Abstract
The hindgut of soil-feeding termites is highly compartmentalized and characterized by pronounced axial dynamics of the intestinal pH and microbial processes such as hydrogen production, methanogenesis, and reductive acetogenesis. Nothing is known about the bacterial diversity and the abundance or axial distribution of the major phylogenetic groups in the different gut compartments. In this study, we showed that the variety of physicochemical conditions is reflected in the diversity of the microbial communities in the different gut compartments of two Cubitermes species (Termitidae: Termitinae). 16S rRNA gene clones from the highly alkaline first proctodeal segment (P1) of Cubitermes orthognathus represented almost exclusively gram-positive bacteria with low G+C content (LGC bacteria). In the posterior gut segments, their proportion decreased progressively, and the clone libraries comprised a variety of phyla, including the Cytophaga-Flexibacter-Bacteroides group, various subgroups of Proteobacteria, and the spirochetes. Phylogenetic analysis revealed that many of the clones clustered with sequences from the guts of other termites, and some even formed clusters containing only clones from C. orthognathus. The abundance and axial distribution of major phylogenetic groups in the gut of Cubitermes ugandensis were determined by fluorescence in situ hybridization with group-specific oligonucleotide probes. While the results were generally in good agreement with those of the clonal analysis, direct counts with probes specific for the Planctomycetales revealed a severe underestimation of representatives of this phylum in the clone libraries. Results obtained with newly designed FISH probes directed against two clusters of LGC clones from C. orthognathus indicated that the clones were restricted to specific gut regions. A molecular fingerprinting analysis published in a companion paper (D. Schmitt-Wagner, M. W. Friedrich, B. Wagner, and A. Brune, Appl. Environ. Microbiol. 69:6018-6024, 2003) corroborated the presence of compartment-specific bacterial communities in the gut of different Cubitermes species.
Humivorous termites represent one of the most abundant and ecologically important groups of soil macroinvertebrates in tropical ecosystems. In contrast to the (phylogenetically) lower termites, which are wood-feeding, the majority of species of the higher termites (family Termitidae) consume soil organic matter in various stages of humification (12). The acquisition of host-derived capacities for efficient digestion of lignocellulose rendered the Termitidae independent of the necessity to harbor symbiotic, cellulolytic flagellates, thereby removing important evolutionary constraints and allowing further diversification of the gut (52). Especially in the true soil feeders, the increase in length, volume, and compartmentalization transformed the hindgut into a series of complex microbial habitats with pronounced axial dynamics of the intestinal pH (7, 18), redox potential (37), and other physicochemical parameters [reviewed by Brune (16) and Brune and Friedrich (17)].
The intestinal tract of soil-feeding termites contains a high density of microbial cells (10), and high concentrations of microbial fermentation products in the individual gut compartments indicate the presence of an active gut microbiota (69; E. Miambi, A. Tholen, H. Boga, and A. Brune, unpublished results). However, in view of the specific morphological and physicochemical adaptations of the digestive system, the need for a specific (unique) gut microbiota in soil-feeding termites has been questioned (6), and it cannot be excluded that the microorganisms in the gut represent a transient microbiota of soil microorganisms proliferating under favorable conditions in certain compartments of the intestinal tract.
On the other hand, microsensor and radiotracer studies have shown that the individual gut compartments harbor specific microbial processes, such as H2 production, methanogenesis, and reductive acetogenesis (63, 70). Since cultivation-based studies covered only a small proportion (0.1%) of the gut microorganisms present (70; E. Miambi, A. Tholen, H. Boga, and A. Brune, unpublished results), little is known about the microbial populations involved in these processes. A recent, cultivation-independent study of the archaeal communities of the soil-feeding termite Cubitermes orthognathus (26) provided the first evidence for pronounced differences among the microbiota not only between the gut and the ingested soil, but also among the different gut compartments.
In the present paper, we describe the bacterial diversity in the gut of C. orthognathus based on a clonal analysis of bacterial 16S rRNA genes. The abundance and axial distribution of the major phylogenetic groups were determined by fluorescence in situ hybridization (FISH) of microbial cells in different gut compartments. Since no living colonies of C. orthognathus were available at this point, the experiments were conducted with the closely related C. ugandensis, which is very similar to C. orthognathus not only in its biology and general morphological features, but also with respect to its physicochemical gut parameters (A. Brune, unpublished results).
In a companion study (64), we compared dominant genotypes among the microbiota in the different gut compartments of several Cubitermes species, including C. orthognathus and C. ugandensis, to describe the changes in bacterial community structure during gut passage and to compare the composition of the gut microbiota in homologous gut compartments of the different species.
MATERIALS AND METHODS
Termites.
C. orthognathus Emerson was collected in grassland near Busia, and C. ugandensis Fuller was collected in a glade of the Kakamega rainforest; both sites are located in the highlands of western Kenya. Species were identified by Julius Muli, National Museums of Kenya; in addition, partial sequences (≈650 bp) of the mitochondrial cytochrome oxidase II (COII) gene were determined by PCR of DNA extracts from termite heads with previously described primers (71). Voucher specimens of soldiers and workers preserved in alcohol are available from the corresponding author.
Whole nests were transported to the laboratory in Nairobi, where the nests were cut into pieces; termites were distributed into polypropylene containers containing fragments of the nest and soil collected at about 3 m from the nest. The termites were allowed a few days to reconstruct and fix the nest fragments in the containers before they were transported to the laboratory in Konstanz, Germany.
The termites were kept at room temperature in the dark. The containers were inspected regularly, and parts of the nest material were removed and replaced with fresh soil; moisture was controlled by spraying the surface of the nest material with water. For the experiments, only worker caste termites were used. DNA was extracted within a week; samples for whole-cell hybridization were prepared about a month after collection. For this purpose, termites were dissected with sterile, fine-tipped forceps, and the guts were separated into six sections (Fig. 1).
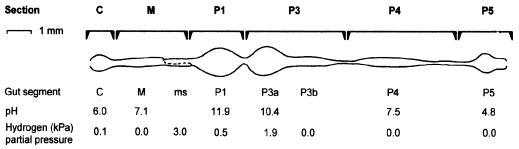
FIG. 1.
Gut morphology of a Cubitermes sp. worker termite. Gut sections were separated at the indicated positions. The gut was drawn in its unraveled state to illustrate the sequence of the individual segments: C, crop; M, midgut; ms, mixed segment; and P1, P3, P4, and P5, the proctodeal segments (53). Hydrogen partial pressure in the individual gut segments was determined with hydrogen-sensitive microsensors for Cubitermes orthognathus (63). The average luminal pH of the major gut segments was determined with glass pH microelectrodes for Cubitermes speciosus (18).
DNA extraction.
