Abstract
Candida tropicalis ATCC 20336 excretes α,ω-dicarboxylic acids as a by-product when cultured on n-alkanes or fatty acids as the carbon source. Previously, a β-oxidation-blocked derivative of ATCC 20336 was constructed which showed a dramatic increase in the production of dicarboxylic acids. This paper describes the next steps in strain improvement, which were directed toward the isolation and characterization of genes encoding the ω-hydroxylase enzymes catalyzing the first step in the ω-oxidation pathway. Cytochrome P450 monooxygenase (CYP) and the accompanying NADPH cytochrome P450 reductase (NCP) constitute the hydroxylase complex responsible for the first and rate-limiting step of ω-oxidation of n-alkanes and fatty acids. 10 members of the alkane-inducible P450 gene family (CYP52) of C. tropicalis ATCC20336 as well as the accompanying NCP were cloned and sequenced. The 10 CYP genes represent four unique genes with their putative alleles and two unique genes for which no allelic variant was identified. Of the 10 genes, CYP52A13 and CYP52A14 showed the highest levels of mRNA induction, as determined by quantitative competitive reverse transcription-PCR during fermentation with pure oleic fatty acid (27-fold increase), pure octadecane (32-fold increase), and a mixed fatty acid feed, Emersol 267 (54-fold increase). The allelic pair CYP52A17 and CYP52A18 was also induced under all three conditions but to a lesser extent. Moderate induction of CYP52A12 was observed. These results identify the CYP52 and NCP genes as being involved in α,ω-dicarboxylic acid production by C. tropicalis and provide the foundation for biocatalyst improvement.
α,ω-Dicarboxylic acids (α,ω-diacids) are versatile chemical intermediates useful as raw materials for the preparation of perfumes, polymers, adhesives, and macrolide antibiotics. Currently, only three diacids, nonanedioic (azelaic), decanedioic (sebacic), and dodecanedioic, are available at a quantity and cost acceptable for commercial applications. Long-chain diacids with more than 12 carbons offer potential advantages over shorter-chain diacids, but their limited commercial availability and high price have prevented widespread growth in many of these applications.
Several species of yeasts belonging to the genus Candida excrete α,ω-diacids as a by-product when cultured on n-alkanes or fatty acids as the carbon source (22). One such yeast, Candida tropicalis ATCC 20336, is the subject of this paper.
In Candida spp., n-alkanes and fatty acids are metabolized by enzymes present in the β-oxidation and ω-oxidation pathways. For alkanes, the first reaction occurs in the ω-oxidation pathway with the formation of the corresponding alcohol. This reaction is catalyzed by a cytochrome P450 hydroxylase complex, which consists of a cytochrome P450 monooxygenase (the CYP protein) and the accompanying NADPH cytochrome P450 reductase (NCP). A terminal carboxy function is ultimately formed from two additional oxidation steps, catalyzed by a fatty alcohol oxidase and a fatty aldehyde dehydrogenase. Fatty acid formed via the ω-oxidation pathway or introduced as a substrate can be oxidized through the same ω-oxidation pathway to the corresponding aliphatic α,ω-dicarboxylic acid. Finally, the diacids as well as the fatty acid substrates can be activated to the corresponding acyl-coenzyme A ester and then metabolized through β-oxidation to yield energy, CO2, and water. In Candida spp., the β-oxidation pathway is localized in the peroxisomes. A necessary step to produce high yields of aliphatic diacids from C. tropicalis ATCC 20336 was to eliminate β-oxidation so as not to consume the desired product. When β-oxidation was blocked by disrupting the POX4 and POX5 genes, encoding acyl-coenzyme A oxidase, the enzyme catalyzing the first step in the pathway, the result was a dramatic increase in the production of diacids. The resultant β-oxidation-blocked strain was designated C. tropicalis ATCC 20962 (13).
Once the β-oxidation-blocked strain was available, the next step toward increasing diacid production was directed toward improving the flow through the ω-oxidation pathway by overexpressing the enzymes constituting the cytochrome P450 hydroxylase complex. Cytochromes P450 (P450s) are terminal monooxygenases of a multicomponent enzyme system. They constitute a superfamily of proteins which exist widely in nature having been isolated from a variety of organisms (9). An alkane-inducible family of P450s, CYP52, has been described in several different Candida species (11, 20). These P450s and their corresponding reductase, NADPH cytochrome P450 reductase (NCP), constitute the hydroxylase complex responsible for the first and rate-limiting step of ω-oxidation of n-alkanes and fatty acids (3, 4). In order to alleviate this rate-limiting step in the conversion of n-alkanes and fatty acids to their corresponding aliphatic diacids, it is necessary to increase the CYP52 protein responsible for the conversion. One method to effect the increase in CYP52 protein would be overexpression of a specific CYP52 gene.
Prior to overexpression, it is imperative to identify the CYP52 enzymes responsible for the ω-oxidation of the target n-alkanes and fatty acids. Quantitative competitive reverse transcription-PCR (QC RT-PCR) was used to identify CYP52 genes induced by selected fatty acid feedstreams. The identity of the CYP52 gene(s) involved in ω-oxidation and the level of mRNA transcription for each CYP52 was determined with this highly specific and sensitive procedure.
This paper describes the isolation and characterization of 10 members of the CYP52 gene family from Candida tropicalis ATCC 20336. A sensitive and specific QC RT-PCR was used to characterize the specific CYP52 mRNA transcription levels in C. tropicalis ATCC 20962 in response to oleic acid, octadecane, and a commercially available fatty acid feedstock. Transcriptional induction patterns identified specific CYP52 genes involved in the conversion of long-chain fatty acids and n-alkanes (>12 carbons) into their corresponding dicarboxylic acid.
The isolation, characterization, and identification of the specific CYP52s from C. tropicalis ATCC 20336 will allow the development of commercially useful strains for the production of aliphatic dicarboxylic acids from n-alkane and fatty acid feedstocks.
MATERIALS AND METHODS
Strains.
Candida tropicalis ATCC 20336, a natural isolate from an oil field, was the wild-type strain used to generate genomic DNA and genomic libraries and was the parent of C. tropicalis ATCC 20962 (ura3A/ura3B pox4A::ura3A/pox4B::ura3A pox5::ura3A/pox5::URA3A). C. tropicalis ATCC 20962 (initially designated strain H5343), an acyl-coenzyme A oxidase-deficient strain blocked for β-oxidation, was used for QC RT-PCR analysis of CYP and NCP gene expression.
Substrate composition.
Pure oleic acid had a composition of 99.45% oleic acid (C18:1), 0.14% C16:0 (palmitic acid), 0.20% C18:2 (linoleic acid), and 0.06% C20:1 (eicosenoic acid). Pure octadecane was 99.73% octadecane with the remainder of the substrate consisting of unidentifiable isomers. Emersol 267 (E267) fatty acid feedstream had the following fatty acid composition: 2.4% C14:0, 0.7% C14:1, 4.6% C16:0, 5.7% C16:1, 5.7% C17:1, 1.0% C18:0, 69.9% C18:1, 8.8% C18:2, 0.3% C18:3, and 0.9% C20:1.
