that only a few bacteria became associated with symbiotic tissue when animals were exposed for a discrete period (3 h) to an inoculum of V. fischeri cells equivalent to natural population levels; nevertheless, all these hosts became infected. When three differentially marked strains of V. fischeri were coincubated with juvenile squid, the number of strains recovered from an individual symbiotic organ was directly dependent on the size of the inoculum. Further, these results indicated that, when exposed to low numbers of V. fischeri, the host may become colonized by only one or a few bacterial cells, suggesting that symbiotic infection is highly efficient.
The luminous bacterium Vibrio fischeri lives in a cooperative association with the Hawaiian bobtail squid, Euprymna scolopes. The squid has developed a specialized light-emitting organ to house the bacteria within the animal's mantle cavity. When juvenile E. scolopes squid hatch, they are symbiont free and must acquire V. fischeri from the surrounding seawater (27). The bacteria enter the nascent organ through six pores which connect to ducts that, in turn, lead to specialized, epithelium-lined crypts (18, 26, 30). Once inside these crypts, the bacteria divide rapidly and fill the light organ with approximately 5 × 105 cells (34). During this colonization process, after a critical cell density is reached, the bacteria induce the lux operon and begin to luminesce. The animal can control the amount of light emitted to the environment and uses this luminescence during its nocturnal activities, probably as a means to disguise itself from predators (42). At dawn, the host expels approximately 95% of the bacteria; the remaining symbionts multiply over the next 12 h, thereby providing the squid again with a source of luminescence by nightfall (34).
The relationship between V. fischeri and its squid host is complex and highly specific. For example, although surrounded by seawater containing hundreds of different bacterial species, juvenile E. scolopes squid become colonized only by V. fischeri (5, 35). Lee and Ruby (19) used a lux gene probe to determine that there were between 5 and 200 CFU of V. fischeri per 100 ml of seawater in E. scolopes habitats. However, subsequent work suggested that the actual number of V. fischeri cells was between 100 and 1,500/ml of seawater, although most of these cells, while symbiotically infective, were in a cryptic state that did not form colonies on typical media (20). Thus, it appeared that juvenile squid are normally colonized in seawater containing a concentration of only a few hundred V. fischeri cells per ml (34), although higher population densities might occur in sediments (20). That V. fischeri cells constitute less than 0.1% of the total bacterial community makes a strong argument for both the specificity and the efficiency of this benign infection process. In addition, successful colonization of the animal by V. fischeri has been shown to trigger a complex developmental program in the light-organ morphology of the juvenile, and this program does not occur if the squid remains uncolonized (11, 18, 30). This close linkage between colonization and juvenile development further emphasizes the importance of the initiation of the relationship between host and symbiont.
To understand the basis for both the efficiency and the specificity of colonization more fully, we developed derivatives of the site-specific transposon mini-Tn7 (22) to create differentially marked strains of V. fischeri. These strains were used to track the progeny of individual cells as they infected the light organ of juvenile E. scolopes. Because in Escherichia coli and other bacterial species Tn7 inserts itself in single copy, and with high fidelity, into a specific and apparently unimportant intergenic region (8, 13, 22, 33, 43), we adapted it as a useful tool to chromosomally mark V. fischeri strains without affecting their biological fitness.
In this study, we determined the efficiency of V. fischeri infection of juvenile squid and used this information, as well as two different mini-Tn7-tagged strains, to track symbiont population dynamics during the early stages of infection. Specifically, we determined (i) the number of cells associated with a juvenile light organ in the first few hours following exposure to V. fischeri, (ii) the influence of inoculum size on the onset and ultimate level of animal luminescence within the first 48 h after inoculation, and (iii) the role that inoculum size plays in determining whether a symbiotic population is derived from a single bacterial cell.
MATERIALS AND METHODS
Bacterial strains and media.
Unless mentioned otherwise, V. fischeri cells were grown with shaking at 28°C in either seawater-tryptone medium (SWT), which contained 5 g of Bacto Tryptone-Peptone (Difco, Inc., Sparks, Md.), 3 g of yeast extract, and 3 ml of glycerol per liter of 70% seawater (5), or LBS, which contained 10 g of Bacto Tryptone-Peptone, 5 g of yeast extract, 50 ml of 1 M Tris base (Sigma Chemical Co., St. Louis, Mo.) at pH 7.5, and 20 g of NaCl per liter (10). Strains of E. coli were grown in either Luria-Bertani medium (7) or brain heart infusion medium (Difco). All strains were stored at −80°C, and inocula for cultures used in each experiment were grown from these frozen stocks. Antibiotics were used, when indicated, at the indicated concentrations: ampicillin, 100 μg/ml; chloramphenicol (CHL), 3 μg/ml for V. fischeri and 20 μg/ml for E. coli; erythromycin (ERY), 3 to 5 μg/ml for V. fischeri and 150 μg/ml for E. coli; kanamycin, 20 μg/ml; and rifampin, 100 μg/ml. For solid media, agar was added to a final concentration of 1.5%. Unless otherwise noted, all chemicals were obtained from Sigma Chemical Co. Oligonucleotides and primers (Table 1) were synthesized by Integrated DNA Technologies (Coralville, Iowa) or Operon Technologies (Alameda, Calif.). All restriction enzymes were obtained from New England BioLabs (Beverly, Mass.), and T4 DNA ligase was obtained from Promega Corp. (Madison, Wis.). Genetic sequencing reactions were performed at the Biotechnology-Molecular Biology Instrumentation Facility at the University of Hawaii, Manoa.