Between 10 and 20 gut sections each were pooled, and DNA was extracted by a direct lysis protocol modified after that of Moré et al. (49) that involves bead beating, as described previously in detail (33). Aliquots (1 g) of soil samples and nest material were extracted with the same procedure. DNA was purified from the supernatant by consecutive ammonium acetate, isopropanol, and ethanol precipitation steps. To remove humic substances, the extracts were passed through spin columns filled with polyvinylpolypyrrolidone as described previously (61). DNA concentrations were determined fluorimetrically with Hoechst dye 33258 and a DyNA Quant 200 fluorimeter (Amersham Pharmacia Biotech, Freiburg, Germany) as recommended by the manufacturer.
PCR amplification.
Bacterial 16S rRNA genes were amplified from DNA extracts of each of the four major hindgut sections (Fig. 1) with primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′; Escherichia coli positions 8 to 27) (22) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′; E. coli positions 1492 to 1512) (76). PCR (30 cycles) was carried out as described previously (33), except that the annealing temperature was 55°C. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany).
Clone libraries.
Clone libraries of 16S rRNA genes were created from the PCR products derived from the four major hindgut sections, as described by Friedrich et al. (26). Briefly, PCR products of approximately 1.5 kb were ligated to the pGEM-T Easy plasmid vector and introduced into E. coli by transformation. Clones were checked for correct insert size by vector-targeted PCR. From each clone library, 30 clones were randomly selected and sequenced.
Phylogenetic analysis.
Sequence data were analyzed with the ARB software package [version 2.5b; O. Strunk and W. Ludwig, Technische Universität München (http://www.arb-home.de)]. The new sequences were added to the ARB database and aligned with the Fast Aligner tool (version 1.03). Alignments were checked and corrected manually where necessary. Clonal 16S rRNA gene sequences were compared to sequences in public databases with Blast (1); 16S rRNA gene sequences with high similarities to those determined in this study were retrieved and added to the alignment. Highly variable regions of the 16S rRNA gene sequences and sequence positions with possible alignment errors were excluded by using only those positions of the alignment that were identical in at least 50% of all sequences.
Framework trees were calculated with fastDNAmL (58), a maximum-likelihood method implemented in ARB, with only almost-full-length sequences (>1,400 bases). Shorter clone sequences (<1,400 bases) were added to these trees with the ARB parsimony tool, which allows the addition of short sequences to existing phylogenetic trees without changing global tree topologies (42). The stability of the branching pattern was tested with the neighbor-joining and maximum-parsimony (DNAPARS) methods included in the PHYLIP package (version 3.573c) (24) implemented in ARB. The reproducibility of the branching pattern was confirmed by bootstrap analysis with 1,000 replicates with the maximum-parsimony algorithm and the program Seqboot implemented in PHYLIP. Sequences were checked for chimerae with the Check_Chimera program (44). In addition, the terminal 400 sequence positions at the 5′ and 3′ ends of the sequences were subjected separately to treeing analysis (fractional treeing) (43); significant differences in phylogenetic placement of these terminal sequence fragments were regarded as indicative of chimera formation.
Rarefaction analysis.
Diversity coverage by the clone libraries was analyzed with the Analytic Rarefaction software (version 1.2; S. M. Holland, University of Georgia, Athens, Ga; http://www.uga.edu/strata/Software.html), as previously described (26).
DAPI staining.
For each gut section, five samples were pooled and homogenized in 0.5 ml of sterile phosphate-buffered saline (32) in sterile 2-ml reaction tubes with plastic pestles (Micropistill sticks; Eppendorf, Hamburg, Germany). For sample fixation, 3 volumes of freshly prepared 4% paraformaldehyde were added and incubated for 1.5 h at 4°C. Fixed cells were washed in phosphate-buffered saline and sedimented by centrifugation, and cell pellets were resuspended in 0.1% sodium pyrophosphate (32) under mild sonication (5 times for 0.5 s). For microscopy, appropriate dilutions of these homogenates were filtered onto white polycarbonate filters, stained with 4′,6′-diamidino-2-phenylindole (DAPI), and air dried, as previously described (30), except that a washing step in 96% ethanol was included before the filter sections were mounted in Citifluor solution. For each homogenate, between 400 and 5,000 cells were counted at 1,000× magnification.
Oligonucleotide probes.
Group-specific probes against major phylogenetic groups were selected in Probebase (http://www.probebase.net/). Oligonucleotide probes were designed and checked for specificity with the respective functions of the ARB software. Probe P1C95 (E. coli target positions 95 to 111; 5′-CGT CCG CCG GTA AGT CAC-3′) was specific for the sequences of six clones with a high degree of similarity, described in the Results section as the P1 cluster. Probe T433 (E. coli target positions 433 to 450; 5′-CAT CCC TCG TCA AAG AAG-3′) and probe T125 (E. coli target positions 125 to 141; 5′-CGA AGG CCA GGT TGC TCA-3′) were specific for the sequences of 10 clones with a high degree of similarity, described in the Results section as the Sporobacter cluster. Helper probes targeting the region opposing the probe target site in the 16S rRNA secondary structure were designed to improve the hybridization signal (27). The sequences of the helper probes were as follows: HelperP1C95 (E. coli target positions 57 to 75), 5′-GTG CTT CCC TCG ACT TGC-3′); HelperT433 (E. coli target positions 480 to 497), 5′-GTT CTC GAG GTA CTG TCA-3′); and HelperT125 (E. coli target positions 221 to 238), 5′-CGC GAG CCC ATC CTT CGG-3′. All oligonucleotide probes were 5′-end labeled with the fluorescent cyanine dye indocarbocyanine, except for probe T125, which was labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (Thermo Hybaid GmbH, Ulm, Germany) and diluted in double-distilled water to a concentration of 50 μg ml−1.
In situ hybridization.
Paraformaldehyde-fixed gut homogenates (see above) were mixed with 0.1% agarose (5 μl each), pipetted onto 10-well (6-mm diameter) Teflon-coated glass slides, and dehydrated as described by Llobet-Brossa et al. (41). Hybridization and counterstaining with DAPI followed the procedures of Glöckner et al. (30). Samples were covered with Citifluor and examined at 1,000× magnification with a Zeiss Axiophot microscope fitted for epifluorescence with filter sets for DAPI (G365, FT395, and LP420; Zeiss), for indocarbocyanine (HQ545/30, HQ610/75, and Q570LP; AHF Analysentechnik, Tübingen, Germany), and for fluorescein (HQ480/40, HQ527/30, and Q505LP; AHF Analysentechnik). For each probe, the equivalent of 100 to 250 cells hybridizing with the universal Bacteria probe mixture were counted. Counting results were verified and corrected by subtracting signals observed with the nonsense probe NONEUB (74).
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the clones obtained in this study were deposited with GenBank under accession numbers AY160775 to AY160876.
RESULTS
The gut of C. ugandensis contained a total of about 108 microorganisms, with high cell counts in all major gut sections (Table 1). Cocci and short rods dominated in all sections; segments P1, P3, and P4 also contained remarkable numbers of spirochetes (up to 4.3 × 106 cells in the P3 section). When gut sections were not homogenized but carefully ruptured with needles, phase-contrast microscopy revealed an abundance of morphologically diverse filamentous microorganisms in the P3 and P4 sections; their number was most probably underestimated because they were apparently fragmented or destroyed during homogenization.
