Abstract
This study examined the natural diversity and distributions of sulfate-reducing bacteria along a natural carbon gradient extending down the shelf-slope transition zone of the eastern Pacific continental margin. Dissimilatory (bi)sulfite reductase gene sequences (dsrAB) were PCR amplified and cloned from five different sampling sites, each at a discrete depth, from two different margin systems, one off the Pacific coast of Mexico and another off the coast of Washington State. A total of 1,762 clones were recovered and evaluated by restriction fragment length polymorphism (RFLP) analysis. The majority of the gene sequences recovered showed site and depth restricted distributions; however, a limited number of gene sequences were widely distributed within and between the margin systems. Cluster analysis identified 175 unique RFLP patterns, and nucleotide sequences were determined for corresponding clones. Several different continental margin DsrA sequences clustered with those from formally characterized taxa belonging to the delta subdivision of the class Proteobacteria (Desulfobulbus propionicus, Desulfosarcina variabilis) and the Bacillus-Clostridium (Desulfotomaculum putei) divisions, although the majority of the recovered sequences were phylogenetically divergent relative to all of the other DsrA sequences available for comparison. This study revealed extensive new genetic diversity among sulfate-reducing bacteria in continental margin sedimentary habitats, which appears to be tightly coupled to slope depth, specifically carbon bioavailability.
Molecular investigations of microbial communities have brought to light extensive and pervasive microbial diversity throughout nature; however, the interacting biological, chemical, and physical forces that control patterns of diversity are not well defined or understood. Carbon bioavailability is, without question, one important factor impacting microbial community structure and function but is often difficult to measure on meaningful scales or manipulate predictably in natural settings. As a result, carbon manipulation studies are often conducted as laboratory enrichments (2, 14, 15, 16, 34) or large-scale field fertilizations (1, 10, 22). These experimental approaches, however, can oversimplify environmental conditions or produce a multitude of treatment effects that complicate interpretation. Shelf-slope transects of the continental margin sediments constitute a natural gradient in carbon quality and quantity, providing an opportunity to address specific questions about the impacts of carbon on microbial community dynamics as they occur under natural environmental conditions.
The continental margins occupy a relatively small fraction of the ocean floor, but they are among the most productive ecosystems known (53). Coastal outwelling, seasonal upwelling, and high rates of planktonic photosynthesis provide for localized carbon enrichment of the margins. However, highly reactive photosynthates or cell lysates are quickly oxidized as fixed carbon descends through the water column (25, 45, 51), progressively enriching the remaining sinking material in more metabolically resistant biomolecules (26). It is estimated that only 10% of the total primary production reaches depths exceeding 100 m and less than 1% reaches a depth of 3,000 m (18, 51). Consequently, recalcitrant carbon compounds are expected to be important substrates for metabolism at greater depths. Changes in carbon supplies should have obvious impacts on microbial growth and metabolism but may also result in discrete shifts in community composition and functionality with increasing depth. The magnitude of this relationship, however, has not been investigated.
Carbon cycling in continental margin sediments is coupled to the reduction of a variety of different electron acceptors, including oxygen, nitrate, sulfate, manganese, and iron (5, 24, 48). Denitrifying bacteria, capable of respiring both O2 and NO3−, play a major role in carbon and nitrogen cycling in this system (9, 11, 12, 18, 20), and microbial investigations conducted thus far have focused almost exclusively on this functional guild (3, 43, 44). In these studies, phylogenetically divergent functional gene sequences from denitrifiers (nirS, nirK, nosZ) were recovered from pure culture isolates and environmental clone libraries. Scala and Kerkhof (44) demonstrated congruency between genetic variability in nosZ and spatial scale, implying that while total denitrifier diversity is high, denitrifying species are apparently restricted by their physiology or ecology to a defined habitat or environmental condition. Those authors attributed these findings to a variety of plausible factors, including perturbation by benthic worms, seasonal fluctuations, and organic carbon. To date, patterns of microbial functional diversity have not been systematically evaluated in the shelf-slope environment. Therefore, it is uncertain whether the findings of Scala and Kerkhof (44) pertain only to denitrifying bacteria in a single margin ecosystem or if they can be generalized to other different metabolic groups and margin systems. Furthermore, the role of carbon in determining patterns of functional diversity remains unclear.
Sulfate reduction is a dominant anaerobic carbon oxidation pathway along the margins (5, 20, 24, 29, 39), accounting for the oxidation of ≥50% of the total organic carbon in some systems (6, 24, 48). As a dominant terminal electron-accepting process, sulfate-reducing bacteria should be particularly sensitive to organic carbon dynamics in the continental margins. In this study, the impact of shelf slope depth and presumptive carbon diagenesis on microbial community structure-function was evaluated by assessing the patterns of genetic diversity of continental margin sulfate-reducing assemblages. Dissimilatory (bi)sulfite reductase (dsrAB) gene sequences from specific depths along two geographically distinct, well-characterized continental margin systems were PCR amplified and sequenced. Extensive new genetic diversity was recovered at all depths and sites, and comparative sequence analysis suggests a predominance of novel and/or deeply branching sulfate reducers that have not been described to date.