TABLE 1.
Bacterial strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence | Source or reference |
|---|---|---|
| V. fischeri | ||
| ES114 | Wild-type light-organ isolate from E. scolopes | 5 |
| ESR1 | Spontaneous Rfr mutant of ES114 | 14 |
| JRM100 to JRM104 | Five ES114 clones with mini-Tn7 insertion; Emr | This study |
| JRM200 | ES114 clone with mini-Tn7 insertion; Cmr | This study |
| E. coli | ||
| BW23474 | Δlac-169 robA1 creC510 hsdR514 uidA(ΔMluI)::pir116 endA(BT33) recA1 | 28 |
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-I rpsE rpoB argE(Am) recA1; lysogenized with λpir | 16 |
| Plasmids | ||
| pEVS94S | R6K ori oriT Emr | 41 |
| pEVS104 | Derivative of pRK2013; conjugal tra and trb genes | 41 |
| pEVS107 | pEVS94S derivative, mini-Tn7; mob; Emr Knr | This study |
| pUXBF13 | R6K ori; tns genes; Apr | 4 |
| pCR2.1 | PCR product cloning vector; ColE1 ori; Apr Knr | Invitrogen Inc. |
| pJRM3 | pEVS107 with Cmr replacing Emr | This study |
| pEVS35 | CmrXbaI fragment cloned into XbaI site of pBC | 39 |
| Primers | ||
| JM1 | 5′ GTTACACGTTACTAAAGGG 3′ | This study |
| JM2 | 5′ ACCAGACCGTTCAGCTGG 3′ | This study |
| JM3 | 5′ GGAACATGTGTGGTATGG 3′ | This study |
| JM4 | 5′ GACAGTCATCTATTCAAC 3′ | This study |
Molecular techniques and genetic manipulations involving Tn7.
To insert the mini-Tn7 into the genome of V. fischeri, a compatible vector was created. The mini-Tn7 vector pEVS107 (Table 1) was derived from pUX-BF5 (4) by replacing the Knr SalI cassette within the transposon with pEVS94S (41), which contains an Emr cassette, linearized with SalI. The Knr cassette was then reintroduced into the vector, replacing a BamHI fragment that is located outside the transposon.
Chromosomal insertion of Emr into V. fischeri ES114 by use of a mini-Tn7 cassette was performed as described previously (9, 43). Briefly, E. coli CC118λpir (16) carrying pEVS104, two strains of E. coli BW23474 (28) carrying either pEVS107 or pUX-BF13 (4), and V. fischeri ES114 (Table 1) were combined in a tetraparental mating and incubated for approximately 12 h at 28°C. This conjugation mixture was then resuspended in LBS and spread onto LBS-agar plates containing ERY. After incubation at 22°C for 24 to 48 h the ES114 colonies that arose were screened for Emr and Kns, and five clones (JRM100 to JRM104) were selected. A second mini-Tn7-containing strain of V. fischeri ES114 was tagged with Cmr as follows. The Cmr gene from pEVS35 (39) was excised on an XbaI fragment and ligated into a derivative of pEVS107 that had previously been digested with EcoRV and allowed to self ligate, resulting in the loss of its Emr gene. After ligation, the resulting mini-Tn7-Cmr plasmid, pJRM3, was transformed into E. coli BW23474 and mobilized by conjugation into V. fischeri ES114 as described above, producing the Cmr-marked strain JRM200. The number and location of six independently arising mini-Tn7-Cmr insertions were determined by Southern blotting and/or PCR amplification. Briefly, 3 μg of genomic DNA was subjected to restriction enzyme digestion, separated by electrophoresis, transferred to a nitrocellulose membrane, and analyzed by Southern hybridization (38). The PCR amplification product from primers JM3 and JM4 (Table 1) was used as a probe. The specific insertion sites of the transposon into the chromosome of V. fischeri were determined by sequencing from either end of the transposon by using primers JM1 and JM2 (Table 1).
Competition assays of Tn7-tagged strains in culture.