TABLE 1.
Enumeration of microbial cells in the different gut sections of C. ugandensisa
| Gut section | Vol (μl) | No. of cellsb (106 cells per gut section)
|
|||||
|---|---|---|---|---|---|---|---|
| Cocci | Short rods | Long rods | Filaments | Spirochetes | Total | ||
| Crop | ND | 0.5 ± 0.2 | 0.4 ± 0.0 | 0.1 ± 0.0 | — | — | 1.0 ± 0.2 |
| Midgut | 0.1 ± 0.0 | 0.4 ± 0.0 | 4.8 ± 0.7 | 2.0 ± 0.2 | 0.1 ± 0.1 | 0.1 ± 0.0 | 7.5 ± 0.5 |
| P1 | 0.8 ± 0.2 | 3.6 ± 1.5 | 16.4 ± 1.7 | 1.7 ± 0.5 | 0.1 ± 0.0 | 0.9 ± 0.2 | 22.7 ± 3.9 |
| P3 | 0.5 ± 0.2 | 4.8 ± 0.6 | 28.1 ± 0.8 | 2.4 ± 0.5 | 0.3 ± 0.1 | 4.3 ± 0.7 | 39.9 ± 1.2 |
| P4 | 0.1 ± 0.0 | 2.1 ± 0.2 | 7.5 ± 0.2 | 0.6 ± 0.1 | 0.1 ± 0.1 | 0.6 ± 0.1 | 11.0 ± 0.7 |
| P5 | 0.2 ± 0.1 | 1.1 ± 0.4 | 4.5 ± 0.2 | 0.1 ± 0.0 | — | 0.1 ± 0.0 | 6.2 ± 0.2 |
| Total gutc | 1.7 ± 0.3 | 12.6 ± 1.7 | 61.8 ± 3.5 | 6.9 ± 1.0 | 0.7 ± 0.1 | 6.1 ± 0.4 | 88.3 ± 6.2 |
Enumeration was done by the DAPI staining technique on polycarbonate filters, differentiated by cell morphotypes (short rods, <5 μm; long rods, 5 to 15 μm; filament, >15 μm). Values are means (± mean deviation) of two independently prepared homogenates. The volumes of the gut sections were estimated by approximation to different geometrical forms (69). Volume estimation values are averages (± standard deviation) of 15 freshly dissected termite guts.
Total, sum of all cells in a section. ND, not determined. —, below the detection limit (<0.1 × 106 cells per gut section).
Sum of cells of a morphotype in all sections.
Clone libraries.
Comparative sequence analysis revealed an incredible phylogenetic breadth of 16S rRNA gene sequences cloned from C. orthognathus. Nearly all clones were affiliated with known taxa of the Bacteria, although some were suspected chimerae and were not analyzed further. Three clones could not be assigned to any of the major phylogenetic groups and were classified as unidentified. The exact number of clones in each clone library is given in Fig. 2.
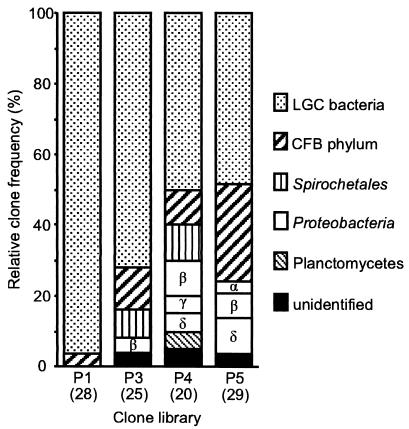
FIG. 2.
Relative clone frequencies in major phylogenetic groups of the clone libraries of the four major hindgut sections of C. orthognathus. The different subphyla of Proteobacteria are differentiated by Greek letters. The number of clones in each clone library is given in parentheses. CFB, Cytophaga-Flexibacter-Bacteroides.
Clones obtained from the first hindgut section (P1) represented almost exclusively gram-positive bacteria with low G+C content (LGC bacteria); only one clone belonged to the Cytophaga-Flexibacter-Bacteroides phylum (Fig. 2). The proportion of LGC clones in the clone libraries of the posterior gut sections decreased progressively, and the clones represented a larger variety of phyla, including different subgroups of the Proteobacteria, the Cytophaga-Flexibacter-Bacteroides phylum, and the spirochetes. The highest phylogenetic diversity was found in the clone library of the P4 section. One-third of the clones from the P5 section were affiliated with the Cytophaga-Flexibacter-Bacteroides phylum. Nevertheless, LGC clones still constituted one-half of the clones in this clone library.
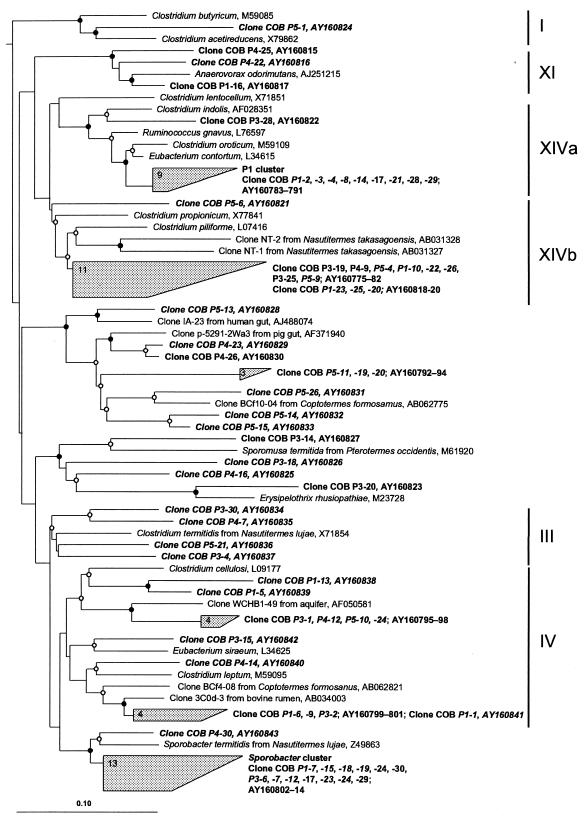
Phylogenetic analysis revealed that many clones grouped in distinct clusters containing only clone sequences derived from termites. This was especially true for the LGC clones (GenBank accession numbers AY160775 to AY160843) (Fig. 3). Nine clones from the P1 clone library formed a distinct cluster within the clostridial cluster XIVa of Collins et al. (19), with 93 to 96% sequence similarity to Eubacterium contortum. Since the cluster contained only clones from this gut section, we will refer to it as the P1 cluster. An even larger cluster (13 clones) consisted almost exclusively of clones from the P1 and P3 clone libraries of C. orthognathus. It was distantly related to clostridial cluster IV (19). Since Sporobacter termitidis (31) was the closest relative of the clones in this cluster (84 to 90% sequence similarity), we will refer to it as the Sporobacter cluster. Both clusters were supported by a reproducible branching pattern in all phylogenetic analyses (including maximum-likelihood, maximum-parsimony, and neighbor-joining algorithms) and by bootstrap values of >90%.