MATERIALS AND METHODS
Sampling sites and geochemical parameters.
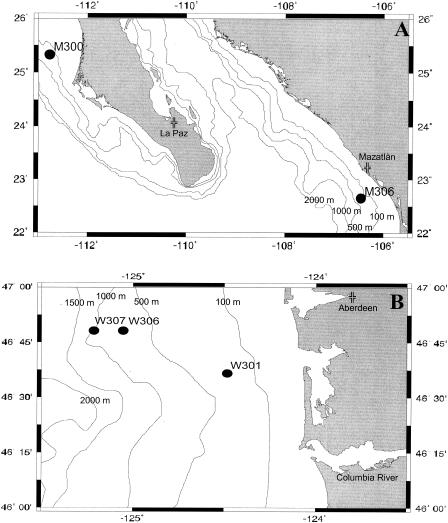
Continental margin sediments were collected from five different sampling stations: two off the Pacific coast of Mexico (sampling stations M300 and M306) and three off the coast of Washington State (W301, W306, and W307). Topographic maps of both margin systems identifying all of the sampling sites are provided in Fig. 1. The Mexican margin is distinguished by a strong oxygen-deficient zone (ODZ), while the more productive Washington margin exhibits profiles that have only the typical North Pacific oxygen minimum, i.e., low concentrations (∼25 μM) of oxygen but not undetectable oxygen. A comprehensive biogeochemical evaluation of these continental margin systems was provided by Hartnett and Devol (24). Briefly, sediment cores, with overlying water, were collected with a Soutar box core and stored at in situ temperatures (approximately 5°C). Weight percent organic carbon was determined on freeze-dried, ground sediment samples by the method of Hedges and Stern (27) with either a Carlo Erba 1106 CHN elemental analyzer or a Leeman Laboratories CHNS elemental analyzer. Standard methods were used to measure water column oxygen concentrations (7). Sulfate reduction rates were integrated over 30-cm sediment cores by measuring reduction of 35SO42− by previously described methods (9, 19). Upon shipment to Oak Ridge National Laboratory, sediment cores were stored at −70°C until used. Measured geochemical parameters were statistically analyzed by principal-components analysis and Pearson correlation coefficients in Systat 10 Statistics 1 (SPSS Inc., Chicago, Ill.).
FIG. 1.
Sampling locations on the continental margins of northwestern Mexico (A) and Washington State (B). The map was created by online map creator (http://www.aquarius.geomar.de/omc/).
Genomic DNA extraction and purification.
Total genomic DNA was extracted from 2 g of sediment from each site by previously described methods (57). High-molecular-weight genomic DNA was further purified by agarose gel electrophoresis (0.8% low-melting-point agarose) and the Wizard DNA Clean-Up Kit (Promega, Madison, Wis.). DNA quantity was determined spectrophotometrically. All nucleic acids were stored at −70°C until used.
RFLP analysis of dsrAB clone libraries.
The dissimilatory (bi)sulfite reductase (dsr) primers used in this study were those of Karkhoff-Schweizer et al. (32). To minimize PCR artifacts, PCR amplification conditions were optimized on the basis of previous suggestions (41). PCR mixtures (20-μl total volume) consisted of 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100, pH 9.0), 1 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 1 μM each primer, 4 μg of bovine serum albumin, and 2.5 U of Taq DNA polymerase. The thermal cycling protocol used included initial denaturation at 94°C for 2 min, followed by 25 cycles of 94°C for 30 s, 58°C for 1 min, and 72°C for 1 min. A final extension step of 72°C for 7 min was also used. Amplimers of the expected size (approximately 1.4 kbp) were excised from 0.8% low-melting-point agarose gels and purified with the Wizard PCR Clean-Up system (Promega) in accordance with the manufacturer's instructions. Amplimers were cloned into the pCR2.1 vector with the TA cloning kit (Invitrogen, Carlsbad, Calif.). The cloned inserts were then amplified for restriction fragment length polymorphism (RFLP) analysis with primers specific to the polylinker of the vector pCRII as described previously (56). The unique dsrAB clone diversity was detected by digestion with the restriction enzymes MspI and RsaI (56). Jaccard coefficients were calculated for all pairwise comparisons of RFLP banding patterns and dendrograms constructed with the unweighted pair group mean average method in Molecular Analyst (version 1.1; Bio-Rad, Hercules, Calif.). Cohesive groupings of highly similar, although not necessarily identical, RFLP banding patterns were identified, and a representative clone was selected for nucleotide sequence determination.
dsrAB gene sequencing and sequence analysis.