The ability of the transposon-tagged strains to grow as well as wild type in liquid medium was determined as follows. V. fischeri strains ES114, JRM100, and JRM200 were grown separately with shaking for 18 h in SWT at 28°C. Approximately equal numbers of cells of each strain were then combined, the mixture was diluted 1:100 in fresh SWT, and the culture was split into three flasks. After approximately 20 h of shaking, 1 ml of each culture was transferred into a new flask containing 15 ml of fresh SWT. To determine the relative ratio of strains in the culture at various times during growth, three 100-μl samples of each flask were removed, serially diluted, and spread onto SWT agar plates. Representative CFU were screened for antibiotic resistance and identified as either strain JRM100, strain JRM200, or strain ES114.
Animal colonization experiments.
Juvenile E. scolopes squid (approximately 1 mm in mantle length) were collected using a plastic eyedropper within minutes of hatching from eggs laid in the laboratory and were held in natural seawater (Waikiki Aquarium, Honolulu, Hawaii) for at least 30 min prior to inoculation with strains of V. fischeri. This water contained fewer than 103 CFU/ml and did not harbor V. fischeri cells in sufficient numbers to colonize the squid (31). To determine the 50% infective dose (ID50), defined as the number of bacteria required for colonization of 50% of the animals, juvenile squid were placed in vials containing 4 ml of seawater inoculated with serial dilutions of a culture of V. fischeri ES114. Samples of the inoculum were spread on agar medium, and the number of CFU that arose at 28°C was counted at 24 h. After a 3-h incubation period at 22°C, the animals were rinsed and placed in vials of natural seawater. The seawater was changed every 12 to 16 h. At 48 h postinoculation the luminescence of each juvenile E. scolopes was measured with a TD20/20 luminometer (Turner Designs, Sunnyvale, Calif.). Because uncolonized squid do not produce light (27), all animals with a luminescence reading significantly above background (i.e., >1.3 × 104 quanta/s) were considered colonized. In other experiments the development of colonization was determined over a 48-h period by using a liquid scintillation counter (TriCarb 2100TR; Packard Inc., Meriden, Conn.) modified to measure the level of light emission, which is an indirect measure of the number of V. fischeri cells in the light organ (27, 29).
Competition of marked V. fischeri strains for host colonization.
To determine whether the transposon-tagged strains have a competitive colonization defect in symbiosis, equal numbers of V. fischeri ES114 and either JRM100 or JRM200 organisms were coinoculated into seawater containing 16 juvenile E. scolopes squid and incubated as described above. The inoculum was spread onto SWT agar plates, and the resulting CFU were screened for antibiotic resistance to determine the ratio of ES114 and the mini-Tn7-marked strains in the inoculum. Approximately 24 h postinoculation, each animal was homogenized in 700 μl of sterile seawater in a 1.5-μl Eppendorf tube, and 100 μl of this homogenate (or serial dilutions of it) was spread on SWT agar to determine the number of V. fischeri CFU per light organ (34). When the juvenile light organ was dissected away from the rest of the body, and both the light organ and the remaining body tissue were homogenized and plated separately, CFU were detected only in the light-organ homogenate, and this homogenate contained the same total CFU as did the whole-animal homogenate (34). Between 100 and 200 of the resulting colonies of V. fischeri were screened for resistance to either CHL or ERY to calculate the proportion of each marked strain in the colonizing population.
In a similar experiment the spontaneously occurring Rfr V. fischeri strain ESR1 (14) was also coinoculated with its wild-type parent as described above to determine its colonization abilities. The proportion of Rfr V. fischeri cells in each light organ at 24 and 48 h postinoculation was compared to that in the inoculum as an indication of the success of V. fischeri ESR1 in competing for colonization of the light organ.
Early kinetics of light-organ colonization.
To determine the efficiency of symbiotic colonization, three sets of 24 animals were incubated with three different concentrations of V. fischeri strain JRM100 as described above. Following the 3-h incubation period, the animals were rinsed twice in 0.22-μm-pore-size-filtered seawater (FSSW) and transferred to vials containing 4 ml of FSSW. Twelve of these animals were placed in sterile 1.5-ml centrifuge tubes and immediately homogenized in 200 μl of FSSW. The whole homogenate was then spread onto SWT agar. The resulting colonies were counted and screened for Emr and colony morphology. V. fischeri strain JRM100 was used so that it could be more easily identified by its antibiotic resistance among any other bacteria that might be present in the animal homogenates. The number of CFU of V. fischeri JRM100 on each plate was a measure of the number of V. fischeri cells that were associated with the animal 3 h after inoculation. The remaining 12 animals in each group were held for an additional 24 h and tested for luminescence to determine the efficiency of colonization at the three different inoculum concentrations used. In related experiments, animals were inoculated with different concentrations of green fluorescent protein (GFP)-labeled V. fischeri cells (29) and observed under confocal scanning laser microscopy at between 3 and 10 h to determine the number and location of bacteria associating with the squid light organ (32). Control experiments showed that all cells in the inoculum expressed GFP (data not shown).