FIG. 3.
Phylogenetic position of clones obtained in this study (shown in bold) affiliated with gram-positive bacteria with low G+C content (LGC bacteria), inferred by maximum-likelihood analysis of 1,318 valid alignment positions. Shorter sequences (shown in italics) were added to the framework tree with the ARB parsimony tool (42). The scale bar indicates approximately 10% sequence difference. All marked nodes were reproducibly present in all phylogenetic analyses (including maximum-likelihood, maximum-parsimony, and neighbor-joining algorithms). Only nodes with bootstrap values (DNAPARS, 1,000 replicates) of >90% (•) and >50% (○) are marked. Clusters of clones containing only sequences from C. orthognathus were grouped; numerals indicate the numbers of clones in a particular cluster. Roman numerals on the right indicate the major clostridial clusters defined by Collins et al. (19). The tree was rooted with Escherichia coli (accession no. J01695), Holophaga foetida (accession no. X77215), Agrobacterium tumefaciens (accession no. M11223), and Verrucomicrobium spinosum (accession no. X90515) as outgroup.
Another large cluster of 11 clones was affiliated with Clostridium piliforme and clostridial clones which have been localized in the alkaline mixed segment of the termite Nasutitermes takasagoensis (72). Although clustering was supported by all phylogenetic analyses, the low bootstrap value (60%) indicates that the exact branching order may change when additional sequences become available. A considerable number of clones (25 clones) were loosely affiliated with bacteria from other clostridial clusters (clusters I, III, IV, XI, XIVa, and XIVb of Collins et al. [19]), many of them including isolates or clone sequences obtained from other termites, or were only distantly related to described members of the Bacillus-Clostridium group (Fig. 3).
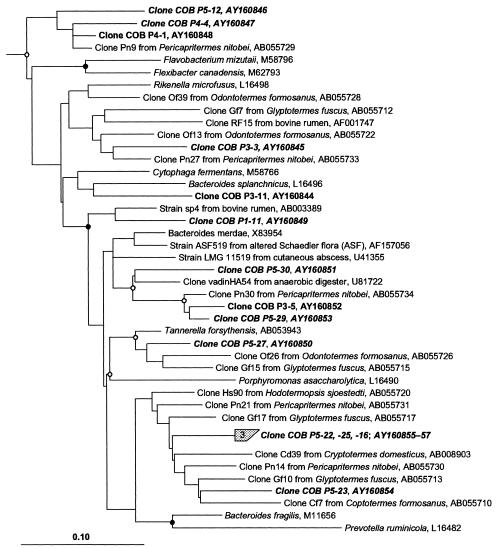
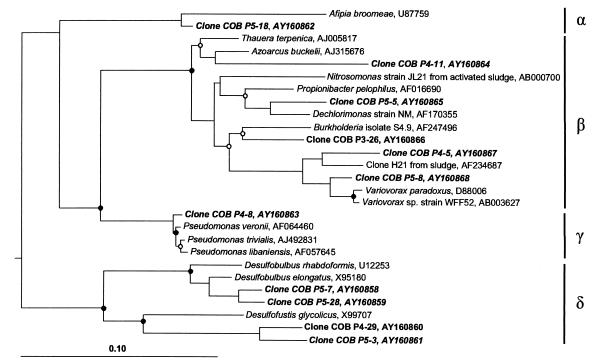
Most of the clones affiliated with the Cytophaga-Flexibacter-Bacteroides phylum (GenBank accession numbers AY160844 to AY160857) (Fig. 4) showed a considerable phylogenetic distance (86 to 97%) from different Bacteroides species; their closest relatives were clones of uncultured bacteria obtained from other termites. Figure 5 shows a phylogenetic tree of the clones affiliated with different subgroups of Proteobacteria (GenBank accession numbers AY160858 to AY160868). There was no obvious clustering except in the case of the clones affiliated with the δ-Proteobacteria, where two clones each were closely or moderately related to representatives of the genera Desulfobulbus and Desulfofustis (94 to 97% and 88 to 89% sequence similarity, respectively).
FIG. 4.
Phylogenetic position of clones obtained in this study (shown in bold) affiliated with the Cytophaga-Flexibacter-Bacteroides phylum, inferred by maximum-likelihood analysis of 1,311 valid alignment positions. For details on tree calculation, evaluation, and notation, see the legend to Fig. 3.
FIG. 5.
Phylogenetic position of clones obtained in this study (shown in bold) affiliated with the Proteobacteria, inferred by maximum-likelihood analysis of 1,348 valid alignment positions. Greek letters represent the different subgroups. For details on tree calculation, evaluation, and notation, see the legend to Fig. 3. The tree was rooted with Thermus aquaticus (accession no. L09663), Aquifex pyrophilus (accession no. M83548), Thermotoga maritima (accession no. M21774), and Verrucomicrobium spinosum (accession no. X90515) as outgroup.
All clones belonging to the Spirochaetales (COB P4-2, P4-15, P3-22, and P3-9; GenBank accession numbers AY160869 to AY160872) clustered within the termite branch of uncultivated treponemes (40) (details not shown). A single clone (COB P4-6; GenBank accession number AY160873) was clearly affiliated with the Planctomycetales but was not closely related to any published sequence (88% sequence similarity to clone PBS-II-13 from anoxic bulk soil [21]). Three clones (COB P3-21, P4-24, and P5-2; GenBank accession numbers AY160874 to AY160876) could not be assigned to existing taxa and remained unclassified.
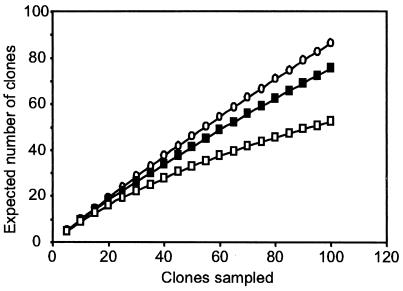
Rarefaction analysis of the results of the clone libraries underlined that the number of clones analyzed was far from sufficient to describe the bacterial diversity in the gut at the species or genus level (Fig. 6). Only at a sequence similarity threshold of 90%, arbitrarily defined as the phylogenetic group level, did the slope of the rarefaction curve indicate reasonable coverage.
FIG. 6.
Rarefaction analysis of all bacterial 16S rRNA gene clones recovered from the hindgut of C. orthognathus. The expected number of clones was calculated from the number of clones analyzed at the species level with 97% sequence similarity (○) and at a sequence similarity level of 95% (▪) and 90% (□), arbitrarily defined as the genus or group level. The slope of the curves indicates whether the diversity was covered (zero or low slope) or whether new taxa can be expected if additional clones were to be analyzed (steep slope).
FISH.