Genomic DNA preparation.
Genomic DNA from C. tropicalis was prepared according to the standard protocol as defined in Current Protocols in Molecular Biology (2), with the following modifications. For spheroplasting, 50 μl of a 10-μg/ml zymolase solution was added to the sorbitol mixture, and the cell suspension was incubated at 37°C for 1.5 h on a rotary shaker (200 rpm). In addition, the DNA concentration was determined by the ratio of the absorbance at 260 nm to that at 280 nm (A260/280).
PCR strategy to isolate the CYP52 and NCP genes.
CYP52 proteins are highly similar and contain several regions of amino acid conservation. Highly conserved regions within the CYP52 proteins include the I-helix and heme-binding (HR2) regions (6). These regions were used as a basis for the design of degenerate CYP PCR primers. In a similar manner, NCP proteins contain several regions of amino acid conservation, including flavin mononucleotide binding regions 1 (FMN1) and 2 (FMN2), the flavin adenine dinucleotide (FAD) region, and the NAD phosphate region (NADPH) (23). With C. tropicalis ATCC 20336 genomic DNA as a template, PCR amplification was performed with degenerate primers for either CYP52 or NCP. PCRs were performed in a Perkin Elmer Thermocycler or a Perkin Elmer 2400 with the AmpliTaq Gold enzyme (Perkin Elmer Cetus) kit according to the manufacturer's specifications. PCRs were analyzed by gel electrophoresis, and products of the predicted sizes were isolated and sequenced. Analysis of the DNA sequence of these PCR products identified novel CYP52 and NCP sequences, which were used as probes to screen a C. tropicalis ATCC 20336 genomic library.
Construction of Candida tropicalis ATCC 20336 genomic libraries.
Over the course of this study, three genomic libraries of C. tropicalis ATCC 20336 were constructed. The first library was prepared for Cognis by Clontech laboratories.
The second and third libraries were constructed at Cognis Corporation with the λZAP Express vector (Stratagene). C. tropicalis ATCC 20336 genomic DNA was partially digested with Sau3A1, and fragments in the range of 6 to 12 kb were purified from an agarose gel after electrophoresis of the digested DNA. These DNA fragments were then ligated to BamHI-digested λZAP Express vector arms and processed according to the manufacturer's recommendation.
Screening genomic libraries.
Clontech genomic library filters were generated according to Clontech recommendations. Colony/Plaque Screen Hybridization Transfer Membrane disks (DuPont NEN Research) were used for lifting bacterial colonies representing the Clontech library. Additional treatment of membranes was as described in the protocol provided by NEN Research Products.
Membranes were dried overnight before hybridizing to oligonucleotide probes prepared with a nonradioactive enhanced chemiluminescence (ECL) 3′ oligolabeling and detection system (Amersham Life Sciences). DNA labeling, prehybridization, hybridization, and detection were performed according to the manufacturer's protocols. The hybridization signal was detected with Hyperfilm ECL (Amersham). Membranes were aligned to plates containing bacterial colonies from which colony lifts were performed, and colonies corresponding to positive signals on X-ray were then isolated and propagated in Luria-Bertani broth. Plasmid DNAs were isolated from these cultures and analyzed by restriction enzyme digestions and by DNA sequencing.
The λZAP Express genomic library was screened by plating a phage suspension containing ≈2.5 × 104 amplified lambda phage according to the manufacturer's recommendation. Magna Lift nylon membranes (Micron Separations, Inc.) were used to prepare library filters.
DNA fragments used as probes were purified from agarose gels with a QIAEX II gel extraction kit (Qiagen Inc.) according to the manufacturer's protocol and labeled with an ECL direct nucleic acid labeling kit (Amersham). The membranes were prehybridized and hybridized with the conditions described for the ECL protocol. Labeled DNA (5 to 10 ng of hybridization solution per ml) was added to the prehybridized membranes, and the hybridization was allowed to proceed overnight. The following day, membranes were washed twice at 42°C for 20 min in a buffer containing either 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (high stringency) or 0.5× SSC (low stringency) plus 0.4% sodium dodecyl sulfate and 360 g of urea per liter. The two washes were performed with shaking (60 rpm) and in a final buffer volume equivalent to 2 ml/cm2 of membrane. This was followed by two 5-min washes in 2× SSC at room temperature in a final volume equivalent to 2 ml/cm2 of membrane.
Hybridization signals were generated with the ECL nucleic acid detection reagent and detected with Hyperfilm ECL (Amersham). Positive plaques were screened secondarily, by similar methods, in order to isolate individual plaques. To convert the λZAP Express plaques to plasmid form, Escherichia coli strains XL1Blue-MRF′ and XLOR were used. The conversion was performed according to Stratagene's protocols for single-plaque excision.
Plasmid DNA isolation.
Plasmid DNA was isolated from E. coli cultures with the Qiagen plasmid isolation kit (Qiagen Inc.) according to the manufacturer's protocols.
DNA sequencing and analysis.
DNA sequencing was performed at Sequetech Corporation (Mountain View, Calif.). DNA sequences were analyzed with the MacVector 7.1 and GeneWorks software packages (Oxford Molecular Group). Phylogenetic analysis was conducted with MacVector 7.1.1. The CYP52 gene sequences were submitted to David Nelson at the University of Tennessee at Memphis and assigned official cytochrome P450 designations based on established criteria set forth by the P450 nomenclature committee (http://drnelson.utmem.edu/CytochromeP450.html).
Gene induction studies.
Fermentations were performed according to an established protocol (1). Briefly, a semisynthetic growth medium containing, per liter, 75 g of glucose (anhydrous), 6.7 g of yeast nitrogen base (Difco Laboratories), 3 g of yeast extract, 3 g of ammonium sulfate, 2 g of monopotassium phosphate, and 0.5 g of sodium chloride and with a final pH of ≈5.2 was used for each fermentation. The fermentor was inoculated with 5 to 10% of an overnight culture of C. tropicalis ATCC 20962 and either pure oleic acid, pure octadecane, or Emersol 267 and a glucose cosubstrate feed were added in a fed-batch mode beginning near the end of exponential growth. It has been suggested that catabolite repression can affect the expression of genes involved in alkane utilization from yeasts (8); therefore, the glucose feed was controlled so as not to allow glucose accumulation in the fermentation media. Caustic was added to maintain the pH in the desired range. Samples for gene induction studies were collected just prior to starting the fatty acid feed (time zero) and over the first 4 h of bioconversion.