To understand phylogenetic diversity, representative dsrAB clones that occurred more than once in a given library, as well as representatives of some of the unique clones as determined by cluster analysis of RFLP banding patterns, were fully sequenced. A cloned insert was PCR amplified as described above and purified with the ArrayIt PCR Purification Kit (TeleChem International Inc., Sunnyvale, Calif.). DNA sequencing was performed with an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster, Calif.) and an ABI PRISM 3700 DNA analyzer (Applied Biosystems). One microliter (about 30 ng) of purified DNA was used for each sequencing reaction. The vector-specific primers TAF (5′-GCCGCCAGTGTGCTGGAATT-3′) and TAR (5′-TAGATGCATGCTCGAGCGGC-3′) were then used for sequencing (41). DNA sequences were assembled and edited with the Sequencher program, version 4.0 (Gene Codes Corporation, Ann Arbor, Mich.). Nucleotide sequences were aligned with relevant GenBank sequences in CLUSTAL W (49) and translated in GCG (Wisconsin Package, version 10; Genetics Computer Group, Madison, Wis.) (17). Deduced amino acid sequences were realigned with CLUSTAL W. Unambiguously aligned sequence regions were used to construct bootstrap-supported (500 resamplings) neighbor-joining phylogenies in MEGA (version 2.1; http://megasoftware.net) from Poisson correction distances to account for multiple substitution events.
Nucleotide sequence accession numbers.
Continental margin cloned sequences are labeled by margin (Mexico = M, Washington = W), site number (M = 300 or 306, and W = 301, 306, or 307), and an arbitrary clone number. All of the dsrAB sequences described in this study have been submitted to GenBank under accession numbers AY337046 to AY337232 as follows: W-301-018, AY337046; W-301-165, AY337047; W-307-046, AY337048; W-307-294, AY337049; M-306-254, AY337050; M-306-256, AY337051; M-300-080, AY337052; M-300-083, AY337053; M-306-285, AY337054; W-301-186, AY337055; W-301-032, AY337056; W-301-233, AY337057; W-301-083, AY337058; W-301-209, AY337059; M-300-115, AY337060; M-300-365, AY337061; W-306-461, AY337062; W-301-238, AY337063; M-300-284, AY337064; W-301-036, AY337065; W-306-517, AY337066; M-300-255, AY337067; M-306-211, AY337068; W-301-262, AY337069; M-306-096, AY337070; M-306-103, AY337071; W-306-487, AY337072; W-301-328, AY337073; M-300-244, AY337074; M-306-024, AY337075; M-306-189, AY337076; M-306-192, AY337077; M-300-167, AY337078; M-300-381, AY337079; M-306-085, AY337080; W-306-449, AY337081; M-306-264, AY337082; W-301-181, AY337083; W-307-245, AY337084; W-306-417, AY337085; W-307-284, AY337086; W-307-290, AY337087; W-306-390, AY337088; W-307-175, AY337089; W-307-077, AY337090; M-300-238, AY337091; W-306-791, AY337092; W-307-231, AY337093; W-301-185, AY337094; W-301-315, AY337095; W-306-401, AY337096; M-306-011, AY337097; W-307-323, AY337098; W-301-008, AY337099; W-301-241, AY337100; W-306-824, AY337101; W-307-106, AY337102; W-307-140, AY337103; W-306-683, AY337104; W-306-739, AY337105; W-307-263, AY337106; W-306-646, AY337107; W-306-731, AY337108; W-301-200, AY337109; W-301-015, AY337110; W-307-344, AY337111; M-306-080, AY337112; W-307-196, AY337113; M-300-263, AY337114; M-306-130, AY337115; W-306-741, AY337116; W-301-134, AY337117; W-307-224, AY337118; W-306-552, AY337119; W-301-207, AY337120; W-306-411, AY337121; W-307-082, AY337122; W-306-570, AY337123; W-307-299, AY337124; W-306-852, AY337125; W-306-647, AY337126; W-306-677, AY337127; W-301-261, AY337128; M-306-199, AY337129; M-300-174, AY337130; M-300-275, AY337131; M-300-270, AY337132; M-306-118, AY337133; M-306-026, AY337134; M-306-194, AY337135; M-300-209, AY337136; M-306-268, AY337137; M-300-087, AY337138; M-306-202, AY337139; M-306-203, AY337140; M-306-157, AY337141; M-300-002, AY337142; M-300-308, AY337143; W-301-122, AY337144; W-301-131, AY337145; M-300-125, AY337146; W-306-636, AY337147; M-306-059, AY337148; W-306-769, AY337149; W-306-776, AY337150; M-306-072, AY337151; W-306-