The clonal analysis provided information only on the diversity and relative composition of the bacterial community in the major gut sections. In order to determine the abundance of bacteria from the different phylogenetic groups, we employed FISH with rRNA-targeted oligonucleotide probes. Since no living colonies of C. orthognathus were available at this point, the experiments were conducted with the closely related C. ugandensis.
Table 2 shows the proportion of microbial cells in gut homogenates that hybridized with domain-specific fluorescently labeled oligonucleotide probes relative to the number of DAPI-stained cells in the gut sections. In all gut sections, a large proportion of cells (25 to 64%) hybridized with a mixture of probes targeting most Bacteria (20), whereas cells hybridizing with a domain-specific archaeal probe (66) were found only in the P3 and P4 sections and were much less abundant.
TABLE 2.
Cell densities in different gut segments of C. ugandensis and proportion of cells hybridizing with domain-specific FISH probes relative to the number of DAPI-stained cells in homogenates of the respective sectionsd
| Gut section | Cell densitya (109 cells ml−1) | Bacteriab (%) | Archaeac (%) |
|---|---|---|---|
| Midgut | 82.5 ± 27.9 | 63.7 ± 12.1 | — |
| P1 | 22.9 ± 1.5 | 24.7 ± 5.9 | — |
| P3 | 54.4 ± 13.4 | 29.9 ± 9.8 | 1.6 ± 1.0 |
| P4 | 85.9 ± 37.7 | 39.6 ± 13.7 | 3.8 ± 2.8 |
Total cell counts (DAPI) per gut segment were converted to cell densities by using the average volume of the respective segment (Table 1).
Mixture of probes EUB338, EUB338-II, and EUB338-III (35% formamide) (20).
Probe ARCH915 (35% formamide) (66). —, below the detection limit (<0.1%).
Values are means ± mean deviations of two independently prepared homogenates.
The distribution of major phyla within the domain Bacteria was assessed with several group-specific probes targeted at those phyla represented in the clone library (Table 3). The largest number of clones in the clone libraries were LGC clones, but group-specific probes targeting all bacteria in the LGC subphylum were not available. Published group-specific probes for LGC bacteria (probes LGC354A to LGC354C) (48) were specifically designed to detect Bacillales and Lactobacillales (Table 3) and were therefore not useful. Since the large 16S rRNA sequence heterogeneity among LGC bacteria did not allow the designing of a probe specific for all Clostridiales, we designed cluster-specific probes against the clones from the P1 cluster and the Sporobacter cluster, which represented the largest monophyletic clone groups recovered from the gut of C. orthognathus (Fig. 3).
TABLE 3.
Abundance of bacterial cells in different gut sections of C. ugandensis hybridizing with various group-specific oligonucleotide probes, analyzed by FISH
| Gut section | Relative abundance (% EUB) ± SDm
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteobacteria
|
CFB phylume | Planctomycetesf | HGC bacteriag | LGC bacteria
|
Totalk | ||||||
| αa | βb | γc | δd | LGC354h | P1 clusteri | Sp. clusterj | |||||
| Midgut | 1.6 ± 0.7 | —l | — | 3.2 ± 1.5 | 1.8 ± 0.1 | 3.9 ± 0.8 | — | 2.2 ± 0.2 | 1.4 ± 0.6 | 0.7 ± 0.5 | 14.8 |
| P1 | — | — | — | — | — | 9.3 ± 8.1 | — | 0.4 ± 0.3 | 4.4 ± 1.9 | 0.6 ± 0.5 | 14.7 |
| P3 | 0.2 ± 0.0 | 1.3 ± 1.5 | 1.0 ± 1.0 | 2.0 ± 0.6 | 3.1 ± 1.8 | 36.0 ± 9.9 | — | 0.3 ± 0.1 | 0.3 ± 0.1 | 12.6 ± 4.9 | 56.8 |
| P4 | 3.5 ± 0.9 | 4.3 ± 2.1 | 3.1 ± 1.7 | 2.8 ± 0.8 | 12.3 ± 6.0 | 9.9 ± 3.9 | 4.1 ± 1.2 | 3.0 ± 0.0 | 0.8 ± 0.5 | 3.8 ± 1.8 | 47.6 |
Probe BET42a (35% formamide) (46), used together with the unlabeled competitor cGAM42a.
Probe GAM42a (35% formamide) (46), used together with the unlabeled competitor cBET42a.
Probe SRB385 (30% formamide) (2).
Mixture of probes CF319a and BAC303 (35% formamide) (45). CFB, Cytophaga-Flexibacter-Bacteroides.
Mixture of probes PLA866 and PLA46 (30% formamide) (51) and unlabeled competitor cPLA886.
Probe HGC69a (30% formamide) (62) for gram-positive bacteria with high G + C content.
Mixture of probes LGC354A, LGC354B, and LGC354C (35% formamide) (48) for the Bacillales and Lactobacillales.
Probe P1C95 (35% formamide) for the P1 cluster (this study), used together with the helper probe HelperP1C95.
Probes T125 and T433 (35% formamide) for the Sporobacter cluster (this study), used together with the helper probes HelperT125 and HelperT433.
Sum of the results obtained with individual probes.
—, below the detection limit (<0.1%).
Values are based on those obtained with the EUB probe mixture (see Table 2) and represent means ± standard deviations of two independently prepared homogenates.
The newly designed probe P1C95 was complementary to almost all clones from the P1 cluster (three exceptions). It was checked for specificity with Clostridium coccoides (DSM 935), whose sequence contains only one mismatch to probe P1C95 but which could be discriminated under the hybridization conditions used (Table 3). The newly designed probe T433 was complementary to almost all clones from the Sporobacter cluster (three exceptions). Since no isolate was available to verify probe specificity, we designed a second probe, T125, which was complementary to a different region of the 16S rRNA molecule of the clones targeted by probe T433. Double hybridization of gut homogenates with probes T433 and T125 showed that both probes usually bound to the same cells, indicating that the probes were specific under these conditions.
The results of the hybridization obtained with these probes confirmed that cells detected with the probe specific for the P1 cluster were mainly located in the P1 section, whereas the majority of cells hybridizing with the probes specific for the Sporobacter cluster were detected in the posterior sections. However, the proportion of cells hybridizing with these cluster-specific probes (relative to those hybridizing with the Bacteria-specific probe mixture) in the P1 and P3 sections (5 and 13%, respectively) was considerably lower than that expected on the basis of the abundance of target clones in the clone libraries (36 and 24%, respectively). For other groups, such as the Cytophaga-Flexibacter-Bacteroides phylum and the different subdivisions of the Proteobacteria, the proportions of cells detected with the group-specific probes were in good agreement with the frequencies of these phylogenetic groups within the clone library of the respective gut sections (Fig. 2).
Surprisingly, a large number of cells hybridized with a mixture of probes targeting the Planctomycetales (Table 3). They represented almost 36% of the cells hybridizing with the Bacteria-specific probe mixture in the P3 section and about 10% in the P1 and P4 sections. Among the clone libraries, however, only that of the P4 section contained a single clone affiliated with this phylum.