Cellular RNA was isolated with the Qiagen RNeasy mini kit (Qiagen Inc.). A 2-ml sample of C. tropicalis culture was collected from the fermentor in a standard 2-ml screw-cap tube. Cell samples were immediately frozen in a dry ice-alcohol bath. To isolate total RNA from the samples, the tubes were allowed to thaw on ice, and the cells were pelleted by centrifugation at 11,000 × g in a microcentrifuge for 5 min at 4°C, and the supernatant was discarded while keeping the pellet cold. The microcentrifuge tubes were filled half full with ice-cold zirconia-silica 0.5-mm-diameter beads (Biospec Products), and the tube was filled to the top with ice-cold RLT lysis buffer. Cell rupture was achieved by placing the samples in a mini bead beater (Biospec Products) and immediately homogenizing at full speed for 2.5 min. The samples were allowed to cool on ice for 1 min, and the homogenization-cooling process was repeated twice. The homogenized samples were microcentrifuged at 11,000 × g for 10 min, and 700 μl of the supernatant was removed and transferred to a new Eppendorf tube. Ethanol (700 μl of 70% ethanol) was added to each sample, followed by mixing by inversion.
Each sample was transferred to a Qiagen RNeasy spin column and processed according to the manufacturer's protocols. RNase-free water (100 μl) was added to the column, followed by centrifugation at 8,000 × g for 15 s to elute the RNA. An additional 75 μl of RNase-free water was added to the column, followed by centrifugation at 8,000 × g for 2 min. In order to remove contaminating DNA, 20 μl of 10× DNase I buffer (0.5 M Tris [pH 7.5], 50 mM CaCl2, 100 mM MgCl2), 10 μl of RNase-free DNase I (Ambion), and 40 units of Rnasin (Promega Corporation) were added to the RNA sample. The mixture was then incubated at 37°C for 30 min. Samples were placed on ice, and 250 μl of lysis buffer RLT (Qiagen) and 250 μl of 100% ethanol were added. The samples were transferred to Qiagen RNeasy spin columns and processed according to Qiagen protocols, beginning with the RPE wash buffer step. One hundred microliters of RNase-free water was added, followed by centrifugation at 8,000 × g for 15 s. Residual RNA was collected by adding an additional 50 μl of RNase-free water to the spin column, followed by centrifugation at 11,000 × g for 2 min. 10 μl of the RNA preparation was removed and quantified by the A260/280 method. RNA was stored at −70°C.
QC RT-PCR protocol.
Unique primers directed to variable regions within target members of the CYP52 gene family were constructed to be specific enough to anneal to the variable region of the target CYP52 gene and its allele without annealing to other nontarget members of the CYP52 family. After conducting PCR with the specific primers for that target CYP52 gene, the reaction product was checked to ensure it represented the unique target gene product or its presumed allelic variant. If not, the reaction conditions were altered in terms of stringency to focus the reaction to the desired CYP52 target.
The competitor DNA template was designed and synthesized as follows. The competitor RNA was synthesized in vitro from a competitor DNA template that has the T7 polymerase promoter and carries a small deletion of about 10 to 25 nucleotides relative to the native target RNA sequence. In each case, the forward primer contains the T7 promoter consensus sequence GGATCCTAATACGACTCACTATAGGGAGG fused to the respective target gene primer. The reverse primer contains the sequence of the original primer followed by 20 bases of upstream target sequence creating a deletion of about 10% of the total product length (ca. 20 bp) between the primer sequence and the upstream target sequence. The forward primer was used with the corresponding reverse primer to synthesize the competitor DNA template. The primer pairs were combined in a standard Taq Gold polymerase PCR according to the manufacturer's recommended conditions (Perkin-Elmer). The PCR mix contained a 250 nM final concentration of each primer and 10 ng of C. tropicalis chromosomal DNA for the template. The reaction mixture was placed in a thermocycler for 25 to 35 cycles with the highest annealing temperature possible during the PCRs to ensure a homogeneous PCR product. The PCR products were either gel purified or filter purified to remove unincorporated nucleotides and primers. The competitor template DNA was then quantified with the A260/280 method.
Competitor template DNA was transcribed in vitro to make the competitor RNA with the Megascript T7 kit (Ambion). Competitor DNA (250 ng) template and the in vitro transcription reagents were mixed according to the directions provided by the manufacturer. The reaction mixture was incubated for 4 h at 37°C. The resulting RNA preparations were then checked by gel electrophoresis. The DNA template was then removed with DNase I as suggested by the manufacturer. The RNA competitor was then quantified by the A260/280 method. Serial dilutions of the RNA (1 ng/μl to 1 fg/μl) were made for use in the QC RT-PCRs.
QC RT-PCRs.
QC RT-PCRs were performed with rTth polymerase (Perkin-Elmer) according to the manufacturer's recommended conditions. The reverse transcription reaction was performed in a 10-μl volume with final concentrations of 200 μM each deoxynucleoside triphosphate, 1.25 units of rTth polymerase, 1.0 mM MnCl2, 1× buffer, 100 ng of total RNA isolated from a fermentor-grown culture of C. tropicalis, and 1.25 μM appropriate reverse primer. To quantitate CYP52 expression in C. tropicalis, an appropriate target gene reverse primer was used. Several reaction mixes were prepared for each RNA sample characterized.
To quantitate CYP52 expression, a series of previously described QC RT-PCR mixes were aliquoted to different reaction tubes. To each tube, 1 μl of a serial dilution containing from 100 pg to 100 fg of target gene competitor RNA per μl was added, bringing the final reaction mixtures up to the final volume of 10 μl. The QC RT-PCR mixtures were mixed and incubated at 70°C for 15 min according to the manufacturer's recommended times for reverse transcription. After incubation, the sample temperature was reduced to 4°C to stop the reaction, and 40 μl of the PCR mix was added to the reaction to bring the total volume up to 50 μl. The PCR mix consists of an aqueous solution containing 0.3125 μM target gene forward primer, 3.125 mM MgCl2, and 1× chelating buffer. The reaction mixtures were placed in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler, and the following PCR cycle was performed: 94°C for 1 min, followed by 94°C for 10 s, followed by 58 to 66°C for 40 s for a total of 17 to 22 cycles. The PCR was completed with a final incubation at 58 to 66°C for 2 min, followed by 4°C. In some reactions where no detectable PCR products were produced, the samples were returned to the thermocycler for additional cycles, and this process was repeated until enough PCR products were produced to quantify by high-pressure liquid chromatography (HPLC). This QC RT-PCR procedure was applied to all the target genes with the primers indicated.
Upon completion of the QC RT-PCRs, the samples were analyzed and quantitated by HPLC. From 5 to 15 μl of the QC RT-PCR mix was injected into a Waters Bio-Compatible 625 HPLC with an attached Waters 484 tunable detector. The detector was set to measure a wavelength of 254 nm. The HPLC contained a DNASep column (Sarasep, Inc.) which was placed within the oven at 52°C. The column was installed according to the manufacturer's recommendation of having 30 cm of heated polyether ether ketone tubing installed between the injector and the column. The system was configured with a Sarasep brand guard column positioned before the injector. In addition, there was a 0.22-μm filter disk just before the column, within the oven.