To estimate the number of V. fischeri cells that serve as the progenitors of the light-organ population at low inoculum levels, juvenile squid were incubated with seawater containing a mixture of V. fischeri strains ES114, JRM100, and JRM200 in roughly equal proportions. In three separate experiments with three different total inoculum concentrations, animals were incubated as described above. Between 24 and 48 h postinoculation, each animal was homogenized and aliquots of the homogenate were diluted in sterile seawater and spread onto SWT agar. Between 100 and 200 CFU from each animal were screened for resistance to CHL, ERY, or neither antibiotic to identify the number of JRM100, JRM200, and ES114 bacteria present in each light organ. The experiment was repeated four times, and the number of animals that carried detectable progeny of one, two, or all three strains from the V. fischeri inoculum was recorded.
RESULTS
Determination of the ID50 and kinetics of early colonization.
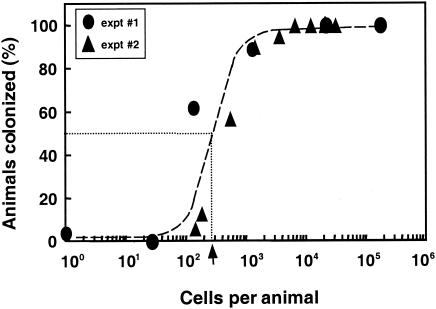
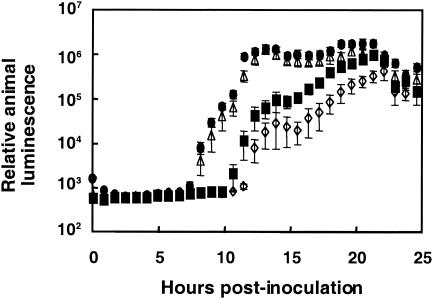
When exposed for as little as 3 h to about 250 V. fischeri cells per squid, an average of 50% of juvenile E. scolopes squid became colonized (Fig. 1), as indicated by the appearance of host luminescence. Colonization was rare at inoculum levels below 100 bacteria per animal, but colonization efficiency rapidly improved as the inoculum size increased, reaching 100% at levels above 1,000 bacteria. Among juveniles that became infected, those exposed to higher doses of V. fischeri cells became luminescent earlier, and reached maximal luminescence sooner, than those animals that were exposed to a lower dose (Fig. 2). However, this enhancement reached a limit, and exposing animals to more than 36,000 cells did not lead to a more rapid colonization. Importantly, all animals became equally luminescent by the end of the experimental period, indicating that, while any of a wide range of doses is sufficient to initiate colonization, the length of time required for onset of host luminescence is inversely correlated with the size of the dose of V. fischeri administered.
FIG. 1.
ID50 of V. fischeri for symbiotic colonization. In two separate experiments juvenile squid were incubated with different numbers of V. fischeri ES114 cells, and the percentage of animals subsequently colonized was determined. The dose at which 50% of the animals became luminescent by 48 h postinoculation, indicated by the dotted lines and black arrow, was determined using the equation y = 0.3735 lnx − 1.794 for experiment 1 (circles) and the equation y = 0.1381 lnx − 0.2459 for experiment 2 (triangles).
FIG. 2.
Kinetics of onset of animal luminescence as a function of V. fischeri dose. Juvenile squid were incubated for 12 h in seawater containing 40 (open diamonds), 580 (closed squares), 36,000 (open triangles), or 800,000 (closed circles) V. fischeri cells, and the development of luminescence by each animal was monitored (n = 12 for each dose). Mean luminescence levels of those animals that became colonized are reported in relative light units. Values below about 103 units indicate no detectable light production. Error bars depict the standard errors of the means.
Effect of mini-Tn7 carriage on V. fischeri ES114.