DISCUSSION
The phylogenetic analysis of 16S rRNA genes recovered from the hindgut of C. orthognathus revealed an enormous diversity of bacteria in the different gut compartments. Almost every clone in the clone libraries represented a new phylotype. Rarefaction analysis revealed that the more than 100 clones analyzed in this study provided reasonable coverage of the major phylogenetic groups among the gut microbiota but were far from being sufficient to describe the bacterial diversity at the species or genus level (Fig. 6).
Also, the results obtained from molecular characterizations of the gut microbiota of other termites (54, 55, 56, 57) lead to the impression that diversity studies in insect guts are a bottomless pit. In a detailed study of spirochetal diversity in the gut of wood-feeding termites (40), almost 100 clones had to be analyzed for reasonable coverage of ribotype diversity within a single bacterial phylum. Clearly, other approaches are necessary to follow spatial or temporal changes in community structure within specific gut microhabitats or to compare the gut microbiota of different termites. In a companion study (64), we took advantage of the sensitivity of the PCR-based approach and the integrative nature of terminal restriction fragment length polymorphism (T-RFLP) fingerprints (47) to combine exact information on the phylogenetic position of a clone or clone group with its relative abundance in the amplified products. However, since both clone libraries and T-RFLP analysis have the limitations inherent to all PCR-based approaches, such as efficiency of DNA extraction, primer selectivity, possibility of PCR bias, and different copy numbers of 16S rRNA genes (23, 68, 73), we employed FISH with group-specific oligonucleotide probes (3) to document the abundance of major phylotypes.
Abundance and distribution of major phylogenetic groups.
The steep increase in gut pH between the midgut and the first hindgut segment (Fig. 1) coincides with a dramatic drop in the density of microorganisms and also in the proportion of DAPI-stained cells hybridizing with the domain-specific probes (Table 2). This and the subsequent increase in cell density and hybridization efficiency are strong evidence that the dynamic change of physicochemical conditions at the midgut-hindgut junction represents a significant barrier for microorganisms passing with the gut contents. This is in agreement with the rapid mineralization of radiolabeled peptidoglycan during gut passage (35) and the decrease in viable counts by two orders of magnitude between the midgut and P1 section observed in the case of the soil-feeding termite Procubitermes aburiensis (10). High lysozyme activities have been reported for the salivary glands and the midgut of several wood-feeding termites (29) but remain to be studied with soil-feeding species.
In general, the hybridization efficiencies obtained in this study (25 to 64%) are in good agreement with values obtained in other studies. Also in Mastotermes darwiniensis, the proportion of cells hybridizing with probe EUB338 relative to DAPI-stained cells was in the same range (32 to 52%) (5). Similar values were reported for hybridization efficiencies in bulk soil (37 to 47%) (77); a recent quantitative review of numerous studies gives an average value of 56% for various aquatic ecosystems and also discusses factors influencing the detection of bacterial cells with FISH (11).
In the case for the Cytophaga-Flexibacter-Bacteroides phylum and the various subgroups of Proteobacteria, the FISH results obtained for C. ugandensis, when expressed relative to the hybridization efficiency obtained with the Bacteria-specific probes mixture (Table 3), are in good agreement with the clone frequencies of the different clone libraries of C. orthognathus (Fig. 2). No specific probe was available for spirochetes, but the abundance of DAPI-stained cells with spirochetal morphology in the P3 and P4 sections of C. ugandensis guts (11 and 6%, respectively) is in good agreement with the relative clone frequency of spirochetes in the clone libraries of C. orthognathus (8 and 10%, respectively).
Although the distribution of the clones affiliated with the P1 cluster and the Sporobacter cluster among the different clone libraries of the different segments of C. orthognathus is supported by the results of the FISH analysis in C. ugandensis, the proportion of cells hybridizing with these cluster-specific probes was considerably lower than that expected on the basis of the abundance of target clones in the respective clone libraries. Such discrepancies may reflect species-specific differences among the gut microbiota of the two termite species. Although the T-RFLP fingerprints of homologous gut sections were quite similar (64), it has to be considered that identical terminal restriction fragments shared between the two termite species do not necessarily mean that the underlying genotypes are identical. Therefore, it is possible that the cluster-specific probes designed on the basis of the sequences obtained with C. orthognathus were inadequate for the detection of all bacteria from the same cluster in C. ugandensis; even in C. orthognathus, the cluster-specific probes would not have covered all clones in the cluster. Moreover, it has to be kept in mind that the clones in the P1 cluster and the Sporobacter cluster were not designed to cover the diversity of LGC bacteria in the respective segments.
The most convincing example for the importance of backing the results of a clonal analysis with FISH, however, is the discrepancy in the relative abundance of the Planctomycetales. In all gut sections tested, a large number of cells hybridized with the planctomycete-specific probe mixture (Table 3), but only the clone library of the P4 section contained a single clone affiliated with the Planctomycetales. The reason for the strong bias against planctomycetes most probably lies in the inadequacy of the Bacteria-specific primers, especially of the forward primer (27F), to amplify planctomycete 16S rRNA gene sequences (21). Although FISH analysis indicates that more than one-third of the bacteria in the P3 section of C. ugandensis may be planctomycetes, their metabolic function remains obscure. To date, most planctomycetes have been isolated from aquatic habitats, but recently they have also been found in soil (75), in the midgut gland of shrimp (28), and in the pig gut (39).
Functional implications.
The largest number of clones in the clone libraries represent gram-positive bacteria with low G+C content affiliated with the clostridia and gram-negative bacteria of the Cytophaga-Flexibacter-Bacteroides phylum. Representatives of these phyla are typical components of the intestinal microflora of mammals, including humans (25, 34, 39, 67), and have also been recovered from wood- and soil-feeding termites, either by cultivation (15) or as 16S rRNA gene fragments (38, 54). The accumulation of butyrate and succinate in various gut sections of C. orthognathus and other soil-feeding termites (69) is in general agreement with the presence of these groups, but it has to be kept in mind that many of the clones obtained from C. orthognathus belong to clusters that are only distantly related to described species.
Studies with hydrogen microsensors have demonstrated that hydrogen accumulates in the anterior hindgut of all Cubitermes spp. investigated so far (37, 63), with highest partial pressures in the mixed segment and the P3 segment (Fig. 1). The results of this study showed that the microbiota in the anterior hindgut is dominated by gut section-specific populations of LGC bacteria that are most closely related to clostridia. Since clostridia are fermentative bacteria and many produce hydrogen during fermentation, it can be speculated that clostridia are a possible source of hydrogen in Cubitermes guts. Many clostridia are also specialized in the fermentation of amino acids and might benefit from the proteolytic activity present in the gut of soil-feeding termites, mainly in the M section comprising the midgut and mixed segment (R. Ji and A. Brune, unpublished results), whose origin (host or microbial) remains to be established.