Two buffers were used to create an elution gradient to resolve and quantitate the PCR products from the QC RT-PCRs. Buffer A consists of 0.1 M triethylammonium acetate and 5% acetonitrile (vol/vol). Buffer B consists of 0.1 M triethylammonium acetate and 25% acetonitrile (vol/vol). The QC RT-PCR samples were injected into the HPLC, and a linear gradient of 75% buffer A-25% buffer B to 45% buffer A-55% buffer B was run over 6 min at a flow rate of 0.85 ml min−1. The amount of each QC RT-PCR product was plotted and quantitated with an attached Waters Corporation 745 data module. The log ratios of the amount of QC RT-PCR mRNA product to competitor QC RT-PCR product, as measured by peak areas, was plotted, and the amount of competitor RNA required to equal the amount of mRNA product was determined. In the case of each of the target genes, the competitor RNA contained fewer base pairs than the native target mRNA, and therefore the competitor PCR product eluted before the native PCR product.
In addition to the labor-intensive HPLC method for QC RT-PCR quantification, agarose gel electrophoresis of the QC RT-PCRs was employed as an alternative method for quantification (15). Similar levels of gene induction were obtained when the HPLC and 4% agarose gel analysis methods were compared with identical QC RT-PCRs.
Nucleotide sequence accession numbers.
GenBank accession numbers have been obtained for CYP52A12 (AY230498), CYP52A13 (AY230499), CYP52A14 (AY230500), CYP52A15 (AY230501), CYP52A16 (AY230502), CYP52A17 (AY230504), CYP52A18 (AY230505), CYP52A19 (AY230506), CYP52A20 (AY230507), and CYP52D2 (AY230503). Sequences used for phylogenetic analysis included Saccharomyces cerevisiae CYP51 (M18109) and C. tropicalis CYP51 (M23673), CYP52C2 (D12718), CYP52C1 (Z13014), CYP52A3 (D12475), CYP52A6 (Z13010), CYP52A4 (X51932), CYP52A1 (M15945), CYP52A5 (D12714), CYP52A2 (M63258), CYP52A7 (Z13011), CYP52A9 (D26160), CYP52A8 (Z13012), CYP52A10 (D12719), CYP52A11 (D26159), CYP52D1 (D12716), CYP52B1 (Z13013), CYP52E1 (X76225), and CYP52E2 (X87640).
RESULTS
Cloning and characterization of C. tropicalis 20336 cytochrome P450 monooxygenase (CYP) and cytochrome P450 NADPH oxidoreductase (NCP) genes.
To clone the CYP52 and NCP genes, several different strategies were employed. Available CYP52 amino acid sequences were aligned, and regions of similarity were observed. These regions corresponded to described conserved regions seen in other cytochrome P450 families (5, 7). Proteins from eight eukaryotic cytochrome P450 families share a segmented region of sequence similarity. One region corresponded to the HR2 domain, containing the invariant cysteine residue near the carboxyl terminus which is required for heme binding, while the other region corresponded to the central region of the I helix, thought to be involved in substrate recognition (6).
Degenerate oligonucleotide primers corresponding to these highly conserved regions of the CYP52 gene family present in Candida maltosa and Candida tropicalis ATCC 750 were designed and used to amplify DNA fragments of CYP genes from C. tropicalis ATCC 20336 genomic DNA. These discrete PCR fragments were then used as probes to isolate full-length CYP genes from the C. tropicalis ATCC 20336 genomic libraries. In a few instances oligonucleotide primers corresponding to highly conserved regions were directly used as probes to isolate full-length CYP genes from genomic libraries. In the case of NCP, a heterologous probe based upon the known DNA sequence for the NCP gene from C. tropicalis 750 was used to isolate the C. tropicalis ATCC 20336 NCP genes.
Cloning of the NCP gene from C. tropicalis ATCC 20336.
The first genomic library was screened to isolate a full-length NCP gene, and three putative NCP clones were obtained. The three clones were determined to be truncated for an NCP open reading frame; however, they were shown to overlap, and a complete NCPA sequence was identified. The NCPA is 4,206 nucleotides and includes a regulatory region and a protein coding region which is 2,037 bp in length. NCPA encodes a putative protein of 679 amino acids that shows extensive homology to NCP proteins from C. tropicalis 750 and C. maltosa.
To clone the second NCP allele, a genomic library was screened with DNA fragments from the NCPA truncated clones. Five clones were obtained, and these were sequenced with the three internal primers used to sequence NCPA. Sequence analysis suggested that four of these clones contained inserts which were identical to the NCPA allele isolated earlier. All four contained a full-length NCPA gene. The fifth clone was very similar to the NCPA allele, especially in the open reading frame region, where the identity was very high. However, there were significant differences in the 5′ and 3′ untranslated regions. This suggested that the fifth clone was the allele to NCPA. A 4.14-kb region of this plasmid was sequenced, and the analysis of this sequence confirmed the presence of the NCPB allele. NCPB encodes a putative protein of 679 amino acids.
Cloning of CYP52A13, CYP52A15, CYP52A16, CYP52A17, and CYP52A18.
Clones carrying five CYP52 genes were isolated from a genomic library with an oligonucleotide probe whose sequence was based upon the amino acid sequence for the highly conserved heme binding region (HR2) present throughout the CYP52 family. Based upon DNA sequence analysis, three of the CYP genes appeared unique, while the remaining two were designated alleles. These five genes were designated CYP52A13, CYP52A15, CYP52A16, CYP52A17, and CYP52A18.
Cloning of CYP52A12 and CYP52A19.
CYP52A12 and CYP52A19 were isolated from a genomic library with PCR fragments as probes. The PCR probe for CYP52A12 was generated with oligonucleotide primers designed to amplify a region from the helix I region to the HR2 region. Primers were designed with all available CYP52 gene sequences available from the National Center for Biotechnology Information. Degenerate forward primers were designed based upon an amino acid sequence (RDTTAG) from the helix I region. Degenerate reverse primers were designed based upon an amino acid sequence (GQQFAL) from the HR2 region. A PCR product of approximately 450 bp was obtained. This product was ligated to the pTAG vector (R&D Systems). Plasmids from several transformants were isolated, and their inserts were characterized. One plasmid contained the PCR clone intact. The DNA sequence of the PCR fragment shared homology with the DNA sequences for the CYP52A1 gene of C. maltosa and the CYP52A3 gene of C. tropicalis 750. When the genomic library was screened with the 450-bp PCR product as a probe, a clone that contained the full-length CYP52A12 gene was isolated.
A similar approach was taken to clone CYP52A19. The design of the forward primer was based upon a sequence conserved near the N terminus of the CYP52A3, CYP52A2, CYP52A17, and CYP52A18 genes from C. tropicalis. The reverse primer was designed based on the highly conserved heme-binding region. Amplification of C. tropicalis ATCC 20336 genomic DNA with these two primers gave a mixed PCR product. When this PCR product was used to screen a genomic library, one clone was identified that contained a full-length CYP52 gene along with 5′- and 3′-flanking sequences. This gene was designated CYP52A19.
Cloning of CYP52D2.
Screening the genomic library with an HR2 degenerate probe yielded a clone that contained a truncated CYP52 gene. A 1.3- to 1.5-kb EcoRI-SstI fragment containing part of the truncated CYP52 gene was isolated and used as a probe to screen the genomic library for a full-length CYP52 gene. One clone containing a full-length CYP gene with extensive 5′- and 3′-flanking sequences was isolated and sequenced. This gene was designated CYP52D2.