512, AY337152; M-306-040, AY337153; M-300-247, AY337154; M-306-041, AY337155; M-306-044, AY337156; W-301-119, AY337157; W-306-700, AY337158; M-300-206, AY337159; W-306-439, AY337160; W-306-563, AY337161; W-307-298, AY337162; W-306-735, AY337163; M-300-089, AY337164; M-300-141, AY337165; M-306-196, AY337166; W-301-087, AY337167; W-301-184, AY337168; W-301-112, AY337169; W-306-762, AY337170; W-306-441, AY337171; M-300-222, AY337172; M-300-225, AY337173; W-306-388, AY337174; W-301-377, AY337175; M-300-004, AY337176; M-300-099, AY337177; M-300-288, AY337178; M-300-374, AY337179; M-300-128, AY337180; M-300-135, AY337181; M-300-012, AY337182; M-300-015, AY337183; M-300-226, AY337184; M-300-373, AY337185; M-300-372, AY337186; M-300-048, AY337187; M-306-064, AY337188; M-300-119, AY337189; M-300-304, AY337190; M-300-307, AY337191; M-300-329, AY337192; M-306-048, AY337193; M-300-227, AY337194; W-307-149, AY337195; W-307-308, AY337196; M-300-249, AY337197; W-301-369, AY337198; M-300-293, AY337199; W-301-107, AY337200; W-301-182, AY337201; W-301-157, AY337202; W-307-341, AY337203; M-300-278, AY337204; W-306-423, AY337205; M-300-094, AY337206; M-300-028, AY337207; M-300-353, AY337208; M-300-214, AY337209; W-307-166, AY337210; W-307-173, AY337211; W-301-179, AY337212; W-301-300, AY337213; W-306-505, AY337214; M-300-100, AY337215; M-300-352, AY337216; M-300-150, AY337217; M-300-232, AY337218; W-307-053, AY337219; W-307-154, AY337220; W-307-020, AY337221; W-307-095, AY337222; W-307-206, AY337223; W-306-734, AY337224; W-307-295, AY337225; M-300-079, AY337226; M-300-356, AY337227; W-307-063, AY337228; W-307-127, AY337229; W-307-073, AY337230; W-307-165, AY337231; W-307-044, AY337232.
RESULTS
Site geochemistry.
The primary difference between the Washington and Mexican continental margins is the impingement of the major ODZ on the latter. The ODZ is a major oceanographic feature of the eastern tropical Pacific Ocean in which dissolved oxygen in the water column is undetectable between about 180 and 600 m. Although there is a low-oxygen zone off the coast of Washington State, dissolved oxygen is always detectable (24). Also associated with ODZs are high sedimentary carbon contents and a major contribution of sulfate reduction to the overall metabolism of the sediments. These conditions are clearly evident in our study sites (Table 1). Both Mexican margin sites had unmeasurable oxygen concentrations in their overlying waters. In addition, associated with all major marine ODZs are elevated carbon contents, and those off the coast of Mexico are no exception. Although the absolute sulfate reduction rates off the coast of Mexico are only about 1.2 mmol/m2/day, they account for about 65% of the total organic carbon oxidation. In contrast, at the shallow station off the coast of Washington, the absolute rate of sulfate reduction is higher than that off the coast of Mexico, but the relative contribution to overall sediment metabolism is significantly less (see reference 24 for a detailed discussion). The high sulfate reduction rate on the Washington shelf (W301) is likely due to the fact that the rain of carbon to the sediments is both greater in absolute amount and higher in quality here than is that to the deeper slope stations (W306 and W307).
TABLE 1.
Mexico margin and Washington margin sampling site characteristics
| Sampling site (location)a | Depth (m) | Carbon (%) | O2 (μmol/liter) | SO4 reduction rate (mmol/m2/day) | nb | No. of unique RFLP patterns (%)c |
|---|---|---|---|---|---|---|
| M300 (25°19.49′ 112°45.82′) | 387 | 9.0 | 0 | 1.19 | 368 | 263 (72) |
| M306 (22°39.60′ 106°27.10′) | 340 | 7.9 | 0 | 1.19 | 278 | 202 (73) |
| W301 (46°36.74′ 124°28.94′) | 119 | 1.5 | 151 | 5.96 | 350 | 180 (51) |
| W306 (46°48.28′ 125°03.23′) | 630 | 2.5 | 27 | 0.66 | 409 | 268 (66) |
| W307 (46°48.10′ 125°12.73′) | 997 | 3.0 | 38 | 0.04 | 357 | 169 (47) |
M, Mexico margin; W, Washington margin. Sampling station latitude and longitude are in parentheses.