Many bacteria from the Cytophaga-Flexibacter-Bacteroides phylum are specialized in the degradation of plant fibers and proteins (4, 65). The results of the clone distribution and T-RFLP and FISH analyses documented an increase in the number of bacteria from the Cytophaga-Flexibacter-Bacteroides phylum toward the posterior hindgut, which is in agreement with the accumulation of acetate, lactate, and succinate in the gut fluid of the P3 gut section of Cubitermes spp. (69) and the absence of reportedly alkaliphilic representatives among the described species in this phylum.
In earlier studies, it had been postulated that filamentous bacteria belonging to the actinomycetes might play an important role in the P3 and P4 sections of Cubitermes severus (8, 9). Actinomycetes isolated from soil-feeding termites have been shown to have cellulolytic and lignin-solubilizing activities (59, 60). However, we obtained no indication that actinomycetes play a major role in the gut of C. orthognathus or C. ugandensis — none of the clone libraries contained clones belonging to gram-positive bacteria with high G+C content, which were detected by direct counts with probe HGC69a only in gut section P4 of C. ugandensis (Table 3). A methodological bias can be excluded because in a separate study, numerous clones of actinobacteria were recovered from the gut of humivorous beetle larvae with the same DNA extraction protocol, PCR conditions, and primer set (M. Egert, T. Lemke, A. Brune, and M. Friedrich, unpublished results).
The percentage of archaeal cells among prokaryotic cells in the gut of C. ugandensis, 1.6% and 3.8% of the number of DAPI-stained cells in sections P3 and P4, respectively (Table 2), is similar to the percentage of archaeal 16S rRNA among prokaryotic 16S rRNAs in the gut of other soil-feeding termite species (3.1% in C. fungifaber and 1.7 to 3.1% in other soil-feeding Termitinae) determined by dot blot hybridization (13). Moreover, the restriction of archaea to the P3 and P4 sections is in agreement with the distribution of F420-fluorescent cells and methane emission rates of the respective gut sections (63). The present results also indicate that archaea related to the Methanosarcinales, which form a population specific for the P1 section of C. orthognathus (26), might not be numerically important. However, direct microscopic observations of nonhomogenized gut sections that were carefully ruptured with needles revealed an abundance of morphologically diverse F420-fluorescent microorganisms in the P3b and P4 segments. Since many of them were filamentous and appeared to be fragmented or destroyed during homogenization (see above), it is likely that they were underestimated by the FISH analysis.
Gut-specific populations.
The results of the clonal analysis document that a large number of clones recovered from the hindgut of C. orthognathus cluster among clones obtained from termite guts; some of them even form monophyletic clusters that contain only clones from this particular termite. One example is the Sporobacter cluster (Fig. 3), which is composed exclusively of clones obtained in this study that were present only in the clone libraries of the P1 and the P3 gut segments. The restriction of bacteria affiliated with this group to these moderately to extremely alkaline segments (Fig. 1) is also supported by the results of the T-RFLP analysis, which demonstrated that the majority of the clones in this cluster were represented by large peaks in the profiles of the respective segments (64).
Clones affiliated with the P1 cluster (Fig. 3) were present exclusively in the clone library of the P1 segment of C. orthognathus, and the P1 segment of C. ugandensis also contained the largest proportion of cells hybridizing with the cluster-specific probe (Table 3). The relative predominance of this group of bacteria in this segment is also supported by the T-RFLP fingerprints presented in the companion paper (64). It seems reasonable to assume that the specificity for these gut regions is related to an adaptation to the high alkalinity of the anterior hindgut (mixed segment to P3a; Fig. 1) of soil-feeding termites. Considering the volume differences between the midgut and P1 segment (Table 1) and the strong drop in cell density between these segments (Table 2), the largest number of these organisms is apparently present in the midgut section, which comprises the alkaline mixed segment. Tokuda et al. observed that the mixed segment of Nasutitermes takasagoensis, a wood-feeding higher termite, contains an apparently specific population of clostridia located between the midgut epithelium and the peritrophic membrane (72). Interestingly, the clonal isolates obtained in their study, NT-1 and NT-2, are not closely related to the clones in the P1 cluster but to many other clones recovered from the P1 of C. orthognathus, affiliated with cluster XIVb (Fig. 3).
Conclusions.
The combined results of the present study and of the molecular fingerprinting analysis described in the companion paper (64), together with the previous characterization of the archaeal community structure in C. orthognathus (26), document clearly that the intestinal tract of Cubitermes spp. is colonized by an assortment of phylotypes characteristic of each compartment. Physicochemical and functional differences among the compartments are reflected in the composition of the microbial community, and contrary to previous assumptions, the existence of a specific microbiota in the gut of soil-feeding termites can now be regarded as firmly established.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft and by the Max Planck Society.
We thank Hamadi Boga (Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya), Lucie Rogo and Nixon Onyimbo (International Centre of Insect Physiology and Ecology, Nairobi), and Wanja Kinuthia and the late Julius Muli (National Museum of Kenya) for their help with termite collection and Karen A. Brune for critically reading the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. [DOI] [PubMed] [Google Scholar]
- 4.Avgustin, G., R. J. Wallace, and H. J. Flint. 1997. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Bacteriol. 47:284-288. [DOI] [PubMed] [Google Scholar]
- 5.Berchtold, M., A. Chatzinotas, W. Schönhuber, A. Brune, R. Amann, D. Hahn, and H. König. 1999. Differential enumeration and in situ localization of microorganisms in the hindgut of the lower termite Mastotermes darwiniensis by hybridization with rRNA-targeted probes. Arch. Microbiol. 172:407-416. [DOI] [PubMed] [Google Scholar]
- 6.Bignell, D. E. 1994. Soil-feeding and gut morphology in higher termites, p. 131-158. In J. H. Hunt and C. A. Nalepa (ed.), Nourishment and evolution in insect societies. Westview Press, Boulder, Colo.