Cloning of CYP52A14 and CYP52A20.
A mixed probe containing CYP52A12, CYP52A13, CYP52A15, CYP52D2, CYP52A17, and CYP52A19 sequences was used to screen the genomic library, and putative positive clones were identified. Seven clones were sequenced with degenerate primers designed from highly conserved regions of the four CYP52 subfamilies, namely CYP52A, B, C, and D. The complete DNA sequence, including regulatory and protein coding regions, of the 10 clones was determined. Two unique CYP52 genes were identified and were designated CYP52A14 and CYP52A20.
Phylogenetic analysis of Candida CYP52 proteins.
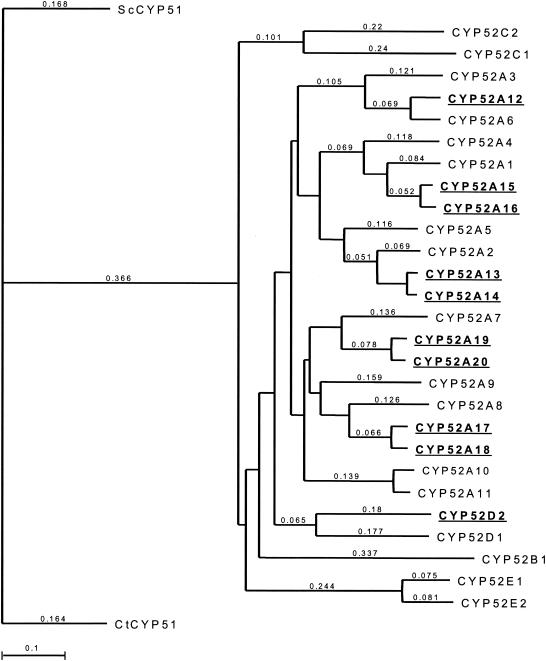
An analysis of the 10 CYP52 deduced amino acid sequences isolated from C. tropicalis ATCC 20336 was conducted with the neighbor-joining method in MacVector 7.1.1 (16). In addition, the deduced amino acid sequences for other currently available CYP52 family sequences from C. maltosa, C. tropicalis ATCC 750, and C. apicola were analyzed for their phylogenetic relationship to the CYP52 sequences of C. tropicalis ATCC 20336. The CYP51 deduced amino acid sequences from Saccharomyces cerevisiae and C. tropicalis were used as a reference for the analysis. The resulting phylogenetic tree is presented in Fig. 1.
FIG. 1.
Phylogenetic tree representing available CYP52 sequences from Candida species. Bold and underlined sequences represent deduced CYP52 proteins from this study. The neighbor-joining method was used to generate the CYP52 relationship (16). Evolutionary distance between members is related to the given values.
The first observation from the analysis indicates that of the 10 CYP52 sequences isolated from C. tropicalis ATCC 20336, four appear to be unique with their corresponding allelic variant. Allelic variants of cytochromes P450 generally have less than 3% dissimilarity at the amino acid level; however, differences in protein specificity and analysis of nontranslated regions can be overriding factors (10). When the CYP52 genes of C. tropicalis ATCC 20336 are compared with their putative allelic variants for amino acid identity and conservative similarity, it is anticipated they exist as allelic variants of the same CYP52 gene (see Table 1). Furthermore, the tree illustrates the relatedness, yet distinct difference, of each C. tropicalis ATCC 20336 CYP52 member with other members of the CYP52 family. As an example, of all the CYP52s previously characterized, CYP52A2 from C. tropicalis ATCC750 is the most closely related to the CYP52 family from C. tropicalis ATCC 20336. Comparison of the amino acid sequence of CYP52A2 to CYP52A13 and CYP52A14 yields 85% identity and 92% conservative similarity for CYP52A13 and 87% identity and 93% conservative similarity for CYP52A14. Based upon these results and the rules for cytochrome P450 nomenclature (9), the CYP52A2 gene from C. tropicalis ATCC 750 would not be considered an allele of either CYP52A13 or CYP52A14. The range of amino acid differences between the CYP52s of C. tropicalis ATCC 20336 and the existing CYP52 family indicates that C. tropicalis ATCC 20336 possesses unique CYP52 proteins different from what is currently known.
TABLE 1.
CYP52 amino acid sequence comparison
| CYP52 amino acid sequence | Putative CYP52 allelic variant | % Amino acid identity between variants | % Amino acid similarity between variants |
|---|---|---|---|
| CYP52A13 | CYP52A14 | 96 | 97 |
| CYP52A15 | CYP52A16 | 95 | 97 |
| CYP52A17 | CYP52A18 | 94 | 97 |
| CYP52A19 | CYP52A20 | 95 | 97 |
Induction patterns of CYP and NCP genes by selected fatty acid and alkane substrates.
Genes whose transcription is turned on by the presence of selected fatty acid or alkane substrates were identified with the QC RT-PCR assay. This assay was used to measure CYP and NCP mRNA synthesis in fermentor-grown cultures of C. tropicalis ATCC 20962 with both fatty acid and alkane feeds. In addition to Emersol 267, a commercial fatty acid feed, pure oleic acid (C18:1) and pure octadecane (C18) substrates were also analyzed. Induced genes were identified by the calculation of their mRNA concentration before and at various times after addition of the target substrate. The log ratio of unknown mRNA product (U) to competitor RNA product (C) is plotted versus the concentration of competitor RNA present in the QC RT-PCRs. The concentration of competitor which results in a log ratio of U/C of zero represents the point where the unknown mRNA concentration is equal to the concentration of the competitor RNA concentration. Primer design allowed different CYP52 genes to be differentially measured with the QC RT-PCR procedure. In the cases where a gene and its allele exist, the primers for detection and competitor synthesis were designed so that both alleles were detected in the resulting analysis.
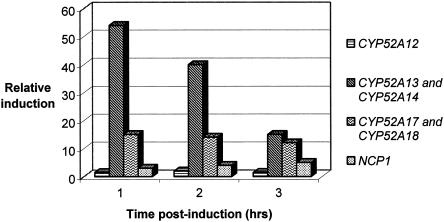
CYP52 and NCP induction in an Emersol 267 fermentation.
The fatty acid composition of Emersol 267 (E267) is representative of a commercial feedstock that could be used in a fermentation process to produce aliphatic dicarboxylic acids. It consists mainly of C18:1 fatty acid (69.9%), C18:2 (8.8%), equal amounts of C16:1 and C17:1 (5.7%), and C16:0 (4.6%).
E267 was used as a substrate in a fermentation and analyzed for CYP52 and NCP mRNA induction. The results are shown in Fig. 2. The gene with the largest increase in transcription in response to this substrate was CYP52A13 and its putative allele, CYP52A14. Within 1 h following substrate addition to the fermentation, mRNA synthesis was induced greater than 50-fold compared to preinduction levels. In contrast, transcription of CYP52A17 and its allelic variant, CYP52A18, was induced approximately 15-fold over the first 3 h following the addition of E267. CYP52A12 was only shown to be induced minimally following E267 addition. The induction of NCPA and NCPB was shown to be three- to fivefold above preinduction levels. No transcriptional induction of CYP52A15 and CYP52A16, CYP52A19 and CYP52A20, and CYP52D2 was observed by QC RT-PCR analysis. If induction occurred, it was below levels detectable by the QC RT-PCR analysis.