The total number of clones per site evaluated by RFLP = n.
The total number of unique RFLP patterns per site-specific clonal library is shown and the percentage of the total is in parentheses.
The geochemical parameters (Table 1) were analyzed by principal-components analysis to assess geochemical variability between sites and to identify the geochemical parameters driving this variability. Overall, all of the sites had equivalent component loading factors for PC1 (0.99), except site W301 (0.54). All of the sites combined could explain 85% of the geochemical variability measured in this study. The source of variability between sites was determined by analysis of the individual geochemical parameters measured. Site depth (0.63) and organic carbon content (0.59) received positive loading factors, while oxygen (−0.96) and sulfate reduction rate (−0.97) received strong negative loading factors for their contributions to overall geochemical variability. Carbon and depth best explained the geochemical variance among sites (65.4%), implying that sedimentary carbon content, which is dictated by depth, is a key difference among sites.
Pearson correlation matrices were constructed to evaluate covariance between geochemical variables. When the two sets of margin data were combined, a strong positive correlation (0.89) was obtained between oxygen concentrations and sulfate reduction rates. However, this relationship was solely driven by high values for both parameters at site W301. Inverse relationships were observed between sulfate reduction rate and depth (−0.77) and between oxygen concentration and organic carbon content (−0.73). When the data were analyzed for each margin system independently, there was a strong positive correlation between site depth and organic carbon content (>0.99).
RFLP analysis of dsrAB clone libraries.
The molecular diversity of dissimilatory (bi)sulfite reductase genes was assessed for five different sampling sites at various depths along two geographically distinct continental margin systems. All of the positive transformants obtained from each site (278 to 409 clones per sampling site) were screened by RFLP analysis; a total of 1,762 clones were evaluated in this study (Table 1).
RFLP analysis revealed extensive genetic diversity of dsrAB genes for all of the sites examined. The majority of the clones had unique RFLP patterns, i.e., 72, 73, 51, 66, and 47% for M300, M306, W301, W306, and W307, respectively (Table 1). The overall clone diversity sampled was considerably lower in the Washington sites because of the dominance of a single RFLP pattern at these locations.
Two RFLP patterns (W-301-015 and W-301-016) were common to all five sampling sites, and one of them (W-301-015) dominated all of the clone libraries (8 to 28% of each site-specific clone library) except M306 (2% of the clone library). A different RFLP pattern (M-306-203) was more frequently recovered at M306, and it accounted for 4% of that clone library. On average, all other different RFLP patterns were amplified much less frequently and made up less than 3% of each site-specific clone library.
Pairwise site comparisons showed that 14 to 22% and 23 to 36% of the total number of clones recovered from any Mexico or Washington site-specific clone library, respectively, were also detected at other sites within the same geographic location (Table 2). The percentage of shared clones between margin systems varied from 6 to 33% (average ± standard deviation [SD], 15% ± 6%) per site library. Averaged, 18% ± 6% of the clones from the Mexican margin were sampled at both Mexico sites, while 30% ± 3% of the Washington margin clones were sampled at multiple locations. Approximately one-third of the W301 clone library was observed at sites M300 and M306. Apparent genotypic similarity between these sites was in large part due to the dominance of a single RFLP pattern in the W301 clone library that was also amplified from both Mexican margin sites.
TABLE 2.
Pairwise matrix describing the genetic overlap between the Mexico and Washington site-specific clonal librariesa
| Siteb | M300 | M306 | W301 | W306 | W307 |
|---|---|---|---|---|---|
| M300 | 111/646 | 164/708 | 124/777 | 110/725 | |
| M306 | 43/628 | 115/687 | 85/635 | ||
| W301 | 216/759 | 210/707 | |||
| W306 | 212/635 |
Each pairwise comparison provides the total number of clones common to both sites/the total number of clones analyzed.
M, Mexico; W, Washington.
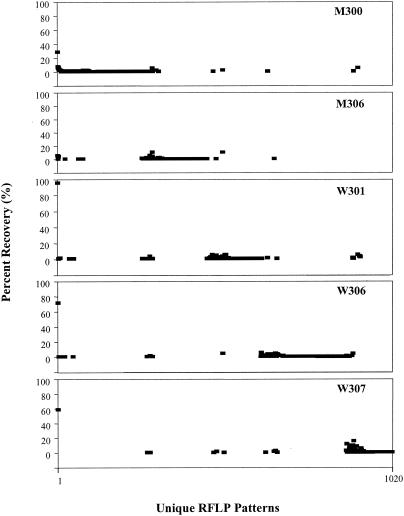
The distributions of the unique dsrAB patterns were evaluated further. Figure 2 shows the distribution and percent recovery of the unique RFLP banding patterns by site and depth. The dominant RFLP banding pattern (the first data point in each panel) recovered repeatedly from most sites accounted for a sizable fraction at each Mexico margin site (<30%) but was amplified more readily in the Washington margin (>60% per site library). The majority of each site-specific clone library, however, was composed of unique RFLP patterns that were amplified more rarely and were not consistently sampled across multiple sites.