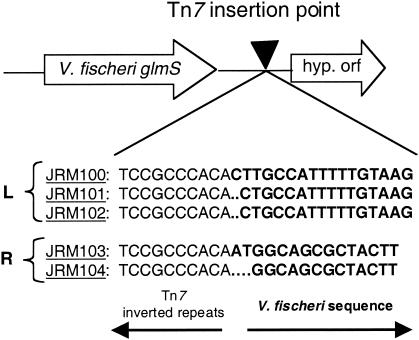
We created genetically marked, but otherwise apparently normal, strains of V. fischeri so that we could describe the nature of the population of bacteria involved in the initiation of squid light-organ colonization. Based on the biology of Tn7 in other systems, we hypothesized that the mini-Tn7-Emr constructs in V. fischeri ES114 would have a single chromosomal insertion site and that the insertion would not affect the ability of bacteria to grow or colonize. To test this hypothesis, the genomic fragment carrying the transposon was cloned and sequenced from each of five independently isolated clones (designated JRM100 to JRM104), revealing that the point of insertion was in essentially the same noncoding, intergenic region (Fig. 3). This region was downstream of an open reading frame encoding a protein with 76% amino acid identity to the glmS product in E. coli and with 88% amino acid identity to the putative glmS product in Vibrio cholerae (2, 15). Similarly, PCR analysis of the insertion in JRM200 confirmed that the Cmr-containing form of the transposon was inserted at the same genomic site (data not shown). Southern analysis confirmed that each strain received a single insertion.
FIG. 3.
Site of mini-Tn7 insertions in the V. fischeri genome. PCR products from insertion strains JRM100, JRM101, and JRM102 were sequenced using a primer extending from the left end of the transposon (labeled “L”), and products from strains JRM103 and JRM104 were sequenced from the right end of the transposon (labeled “R”). The resulting sequences were aligned, and the predicted insertion points are indicated.
To determine whether carriage of the insertion created a significant growth burden, we examined the effect of the transposon on cells growing in culture. Two Tn7-containing strains (JRM100 and JRM200) and their wild-type parent (ES114) were coinoculated into SWT, and the ratio of the strains to each other was determined during several rounds of subculturing. The starting ratio of strains was maintained throughout the 44 h of batch culture (data not shown), indicating that over this period neither the relative growth rate nor survival in stationary phase was significantly affected in the transposon-bearing strains.
Light-organ colonization by Tn7 strains of V. fischeri.
To examine whether insertion of the mini-Tn7 affected symbiotic competence, wild-type V. fischeri was coinoculated with either strain JRM100 or strain JRM200 into seawater containing juvenile E. scolopes. After 24 h, the mini-Tn7 insertion mutants had colonized the light organ to the same degree as had the wild type, as indicated by a relative competitive index value of close to 1.0 (Table 2). We also examined the colonization ability of V. fischeri ESR1 (14), a spontaneously occurring Rfr mutant of V. fischeri ES114, to determine whether it could also be used as a marked strain in multistrain colonization experiments. However, unlike the mini-Tn7-marked strains, ESR1 comprised a significantly smaller portion of the squid light-organ population than it did of the initial inoculum (Table 2), demonstrating that ESR1 could not compete effectively with its wild-type parent during colonization. For this reason V. fischeri ESR1 was not used in the multistrain infection experiments described below.
TABLE 2.
Relative colonization abilitiesa of V. fischeri strains carrying antibiotic resistance
Colonization ability was measured during competition with the parent wild-type strain ES114.
RCI, percentage of a given strain in the light organ divided by percentage of that strain in the inoculum at 24 or 48 h postinoculation.
ND, not determined.
The ability to colonize the light organ was significantly less than that of wild-type V. fischeri (P < 0.05).
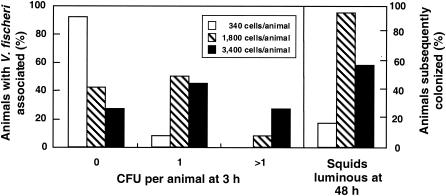
Having determined that transposon carriage did not apparently affect colonization ability, we used JRM100 to investigate how many bacterial cells become associated with the juvenile light organ during the first few hours of the infection process. In a representative experiment, when 36 juvenile squid were exposed to either 340, 1,800, or 3,400 V. fischeri cells for 3 h and then washed extensively, 35 of the animals had between zero and four bacterial cells associated with their tissue. Of the animals that were exposed to 1,800 or 3,400 cells, more than half (60 to 92%) eventually became colonized, while only a few (17%) of the animals exposed to 340 cells were successfully infected (Fig. 4). At each inoculum level the percentage of animals that had at least one cell associated with light-organ tissue at 3 h postinoculation was correlated with the percentage of animals that eventually became colonized.
FIG. 4.
Evidence of the association of V. fischeri cells with host tissue 3 h after inoculation. Three sets of 24 animals were exposed to three different doses of V. fischeri cells: 340, 1,800, and 3,400 per animal. The number of V. fischeri cells (as determined by CFU) associated with light-organ tissue at 3 h postinoculation was determined (left panel) and compared to the percentage of animals that eventually became colonized at these inoculation doses (right panel).
We used confocal microscopy to examine the early stages of infection by GFP-labeled V. fischeri cells. When juvenile squid were exposed to doses below 1,000 CFU/ml, we only rarely observed aggregates containing over five bacteria associated with the mucus or light-organ tissue, a value that compared well with CFU determinations (data not shown). However, animals of the same cohort that had been transferred into natural seawater after exposure to the V. fischeri inoculum for either 3 or 12 h became fully colonized. Therefore, at the lower doses (below 1,000 CFU/ml) described in this study, it appears that bacterial cells are infecting as individuals or a few cells and not aggregating into large groups prior to entering the light organ.