FIG. 2.
CYP52 and NCP transcriptional induction in the presence of Emersol 267.
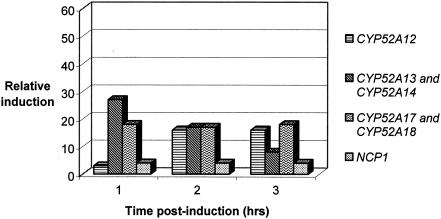
CYP52 and NCP induction in a pure oleic acid fermentation.
Induction of transcription in the presence of pure oleic acid is shown in Fig. 3. As was the case for the E267 fermentation, CYP52A13 and CYP52A14 had the highest levels of mRNA induction following substrate addition. Transcription was induced 27-fold over preinduction levels at 1 h following substrate addition and 17-fold and 8-fold at 2 and 3 h postinduction, respectively. The result for CYP52A17 and CYP52A18 was also similar to the E267 fermentation in that the mRNA levels were approximately 18-fold above preinduction levels, and this pattern continued for the duration of the analysis. In contrast to the E267 fermentation, CYP52A12 mRNA was induced 3-fold at 1 h, 16-fold at 2 h, and 16-fold at 3 h. NCP mRNA was induced 4-fold for each time point over the analysis period. As was in the case of the E267 fermentation, no detectable levels of transcription were observed for CYP52A15 and CYP52A16, CYP52A19 and CYP52A20, and CYP52D2.
FIG. 3.
CYP52 and NCP transcriptional induction in the presence of pure oleic acid.
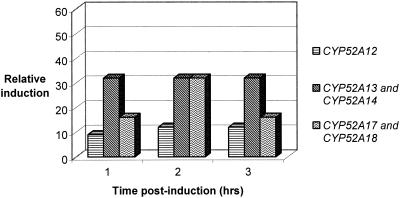
CYP52 and NCP Induction in a pure octadecane fermentation.
In fermentations with pure octadecane, transcriptional induction of CYP52A12, CYP52A13 and CYP52A14, and CYP52A17 and CYP52A18 was observed (Fig. 4). Since no variation in NCP mRNA synthesis was anticipated, no measurement was made. Once again, the highest level of mRNA synthesis was observed for CYP52A13 and CYP52A14, with a 32-fold increase over the first 3 h postinduction, while mRNA synthesis for CYP52A17 and CYP52A18 was also induced at levels ranging from 16-fold (1 h and 3 h postinduction) to 32-fold (2 h). CYP52A12 mRNA was induced 9-fold at 1 h postinduction and increased to 12-fold induction for the next two time points. No detectable level of transcription was observed for CYP52A15 and CYP52A16, CYP52A19 and CYP52A20, and CYP52D2.
FIG. 4.
CYP52 transcriptional induction in the presence of pure octadecane.
DISCUSSION
Biocatalysis offers the chemical industry a green approach to produce both existing and novel specialty chemicals, but historically, high processing costs have limited commercialization. One approach to assist in overcoming these limits is to improve biocatalyst productivity.
In order to generate an improved Candida tropicalis strain for the production of long-chain dicarboxylic acids by fermentation, it was first necessary to isolate and identify the genes responsible for the oxidation of fatty acid and alkane feed stocks. This paper discusses the characterization of CYP52 genes involved in the oxidation of oleic acid and octadecane.
Candida tropicalis ATCC 20336 was chosen as the basis for development of an industrial production strain because of its ability to oxidize long-chain (>C12) fatty acids and alkanes (12, 14). Based on previous studies in C. tropicalis (17, 21) and C. maltosa (19), it was predicted that the CYP52 family would contain numerous members. This was confirmed in this study by the isolation of 10 CYP52 genes. Moreover, since C. tropicalis is diploid, it was anticipated that each isolated gene would have a corresponding allelic variant; however, this turned out not to be the case. While we had success in isolating four CYP52 genes and their corresponding alleles, there were two distinct genes, CYP52A12 and CYP52D2, for which no allelic variant was isolated despite numerous attempts. Because of the highly recombinant nature of Candida species and selective pressure, it is possible that the alleles of these CYP52 genes have been lost during evolution. It is interesting that one of these two genes, CYP52A12, was shown to be induced by fatty acid and alkane substrates while the other, CYP52D2, was not.
If one compares the amino acid sequences deduced for the 10 CYP52 genes to other members of known Candida CYP52 families, it is clear that the genes in this study are most closely related to those of other C. tropicalis species (see Fig. 1). However, within the C. tropicalis ATCC 20336 family itself there exist putative allelic variants. Allelic variants of cytochromes P450 generally have less than 3% dissimilarity at the amino acid level; however, differences in protein specificity and analysis of nontranslated regions can be overriding factors (9). Based on the deduced amino acid sequence information, CYP52A13 and CYP52A14, CYP52A15 and CYP52A16, CYP52A17 and CYP52A18, and CYP52A19 and CYP52A20 exist as putative allelic variants. It will be necessary to determine the protein specificity of these P450s in order to determine their true allelic nature.
While clearly related to CYP52 members from the C. maltosa family, only in one instance was a gene from C. tropicalis ATCC 20336 (CYP52D2) most closely related to a C. maltosa CYP52 (CYP52D1). No ortholog to this gene has been identified in other C. tropicalis strains.
One approach to identify genes important for fatty acid and alkane oxidation is to monitor transcriptional induction for selected substrates. This method involves the isolation of total cellular RNA from cultures of C. tropicalis and the quantification of a specific mRNA within that sample through the design and use of sequence-specific QC RT-PCR primers and an RNA competitor. Quantification is achieved through the use of known concentrations of highly homologous competitor RNA in the QC RT-PCRs. By the addition of various amounts of a competitor RNA molecule to the sample reaction, one can determine the concentration of the RNA molecule of interest (in this case the mRNA transcripts of the CYP and NCP genes). The resulting QC RT-PCR amplified cDNAs are separated and quantitated through the use of ion pairing reverse phase HPLC or agarose gel electrophoresis. The levels of specific mRNA transcripts were assayed over time in response to the addition of fatty acid and/or alkane substrates to the growth medium of fermentation grown C. tropicalis cultures for the identification and characterization of the genes involved in the oxidation of these substrates. This approach has identified the CYP52 and NCP genes involved in the oxidation of various substrates based upon their transcriptional regulation; however, the corresponding CYP52 enzyme specificity associated with the different substrates remains to be characterized.