FIG. 2.
Spatial distributions of unique RFLP banding patterns (1,020 total, x axis) sampled from the Mexico (M) and Washington (W) margins. All unique RFLP patterns were ordered sequentially by site along the x axis (i.e., M300, M306, W301, W306, and W307). Percent recovery (y axis) of each unique RFLP pattern was calculated by site. The dominant RFLP pattern recovered from all of the sites is represented as the first data point in each panel.
Sequence analysis.
Altogether, 175 representative dsrAB clones were sequenced. Comparative sequence analyses were conducted on both nucleotide sequences (on average, 531 and 738 comparable positions for dsrA and dsrB, respectively) and translated sequences (on average, 177 and 246 inferred amino acid residues for DsrA and DsrB, respectively) for each subunit independently and combined. The resulting subunit phylogenies were largely congruent, differing only in the exact placement of individual sequences within low-order branch clusters (data not shown). Strong parallels across different sulfite reductase subunit gene phylogenies has been documented in several previous studies (8, 33, 50) and provides confidence that each dsrAB amplimer described here was most likely recovered from a single source and was not a PCR artifact (41). It should be noted that comparisons of longer sequence tracks did result in greater reproducibility and some apparent improvement in low-order branch resolution (dsrAB ≥ dsrB > dsrA and DsrAB ≥ DsrB > DsrA). However, these fine-scale differences did not alter the outcome of any of the sequence comparisons conducted here (data not shown). On the basis of the overwhelming consistency of sulfite reductase subunit gene phylogenies and to maximize the utility of the National Center for Biotechnology Information database, the detailed phylogenetic analyses and discussion hereafter pertain exclusively to dsrA sequences encoding the protein DsrA (134 comparable positions with a sequence mask).
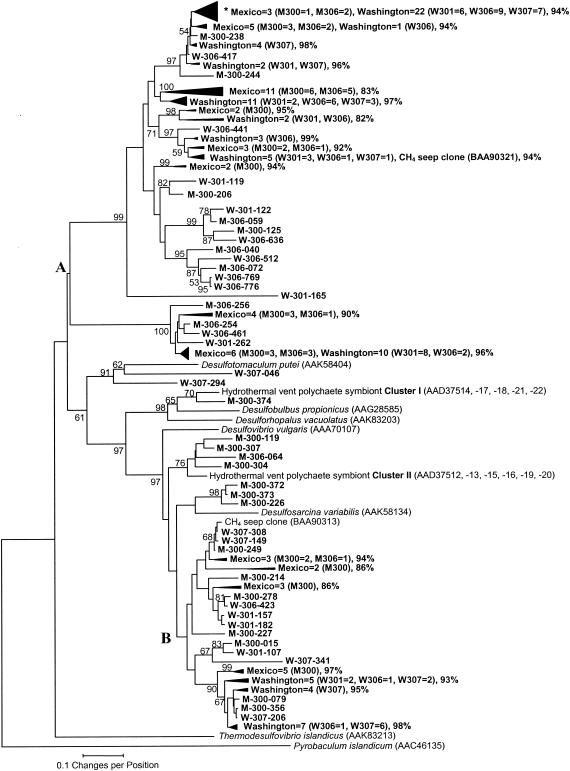
The majority of continental margin sulfite reductase gene sequences were considerably divergent from DsrA sequences from all known divisions of sulfate reducers (Fig. 3). The majority of clone sequences were either deeply branching with no apparent close relatives (DsrA sequences denoted by node A) or formed a previously unrecognized line of descent seemingly affiliated with the delta subdivision of the class Proteobacteria (DsrA sequences are denoted by node B). Few sequences, however, were more strongly related to sulfate-reducing bacteria belonging to the Firmicutes and delta Proteobacteria divisions (Fig. 3). These results suggest that these continental margin sedimentary habitats could harbor novel sulfate-reducing bacteria.
FIG. 3.
Phylogenetic analysis of recovered continental margin dissimilatory (bi)sulfite reductase subunit A protein coding sequences and selected sequences recovered from other different environmental sources and formally described species. Collapsed sequence clades were defined by branching order, and the clone composition and genetic identities of each are provided. The collapsed sequence clade marked by an asterisk indicates the position of clones having the numerically dominant RFLP pattern. The deeply branching sequence lineage is designated at the unifying branch node marked A, and divergent sequences showing an apparent distant relationship to the delta subdivision of the class Proteobacteria are indicated by the node designation B. Continental margin cloned sequences are labeled by margin (Mexico = M; Washington = W), site number (M = 300 or 306; W = 301, 306, or 307), and an arbitrary clone number. The percentage of 500 bootstrap samples that supported each branch is shown. Bootstrap values of <50% are not shown.