Simultaneous colonization with three V. fischeri strains.
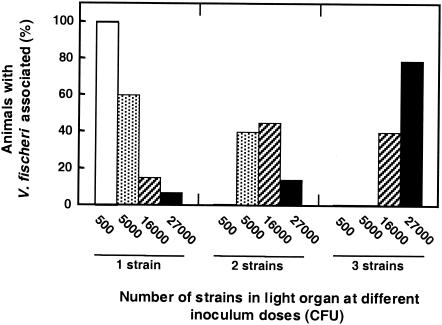
An objective of this study was to describe the numbers and composition of the founding population of bacteria that can colonize the light organ. The experimental results described above suggested (i) that only a small number of bacterial cells were necessary for a successful colonization and (ii) that the number of cells that participated in the colonization was dependent on inoculum density, at least at low inoculum concentrations. To determine whether a single cell, or small group of cells, could indeed be the sole symbiotic founder in a juvenile light organ, we designed the following experiment. Groups of E. scolopes squid were exposed to a mixed inoculum of V. fischeri containing equal cell numbers of wild type (ES114) and two differentiable Tn7-carrying strains (JRM100 and JRM200), but at different total inoculum concentrations. We reasoned that, if the light organ could be populated by a small initial population, or even a single colonizing bacterium, it would be possible to find light organs inhabited exclusively by only one of the three marked strains, even though all three were in the inoculum. We found that, when animals were exposed to large numbers of V. fischeri cells in the mixed inoculum, light-organ populations would consist of two or more strains. However, when exposed to a small inoculum, the animals were most often colonized by only one of the three strains (Fig. 5), which suggested colonization by a single cell or a small number of cells.
FIG. 5.
Effect of the total dose on the frequency of colonization by one, two, or three strains of V. fischeri from a mixed inoculum. Animals were exposed to the following approximate doses of V. fischeri cells per animal: 500 (open bars; n = 8), 5,000 (stippled bars; n = 15), 16,000 (striped bars; n = 20), and 27,000 (solid bars; n = 14). All doses contained a mixed inoculum of approximately equal proportions of V. fischeri strains ES114, JRM100, and JRM200. The number of different strains detected in each light organ was determined 24 h postinoculation.
DISCUSSION
All horizontally transmitted microbial associations, whether beneficial or pathogenic, must develop mechanisms that ensure or enhance infection of the host. Often there is an attrition of the inoculum by host defenses, such as that encountered by V. cholerae cells during their passage through the stomach (6). Unfortunately, even for well-studied pathogeneses, the number of cells that survive and initiate the infection has been difficult to determine, and it is usually impossible to visualize and count the founding bacterial population in host tissue. In this study we investigated the number and composition of V. fischeri cells taking part in the initiation of light-organ colonization in E. scolopes. Using genetic and microscopic approaches, we determined that the efficiency of colonization is dependent on the number of V. fischeri bacteria present in the inoculum and that the symbiont population can be founded by a few cells of V. fischeri.
Mini-Tn7-marked V. fischeri strains.
We used a mini-Tn7 to differentially mark V. fischeri strains with two antibiotic resistance cassettes and tracked their fate during the colonization process. We demonstrated that both the growth rate and the colonization efficiency (Table 2) of the V. fischeri mini-Tn7 insertion strains that we created were comparable to those of the wild-type parent. We also showed that, as in E. coli, the site of mini-Tn7 insertion in V. fischeri is an intergenic region (Fig. 3), located downstream of a glmS-like gene (8, 13, 22). Because glmS encodes glucosamine-fructose-6-phosphate aminotransferase, an important enzyme in cell wall synthesis, it has been conserved among many gram-negative bacteria (22). In previous work the transposon Tn7 has been used to insert reporter constructs such as the gene for lux (luminescence) (36) or GFP (17) into the genomes of several species of gram-negative bacteria. The ability of Tn7 to integrate into the genome of V. fischeri strain ESR1 was demonstrated previously (43), and it has been shown to integrate into Vibrio anguillarum as well (21). The transposon has also recently been used for genetic complementation of chromosomal mutations (9), and similar derivatives can now be used to replace the carriage of other markers (e.g., GFP [32]) on plasmids, whose maintenance requires continuous selection with antibiotics that can lower the rate of normal colonization (data not shown).
Determination of the ID50.