Fermentations were conducted with three different substrates to monitor transcriptional induction of the CYP52 genes from C. tropicalis ATCC 20336 with QC RT-PCR. A mixed commercial feedstream, E267, pure oleic acid, and pure octadecane were used in three separate experiments. The E267 fermentation was performed twice with identical fermentation conditions, and the maximum observed variance of induction values for any analyzed gene between the two fermentations was 14.6%. Because of the prohibitive cost of with pure substrates (oleic acid and octadecane) in the quantities necessary to achieve a commercially comparable substrate concentration, only one fermentation with each pure substrate was conducted and analyzed by QC RT-PCR. The results of the QC RT-PCR experiments provide a qualitative assessment of the CYP52 genes that are induced and a quantitative indication of the trend and level of CYP52 induction in response to the substrates.
For all three substrates, no induction of CYP52A15 and CYP52A16 or of CYP52D2 was ever observed. For E267, minimal induction of CYP52A19 and CYP52A20 was observed (data not shown); however, the level of induction was only slightly above preinduction levels.
The mRNAs detected from the three different fermentations were consistent in the pattern of induction but not in the magnitude. Of the remaining CYP52 genes, CYP52A13 and the putative allele CYP52A14, showed the highest overall level of induction. For the pure oleic acid and octadecane substrates, 27- and 32-fold increases, respectively, were observed at 1 h postinduction. Slightly higher induction levels were observed for E267 with a 54-fold increase at 1 h postinduction. This result may indicate that CYP52A13 and CYP52A14 are responsive to a broader range of fatty acid substrates found in E267 but absent in pure oleic acid. Following the initial induction peak at 1 h, levels of transcriptional induction for both E267 and pure oleic acid fermentations began to decrease as the result of a possible regulatory response, but mRNA could still be detected at 3 h postinduction. Continued transcription could simply reflect an increased need for CYP52 involvement in the ω-oxidation pathway. A similar induction pattern was observed with CYP52A12 with pure octadecane as a substrate.
CYP52A17 and CYP52A18 were found to be induced 16- and 18-fold for the pure oleic and octadecane fermentations, respectively, at 1 h postinduction. Similar magnitudes of induction were seen at subsequent time points following induction. In contrast to the increased induction of CYP52A13 and CYP52A14 for the E267 feedstock, no increase in induction of CYP52A17 and CYP52A18 was observed. Based solely on these induction results, one might infer that CYP52A17 and CYP52A18 have more specificity for C18 based substrates. However, enzymatic analyses of heterologously expressed CYP52A17 indicate it possesses broad specificity (2a). The induction level of CYP52A17 and CYP52A18 was maintained for all sample points over the three fermentations, while induction of CYP52A13 and CYP52A14 peaked quickly (first 1 to 2 h) and then decreased in response to some type of regulatory control. The consistency of the induction of CYP52A17 and CYP52A18 may reflect their importance in diacid production.
No significant induction of CYP52A12 was observed for the E267 fermentation, or at 1 h after induction with pure oleic acid. However, CYP52A12 was induced to maximum levels of 16-fold and 12-fold at 2 h postinduction for pure oleic acid and octadecane, respectively. Once again, the pattern of induction was such that no reduction in the level of transcription was observed.
The results of this study are in contrast to previous work characterizing CYP52 transcription in Candida species (20). In C. tropicalis ATCC750, CYP52A1 was shown to be induced at the highest level when grown on oleic acid as a sole carbon source compared to CYP52A2, CYP52A6, and CYP52A8 (20). Amino acid sequence analysis indicates that CYP52A1 is most closely related to CYP52A15 (this study) and CYP52A16 (this study), which exhibited no transcriptional induction in the presence of the oleic acid and octadecane feed stocks. Similarly, CYP52A6 from C. tropicalis ATCC750 (most closely related to CYP52A12, see Fig. 1) and CYP52A8 from C. tropicalis ATCC750 (most closely related to CYP52A17 and CYP52A18, this study; see Fig. 1), showed very minimal induction in response to oleic acid. However, CYP52A12, CYP52A17, and CYP52A18 (this study) showed modest levels of transcriptional induction in response to oleic acid and octadecane. Only in the case of CYP52A2 from C. tropicalis ATCC750 (most closely related to CYP52A13 and CYP52A14; this study, see Fig. 1) were similar induction responses observed.
Blocking β-oxidation in C. tropicalis ATCC20336 produced an increased yield in diacid production, but the increase was not sufficient to meet the needs of commercialization. Previous attempts to improve the diacid productivity of C. tropicalis ATCC 20336 by overexpressing the CYP52A1 gene from C. tropicalis ATCC 750 have been attempted without success (14). This result is not surprising, as our transcriptional analysis shows that CYP52A1 would not be the gene of choice. The transcriptional response of CYP52A1 would be expected to be similar to that of CYP52A15 and CYP52A16 (this study); however, neither of these two genes exhibited any detectable transcriptional response to oleic acid or octadecane. These results support the uniqueness of certain CYP52s from C. tropicalis ATCC 20336 and their use as targets for biocatalyst improvement and emphasizes the difficulty of comparing CYP52 genes between Candida strains.
Transcription of NADPH cytochrome P450 reductase, NCP, was also measured during fermentation with fatty acid substrates. In all cases, the levels of transcriptional induction of the NCPA and NCPB genes were shown to be maximally induced between 3- and 5-fold within 1 h postinduction. This level of induction was maintained over the measurement period following substrate addition. In all fermentations analyzed to date, this pattern of induction and magnitude were consistent with these results; therefore, no attempts were made to measure NCP induction in the pure octadecane fermentation.
If relative induction values could be directly compared between CYP52 and NCP, it is possible that NCP may be a rate-limiting factor in the conversion of oleic acid and octadecane to their corresponding dicarboxylic acid. One study in an in vitro system suggests an optimal molar ratio of 3:1 for NCP to CYP52 (18); however, it is not clear that these values translate into comparable in vivo ratios. Induction at the mRNA level does not indicate an amount or stability of actual protein for the corresponding CYP52 or NCP gene. It is possible that a gene induced at lower levels may encode a more stable and active enzyme, therefore, be more important as a biocatalyst than an enzyme from a CYP52 or NCP gene that is induced to higher levels but is less stable or that has a lower specific activity. It will be necessary to perform in vitro protein assays for specific activity and protein turnover numbers to fully address this issue.
This study has presented the isolation of a large family of alkane-inducible cytochrome P450 genes (CYP52) and their corresponding NADPH cytochrome P450 reductase (NCP) from Candida tropicalis ATCC 20336. Of these 10 DNA sequences, four genes were isolated with a corresponding allele, CYP52A13 and CYP52A14, CYP52A15 and CYP52A16, CYP52A17 and CYP52A18, and CYP52A19 and CYP52A20. Two of the 10 sequences, CYP52A12 and CYP52D2, were never found to have a corresponding allele following exhaustive attempts to isolate them. Moreover, this study has further demonstrated the ability to differentiate induction of individual CYP52 genes from a large family of highly similar genes in response to a given substrate. In addition, this technique has elucidated the variation in transcriptional induction for various substrates that can exist among a closely related yet dissimilar family of CYP52 proteins.