Two Washington sequences (W-307-046 and W-307-294) clustered with DsrA of Desulfotomaculum putei; however, the sequence similarity scores were weak (55 and 61%, respectively) over the sequence region compared. Both W307 sequences also clustered with DsrA from other members of the Bacillus-Clostridium division (Desulfotomaculum spp., Desulfosporosinus orientis, Desulfitobacterium dehalogenans, and Desulfitobacterium hafniense), but sequence identity scores were unchanged (data not shown). The consistency of this clustering pattern suggests that these two clone sequences may be affiliated with the Bacillus-Clostridium division but could correspond to new gram-positive sulfate-reducing taxa (58% sequence identity to each other).
Five of the Mexico sequences (M-300-374, M-306-064, M-300-119, M-300-307, and M-300-304) formed two cohesive clusters (labeled I and II) with DsrA sequences from presumed symbionts of the hydrothermal vent polychaete annelid Alvinella pompejana (sampled at a depth of 2,620 m on the East Pacific Rise; 13) (Fig. 3). Sequence M-300-374 was 88 to 92% similar to the symbiont DsrA sequences in cluster I. DsrA from Desulfobulbus propionicus was 79% identical to M-300-374; however, this level of sequence similarity likely signifies at least a different genus. The remaining four sequences formed a consistent cluster (II) with the other symbiont DsrA sequences (88% overall homology). Each of these Mexico sequences was 81 to 90% identical to any symbiont sequence within cluster II, and no known sulfate reducer sequence showed significant homology to these sequences.
Three Mexico sequences (M-300-372, M-300-373, and M-300-226; 95% homology) and the divergent lineage of sequences marked by node B (81% overall homology) clustered most closely with DsrA of Desulfosarcina variabilis. Sequences M-300-372 and M-300-226 were 74% similar to DsrA of D. variabilis, while M-300-373 was 77% similar. Divergent sequences marked by node B were ≤81% identical to DsrA of D. variabilis. A single deep-sea cold CH4 seep DsrA sequence (BAA90313) showed significant homology (≥98.5% identity) to continental margin sequences W-307-308, M-300-249, and W-307-149 within the node B lineage. All of these continental margin sequences were equally homologous to other different sulfate reducer genera belonging to the same phylogenetic lineage as D. variabilis (i.e., Desulfofaba gelida, Desulfobotulus sapovorans, and Desulfococcus multivorans; data not shown), and this grouping pattern was consistent.
The great majority of the continental margin DsrA sequences recovered (75% of the sequenced clones) were deeply branching and showed no apparent relationship to known sulfate reducer species (node A sequence lineage, Fig. 3). Clones corresponding to the dominant RFLP pattern recovered from all of the sites and depths belonged to this lineage; the collapsed sequence clade containing these sequences is marked in Fig. 3. A single deep-sea cold CH4 seep sequence (BAA90321) showed considerable sequence homology to several Washington clone sequences within this deep lineage (95.3% identity to W-307-298 and 96.6% identity to W-306-762). The sequences to the right of node A were considerably divergent (≤43% identity) from DsrA of Desulfotomaculum putei, Desulfobulbus propionicus, Desulforhopalus vacuolatus, and Desulfovibrio vulgaris. Sequence comparisons to other different siroheme-containing redox enzymes (i.e., nitrite reductase and the “reverse” sulfite reductase of Allochromatium vinosum; data not shown) were made, but the sequence identity scores were ≤20%. The continental margin DsrA sequences in node A may be representative of a previously unrecognized deep-branching sulfate reducer lineage. Phylogenetic reconstructions with these sequences were consistently rooted by DsrA from Thermodesulfovibrio islandicus, belonging to the Nitrospira division, and Pyrobaculum islandicum, a hyperthermophilic archaeon.