V. fischeri is effective at colonizing the light organ of E. scolopes, requiring a dose of only about 250 cells for 3 h to infect 50% of its squid hosts (Fig. 1). Such a low ID50 had been suggested from previous work (25, 34, 40) and indicates that colonization is an efficient process. This ID50 is especially remarkable because the dose is not directly introduced into host tissue and is present in the surrounding seawater for only a limited time. In comparison, the analogous 50% lethal dose, which is the bacterial dose that causes death in 50% of infected hosts, is typically much higher in other Vibrio species. For instance, Vibrio parahaemolyticus and Vibrio alginolyticus, two pathogens of shellfish, have reported 50% lethal doses on the order of 105 bacteria per g of host tissue (23, 24). This dose is 100-fold higher than what is required for V. fischeri colonization and suggests an unusually efficient mechanism of symbiotic infection by these bacteria. Such a mechanism is not unexpected, considering that the natural concentration of V. fischeri cells in the seawater inhabited by E. scolopes is usually well below a thousand cells per ml of seawater (20). Similarly, while potential hosts resist infection by pathogenic vibrios, E. scolopes has evolved a morphology and behavior that encourage V. fischeri colonization (42).
V. fischeri colonization dynamics.
Previous studies of the initiation of colonization (31, 32) have revealed that the juvenile squid light organ secretes a mucus layer that preferentially entraps V. fischeri cells present in the ambient seawater by a process that is surprisingly like that occurring between rhizobia and their leguminous hosts (12). In the squid, the action of surface cilia removes approximately 1% of the V. fischeri bacteria from the water (S. Nyholm, personal communication), forming bacterial aggregates in the mucus that, depending on the dose, contain a few to hundreds of bacteria. These aggregates eventually migrate to pores on the light-organ surface and through ducts that lead to symbiotic crypts of the light organ. Because the ducts are sites of a potential gauntlet of host-produced oxidative stress (37), the question arose as to how many of the inoculating bacteria survive the initial colonization events and become the progenitors of the resulting population of symbionts.
In the work described here mini-Tn7-tagged V. fischeri cells were used to examine the early colonization events during the initiation of symbiosis. We focused on low inoculum levels of a few hundred or thousand V. fischeri cells, levels at which the rate of colonization appeared to be limited by dose (Fig. 1 and 2). Consistent with previous findings (34), the small number of CFU (one to four) that arose from homogenates of juvenile squid suggested that only a few cells had become associated with the light organs under these inoculation conditions. Nevertheless, such animals typically became subsequently colonized, suggesting that only a few V. fischeri cells needed to be present to initiate a successful colonization. Direct visualization of V. fischeri cells in contact with host tissue correlated with the number of CFU detected by plating, arguing against the possibilities that either (i) the bacteria might be present in larger numbers but are in aggregates that are not easily separated when spread on agar or (ii) many of the cells have entered a viable but nonculturable state (20).
When animals are exposed to a dose of over 10,000 V. fischeri cells per ml, aggregates containing hundreds of these bacteria are seen (32). At such large doses symbiotic luminescence is detected 3 to 4 h sooner than in animals infected at or around the ID50 (Fig. 2). Based on a doubling time in the light organ of about 30 min (34), the number of cells initiating colonization at these large doses is likely to be in the hundreds. That is, it would take an inoculum of at least 256 cells to be eight generations (i.e., 4 h) ahead of an inoculum as small as one cell. The ability to infect the squid with hundreds of cells is experimentally useful because this dose is sufficiently high to make feasible in vivo expression technology approaches for identifying host-induced bacterial genes (3). Traditional genetic studies have already revealed a number of activities, e.g., flagellar motility (14, 29) and gene regulation (9), as well as gene products, e.g., an outer membrane protein (1) and a pilus (40), that are required for normal initiation of symbiosis. Future microarray-based studies are likely to identify other bacterial activities that may underlie the high symbiotic infectivity of V. fischeri.
Taken together, the data presented here indicate that V. fischeri cells, even at the low concentrations that may be found in nature, are highly efficient at colonization, due to a combination of intrinsic properties of the bacterium and selective properties of the host (42). As a result only a few cells need to be in immediate proximity to the light-organ tissue in order for a full colonization to occur. To understand the mechanisms responsible for this efficiency, future investigations will focus on bacterial traits that underlie the species specificity that is characteristic of this association, uncovering those genes whose functions are important in the earliest stages of contact between bacterium and squid host tissue.
Acknowledgments
We thank J. Grad and L. Sycuro as well as C. Lupp, M. McFall-Ngai, P. Patek, and A. Alvarez for experimental advice. Advice on statistical treatments was provided by A. Taylor. We thank T. Koropatnick and C. Yap for maintenance of the animal breeding facility.
This work was funded by National Institutes of Health grant RR12294 to E. G. Ruby and M. McFall-Ngai and National Science Foundation grant IBN 02-11673 to M. McFall-Ngai and E. G. Ruby. Access to the V. fischeri genomic sequence was made possible by Integrated Genomics, Inc., and a grant from the W. M. Keck Foundation.