In order to begin to genetically construct a better C. tropicalis biocatalyst for the production of long-chain dicarboxylic acids, it was necessary to identify what CYP52 genes may be involved in substrate conversion. From this study, it is concluded that CYP52A13 and CYP52A14, CYP52A17 and CYP52A18, and CYP52A12 along with the corresponding reductase, NCP, are candidates for manipulation in C. tropicalis ATCC 20336 for creating a better biocatalyst for producing long-chain dicarboxylic acids from fatty acid and alkane feedstreams.
Acknowledgments
We thank Robert Frayer and Cathy Cornett for their efforts in strain construction and gene isolation.
This work was supported by U.S. Department of Energy (project no. DE-FC36-95GO10099) and the National Institute of Standards and Technology Advanced Technology Program (70NANB8H4033).
REFERENCES
- 1.Anderson, K. W. May 2003. Fermentation process. Cognis Corporation. U.S. Patent 6,569,670.
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., Hoboken, N.J.
- 2a.Eschenfeldt, W. H., Y. Zhang, H. Samaha, L. Stohls, L. D. Eirich, C. R. Wilson, and M. I. Donnelly. 2003. Transformation of fatty acids catalyzed by cytochrome P450 monooxygenase enzymes of Candida tropicalis. 69:5792-5799. [DOI] [PMC free article] [PubMed]
- 3.Gilewicz, M., M. Zacek, J. C. Bertrand, and E. Azoulay. 1979. Hydroxylase regulation in Candida tropicalis grown on alkanes. Can. J. Microbiol. 25:201-206. [DOI] [PubMed] [Google Scholar]
- 4.Gmunder, K., O. Kappeli, and A. Fiechter. 1981. Chemostat studies on the hexadecane assimilation by the yeast Candida tropicalis: regulation of cytochromes and enzymes. Eur. J. Appl. Microbiol. Bio/Technol. 12:135-142. [Google Scholar]
- 5.Goeptar, A. R., H. Scheerens, and N. P. Vermeulen. 1995. Oxygen and xenobiotic reductase activities of cytochrome P450. Crit. Rev. Toxicol. 25:25-65. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh, O., Y. Tagashira, T. Iizuka, and Y. Fujii-Kuriyama. 1983. Structural characteristics of cytochrome P-450. Possible location of the heme-binding cysteine in determined amino-acid sequences. J. Biochem. (Tokyo) 93:807-817. [DOI] [PubMed] [Google Scholar]
- 7.Kalb, V. F., and J. C. Loper. 1988. Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc. Natl. Acad. Sci. 85:7221-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mauersberger, S., W. H. Schunck, and H. H. Muller. 1981. The induction of Lodderomyces elongisporus. Z. Allg. Mikrobiol. 21:313-321. [DOI] [PubMed] [Google Scholar]
- 9.Nebert, D. W., D. R. Nelson, M. J. Coon, R. W. Estabrook, R. Feyereisen, Y. Fujii-Kuriyama, F. J. Gonzalez, F. P. Guengerich, I. C. Gunsalus, and E. F. Johnson. 1991. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 10:1-14. (Erratum, 10:397-398.) [DOI] [PubMed] [Google Scholar]
- 10.Nelson, D. R., L. Koymans, T. Kamataki, J. J. Stegeman, R. Feyereisen, D. J. Waxman, M. R. Waterman, O. Gotoh, M. J. Coon, R. W. Estabrook, I. C. Gunsalus, and D. W. Nebert. 1996. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6:1-42. [DOI] [PubMed] [Google Scholar]
- 11.Ohkuma, M., S. Muraoka, T. Tanimoto, M. Fujii, A. Ohta, and M. Takagi. 1995. CYP52 (cytochrome P450alk) multigene family in Candida maltosa: identification and characterization of eight members. DNA Cell Biol. 14:163-173. [DOI] [PubMed] [Google Scholar]
- 12.Okino, N. 1988. Production of macrocyclic musk compounds via alkanedioic acids produced from n-alkanes, p. 753-760. In B. M. Lawrence, B. D. Mookherjee, and B. J. Willis (ed.), Flavors and fragrances: a world perspective. Proceedings of the 10th International Conference of Essential Oils, Flavors and Fragrances. Elsevier Science Publishers BV, Amsterdam, The Netherlands.
- 13.Picataggio, S., K. Deanda, and J. Mielenz. 1991. Determination of Candida tropicalis acyl coenzyme A oxidase isozyme function by sequential gene disruption. Mol. Cell. Biol. 11:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picataggio, S., T. Rohrer, K. Deanda, D. Lanning, R. Reynolds, J. Mielenz, and L. D. Eirich. 1992. Metabolic engineering of Candida tropicalis for the production of long-chain dicarboxylic acids. Bio/Technology (N.Y.) 10:894-898. [DOI] [PubMed] [Google Scholar]
- 15.Raeymaekers L. 1999. General principles of quantitative PCR, p. 31-41. In B. Kochanowski and U. Reischl (ed.), Quantitative PCR protocols. Humana Press, Totowa, N.J.
- 16.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 17.Sanglard, D., C. Chen, and J. C. Loper. 1987. Isolation of the alkane inducible cytochrome P450 (P450alk) gene from the yeast Candida tropicalis. Biochem. Biophys. Res. Commun. 144:251-257. [DOI] [PubMed] [Google Scholar]
- 18.Scheller, U., T. Zimmer, D. Becher, F. Schauer, and W. H. Schunck. 1998. Oxygenation cascade in conversion of n-alkanes to alpha, omega-dioic acids catalyzed by cytochrome P450 52A3. J. Biol. Chem. 273:32528-32534. [DOI] [PubMed] [Google Scholar]
- 19.Schunck, W. H., E. Kargel, B. Gross, B. Wiedmann, S. Mauersberger, K. Kopke, U. Kiessling, M. Strauss, M. Gaestel, and H. G. Muller. 1989. Molecular cloning and characterization of the primary structure of the alkane hydroxylating cytochrome P-450 from the yeast Candida maltosa. Biochem. Biophys. Res. Commun. 161:843-850. [DOI] [PubMed] [Google Scholar]
- 20.Seghezzi, W., C. Meili, R. Ruffiner, R. Kuenzi, D. Sanglard, and A. Fiechter. 1992. Identification and characterization of additional members of the cytochrome P450 multigene family CYP52 of Candida tropicalis. DNA Cell Biol. 11:767-780. [DOI] [PubMed] [Google Scholar]
- 21.Seghezzi, W., D. Sanglard, and A. Fiechter. 1991. Characterization of a second alkane-inducible cytochrome P450-encoding gene, CYP52A2, from Candida tropicalis. Gene 106:51-60. [DOI] [PubMed] [Google Scholar]
- 22.Shiio, I. 1971. Microbial production of long-Chain dicarboxylic acids from n-alkanes. Agric. Biol. Chem. 35:2033-2042. [Google Scholar]
- 23.Sutter, T. R., D. Sanglard, J. C. Loper, and D. Sangard. 1990. Isolation and characterization of the alkane-inducible NADPH-cytochrome P-450 oxidoreductase gene from Candida tropicalis. Identification of invariant residues within similar amino acid sequences of divergent flavoproteins. J. Biol. Chem. 265:16428-16436. [PubMed] [Google Scholar]