- 7.Bignell, D. E., and P. Eggleton. 1995. On the elevated intestinal pH of higher termites (Isoptera: Termitidae). Insect. Soc. 42:57-69. [Google Scholar]
- 8.Bignell, D. E., H. Oskarsson, and J. M. Anderson. 1979. Association of actinomycete-like bacteria with soil-feeding termites (Termitidae, Termitinae). Appl. Environ. Microbiol. 37:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bignell, D. E., H. Oskarsson, and J. M. Anderson. 1980. Colonization of the epithelial face of the peritrophic membrane and the ectoperitrophic space by actinomycetes in a soil-feeding termite. J. Invert. Pathol. 36:426-428. [Google Scholar]
- 10.Bignell, D. E., H. Oskarsson, and J. M. Anderson. 1980. Distribution and abundance of bacteria in the gut of a soil-feeding termite Procubitermes aburiensis (Termitidae, Termitinae). J. Gen. Microbiol. 117:393-403. [DOI] [PubMed] [Google Scholar]
- 11.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells with fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 12.Brauman, A., D. E. Bignell, and I. Tayasu. 2000. Soil-feeding termites: biology, microbial associations and digestive mechanisms, p. 233-259. In T. Abe, D. E. Bignell, M. Higashi (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Brauman, A., J. Dore, P. Eggleton, D. Bignell, J. A. Breznak, and M. D. Kane. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol. Ecol. 35:27-36. [DOI] [PubMed] [Google Scholar]
- 14.Breznak, J. A., and J. Switzer Blum. 1991. Mixotrophy in the termite gut acetogen, Sporomusa termitida. Arch. Microbiol. 156:105-110. [Google Scholar]
- 15.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 16.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 17.Brune, A., and M. Friedrich. 2000. Microecology of the termite gut: structure and function on a microscale. Curr. Opin. Microbiol. 3:263-269. [DOI] [PubMed] [Google Scholar]
- 18.Brune, A., and M. Kühl. 1996. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J. Insect Physiol. 42:1121-1127. [Google Scholar]
- 19.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 20.Daims, H., A. Brühl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 21.Derakshani, M., L. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrelly, V., V. Rainey, and V. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 1989. PHYLIP, phylogeny inference package version 3.57c. Cladistics 5:164-166. [Google Scholar]
- 25.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita, A., and T. Abe. 2002. Amino acid concentration and distribution of lysozyme and protease activities in the guts of higher termites. Physiol. Entomol. 27:76-78. [Google Scholar]
- 30.Glöckner, F. O., R. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K. H. Trebesius, and K. H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 31.Grech-Mora, I., M.-L. Fardeau, B. K. C. Patel, B. Ollivier, A. Rimbault, G. Prensier, J.-L. Garcia, and E. Garnier-Sillam. 1996. Isolation and characterization of Sporobacter termitidis gen. nov., sp. nov., from the digestive tract of the wood-feeding termite Nasutitermes lujae. Int. J. Syst. Bacteriol. 46:512-518. [Google Scholar]
- 32.Hahn, D., R. Amann, W. Ludwig, A. D. L. Akkermans, and K. H. Schleifer. 1992. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labeled oligonucleotides. J. Gen. Microbiol. 138:879-887. [DOI] [PubMed] [Google Scholar]
- 33.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 35.Ji, R., and A. Brune. 2001. Transformation and mineralization of 14C-labeled cellulose, peptidoglycan, and protein by the soil-feeding termite Cubitermes orthognatus. Biol. Fertil. Soils 33:166-174. [Google Scholar]
- 36.Kane, M. D., A. Brauman, and J. A. Breznak. 1991. Clostridium mayombei sp. nov., an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite, Cubitermes speciosus. Arch. Microbiol. 156:99-104. [Google Scholar]
- 37.Kappler, A., and A. Brune. 2002. Dynamics of redox potential and changes in redox state of iron and humic acids during gut passage in soil-feeding termites (Cubitermes spp.). Soil Biol. Biochem. 34:221-227. [Google Scholar]
- 38.Kudo, T., M. Ohkuma, S. Moriya, S. Noda, and K. Ohtoko. 1998. Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation. Extremophiles 2:155-161. [DOI] [PubMed] [Google Scholar]
- 39.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 41.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 44.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Jr. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 46.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 47.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 48.Meier, H., R. Amann, W. Ludwig, and K. H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 49.More, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neef, A. 1997. Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Doctoral thesis. Technische Universität München, Munich, Germany.
- 51.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 52.Noirot, C. 1992. From wood- to humus-feeding: an important trend in termite evolution, p. 107-119. In J. Billen (ed.), Biology and evolution of social insects. Leuven University Press, Leuven, Belgium.
- 53.Noirot, C. 2001. The gut of termites (Isoptera). Comparative anatomy, systematics, phylogeny. II. Higher termites (Termitidae). Ann. Soc. Entomol. Fr. (N. S.) 37:431-471. [Google Scholar]
- 54.Ohkuma, M., S. Noda, Y. Hongoh, and T. Kudo. 2002. Diverse bacteria related to the Bacteroides subgroup of the Cytophaga-Flexibacter-Bacteroides phylum within the gut symbiotic communities of various termites. Biosci. Biotechnol. Biochem. 66:78-84. [DOI] [PubMed] [Google Scholar]
- 55.Ohkuma, M., T. Iida, and T. Kudo. 1999. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181:123-129. [DOI] [PubMed] [Google Scholar]
- 56.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohkuma, M., S. Noda, and T. Kudo. 1999. Phylogenetic relationships of symbiotic methanogens in diverse termites. FEMS Microbiol. Lett. 171:147-153. [DOI] [PubMed] [Google Scholar]
- 58.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences with maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 59.Pasti, M. B., A. L. Pometto III, M. P. Nuti, and D. L. Crawford. 1990. Lignin-solubilizing ability of actinomycetes isolated from termite (Termitidae) gut. Appl. Environ. Microbiol. 56:2213-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasti, M. B., and M. L. Belli. 1985. Cellulolytic activity of actinomycetes isolated from termite (Termitidae) gut. FEMS Microbiol. Lett. 26:107-112. [Google Scholar]
- 61.Ramakrishnan, B., T. Lueders, R. Conrad, and M. Friedrich. 2000. Effect of soil aggregate size on methanogenesis and archaeal community structure in anoxic rice field soil. FEMS Microbiol. Ecol. 32:261-270. [DOI] [PubMed] [Google Scholar]
- 62.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K. H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content with 23S rRNA-targeted oligonucleotides. Microbiology 140:2849. (Erratum, Microbiology 141: 1267, 1995.) [DOI] [PubMed] [Google Scholar]
- 63.Schmitt-Wagner, D., and A. Brune. 1999. Hydrogen profiles and localization of methanogenic activities in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4490-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). 69:AEM 392-03. [DOI] [PMC free article] [PubMed]
- 65.Shah, H. N. 1992. The genus Bacteroides and related taxa, p. 3593-3607. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, New York, N.Y.
- 66.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, England.
- 67.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tholen, A. 1999. Der Termitendarm als strukturiertes Ökosystem: Untersuchung der Mikrobiota und der Stoffflüsse im Darm von Reticulitermes flavipes und Cubitermes spp. Ph.D. thesis. University of Konstanz, Konstanz, Germany.
- 70.Tholen, A., and A. Brune. 1999. Localization and in situ activities of homoacetogenic bacteria in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4497-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson, G. J., L. R. Miller, M. Lenz, and R. H. Crozier. 2000. Phylogenetic analysis and trait evolution in Australian lineages of drywood termites (Isoptera, Kalotermitidae). Mol. Phylogenet. Evol. 17:419-429. [DOI] [PubMed] [Google Scholar]
- 72.Tokuda, G., I. Yamaoka, and H. Noda. 2000. Localization of symbiotic clostridia in the mixed segment of the termite Nasutitermes takasagoensis (Shiraki). Appl. Environ. Microbiol. 66:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 74.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 75.Wang, J., C. Jenkins, R. I. Webb, and J. A. Fuerst. 2002. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 68:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zarda, B., D. Hahn, A. Chatzinotas, W. Schönhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]