REFERENCES
- 1.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Angelichio, M. J., and A. Camilli. 2002. In vivo expression technology. Infect. Immun. 70:6518-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cash, R. A., S. I. Music, J. P. Libonati, M. J. J. Snyder, R. P. Wenzel, and R. B. Hornick. 1974. Response of man to infection with Vibrio cholerae. I. Clinical, serologic, and bacteriologic responses to a known inoculum. J. Infect. Dis. 129:45-52. [DOI] [PubMed] [Google Scholar]
- 7.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.DeBoy, R. T., and N. L. Craig. 2000. Target site selection by Tn7: attTn7 transcription and target activity. J. Bacteriol. 182:3310-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLoney, C. R., T. M. Bartley, and K. L. Visick. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J. Bacteriol. 184:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlap, P. V. 1989. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J. Bacteriol. 171:1199-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, J. S., and M. J. McFall-Ngai. 1998. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev. Genes Evol. 208:295-303. [DOI] [PubMed] [Google Scholar]
- 12.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gay, N. J., V. L. Tybulewicz, and J. E. Walker. 1986. Insertion of transposon Tn7 into the Escherichia coli glmS transcriptional terminator. Biochem. J. 234:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, B., L. E. Jensen, and O. Nybroe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187-195. [DOI] [PubMed] [Google Scholar]
- 18.Lamarcq, L. H., and M. J. McFall-Ngai. 1998. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect. Immun. 66:777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. H., and E. G. Ruby. 1992. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 58:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K. H., and E. G. Ruby. 1995. Symbiotic role of the viable but nonculturable state of Vibrio fischeri in Hawaiian coastal seawater. Appl. Environ. Microbiol. 61:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemos, M. L., and J. H. Crosa. 1992. Highly preferred site of insertion of Tn7 into the chromosome of Vibrio anguillarum. Plasmid 27:161-163. [DOI] [PubMed] [Google Scholar]
- 22.Lichtenstein, C., and S. Brenner. 1982. Unique insertion site of Tn7 in the Escherichia coli chromosome. Nature 297:601-603. [DOI] [PubMed] [Google Scholar]
- 23.Liu, P. C., Y. C. Chen, C. Y. Huang, and K. K. Lee. 2000. Virulence of Vibrio parahaemolyticus isolated from cultured small abalone, Haliotis diversicolor supertexta, with withering syndrome. Lett. Appl. Microbiol. 31:433-437. [DOI] [PubMed] [Google Scholar]
- 24.Liu, P. C., Y. C. Chen, and K. K. Lee. 2001. Pathogenicity of Vibrio alginolyticus isolated from diseased small abalone Haliotis diversicolor supertexta. Microbios 104:71-77. [PubMed] [Google Scholar]
- 25.Lupp, C., R. E. Hancock, and E. G. Ruby. 2002. The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179:57-65. [DOI] [PubMed] [Google Scholar]
- 26.McFall-Ngai, M. J., and M. K. Montgomery. 1990. The anatomy and morphogenesis of the adult bacterial light organ of Euprymna scolopes Berry (Cephalapoda: Sepiolidae). Biol. Bull. 179:332-339. [DOI] [PubMed] [Google Scholar]
- 27.McFall-Ngai, M. J., and E. G. Ruby. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491-1494. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., W. Jiang, and B. L. Wanner. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138:1-7. [DOI] [PubMed] [Google Scholar]
- 29.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery, M. K., and M. McFall-Ngai. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719-1729. [DOI] [PubMed] [Google Scholar]
- 31.Nyholm, S. V., B. Deplancke, H. R. Gaskins, M. A. Apicella, and M. J. McFall-Ngai. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppon, J. C., R. J. Sarnovsky, N. L. Craig, and D. E. Rawlings. 1998. A Tn7-like transposon is present in the glmUS region of the obligately chemoautolithotrophic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 180:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 35.Ruby, E. G., and K. H. Lee. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen, H., S. E. Gold, S. J. Tamaki, and N. T. Keen. 1992. Construction of a Tn7-lux system for gene expression studies in gram-negative bacteria. Gene 122:27-34. [DOI] [PubMed] [Google Scholar]
- 37.Small, A. L., and M. J. McFall-Ngai. 1999. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J. Cell. Biochem. 72:445-457. [PubMed] [Google Scholar]
- 38.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 39.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabb, E. V., and E. G. Ruby. 2003. Contribution of pilA to competitive colonization of Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 69:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 42.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In P. E. Stanley (ed.), Bioluminescence and chemiluminescence. John Wiley & Sons, Chichester, West Sussex, United Kingdom.