DISCUSSION
The continental margins are major depositional environments for organic carbon on a global scale (40, 52), but the ultimate fate of this carbon is largely determined by the activities of the native microbiota. The relative contribution of sulfate reduction to the overall carbon oxidation in the margins is significant (24, 30) but varies with increasing depth and distance offshore (24). Although the deeper sampling sites on each margin had higher carbon concentrations, sulfate reduction rates decreased with increasing depth, reaching nearly undetectable levels at 997 m (W307). Several previous studies have shown that as organic carbon descends through the water column, labile compounds are preferentially oxidized, leaving behind recalcitrant carbon compounds that are less susceptible to enzymatic degradation (18, 45, 51). Thus, both the absolute amount of carbon reaching the sediment floor and the overall bioavailability decrease as the water depth increases (24-26). Therefore, we presume that overall carbon bioavailability decreases with increasing slope depth despite the increased concentration and that accumulated carbon compounds were largely refractory and an inadequate carbon and energy source for sulfate reducer metabolism. Moreover, the highest rates of sulfate reduction were measured at the shallowest depth examined (W301 at 119 m), where labile carbon supplies likely reach the sediment floor and are turned over quickly (25, 51). Relative trends in interstitial pore water parameters (NH4+, PO43−, dissolved iron, and manganese) have been shown previously to be similar between the Mexican and Washington margin systems, although absolute concentrations and overall productivity differ (23, 24). Other environmental factors not measured in this study could vary with increasing slope depth and contribute significantly to the patterns observed here; however, carbon flux and compound bioavailability appear to be important variables. The continental margin provides a natural experimental system with which to examine the relative impacts of carbon on sulfate reducer diversity and distributions.
Contrary to conventional expectations for “strict” anaerobes, sulfate reduction rates appeared to parallel oxygen concentrations along the Washington margin shelf-slope transition zone. Relatively few data points are presented here, but the overall trend is supported by the comprehensive data set of Hartnett and Devol (24). The highest rates of SO42− reduction were measured at the most oxic site examined (W301), and activity at this location was markedly higher than that at both sites occurring within the ODZ of the Mexican margin system. As is often typical for highly productive shallow marine sediments, the demands of oxygenic respiration often exceed diffusive supplies, leading to the depletion of oxygen within the upper few millimeters to centimeters of surface sediment (4, 24). High rates of aerobic respiration likely produce anaerobic microenvironments within oxic surface layers, as well as drive deeper sediments anoxic. Still, several recent studies have demonstrated that oxygen may not be a strong deterrent for some sulfate-reducing taxa (21, 36-38) or inhibitory to sulfate reduction activity (6, 31). In the continental margins studied, it is likely that the labile carbon supply, which diminishes with depth, is a primary controlling factor for sulfate reduction. Large labile carbon fluxes to the shallow sediments overpower the oxygen and other higher-order electron acceptors, leaving conditions favorable for sulfate reduction.
Several different continental margin DsrA clones showed presumptive membership to the Firmicutes and delta Proteobacteria divisions by consistently clustering with sequences from known taxa. On the basis of the genus level dsrA protein-coding sequence similarity scores of known taxa (i.e., Desulfotomaculum spp., Desulfosarcina spp., and Desulfobulbus spp.), the different continental margin sequences recovered could represent novel species but most likely new genera within different formally characterized lineages. Interestingly, the Desulfotomaculum-like sequences were recovered from Washington margin site W307, the deepest and likely the most carbon-stressed site included in this study. Several other molecular investigations have reported Desulfotomaculum spp. thriving under a variety of harsh environmental conditions, including uranium mine tailings (8) and heavy-metal-contaminated estuarine sediments (47). The extensive physiological capabilities, namely, spore production and utilization of a great many different electron donors and acceptors (54), of the Desulfotomaculum spp. seemingly permit adaptation to anthropogenically impacted or otherwise challenging environmental conditions (e.g., reference 46).
Desulfosarcina- and Desulfobulbus-like sequences were frequently recovered from sites M300 and W307, but few corresponding sequences were also recovered from other different sampling sites. These genera have been shown previously to numerically dominate freshwater (35) and vegetated marine sediments (28, 42). Strains within these different genera are capable of oxidizing a great variety of different electron donors completely to CO2 (55), which would clearly provide a competitive advantage in environments where a broad range of carbon compounds are readily available. Sites M300 and W307 had higher carbon contents than all of the other sites examined within each respective margin system, although these carbon supplies are expected to be largely refractory. Assuming physiological likeness to the closest phylogenetic relatives, metabolic versatility may permit ample oxidation of refractory carbon compounds to support the growth of these organisms. Nonetheless, until these organisms are cultivated, extrapolation of physiological capabilities from comparative gene sequence analyses should be considered extremely tenuous.
The most relevant environmentally recovered database sequences for comparison were those cloned from putative symbionts of a deep-sea hydrothermal vent polychaete annelid (13) and a deep-sea cold CH4 seep (T. Fukuba and T. Naganuma, direct sequence submission to GenBank, 2000). Different CH4 seep cloned sequences corresponded to both Washington and Mexico cloned sequences, while all of the hydrothermal vent polychaete symbiont sequences clustered exclusively with Mexican margin sequences. The latter sequence pairing was somewhat unexpected considering the sharp physicochemical distinctions between these systems; the only identifiable commonality was the close geographic proximity of the Mexican margin and the deep-sea hydrothermal vent system (East Pacific Rise). This finding seemingly corroborates a conclusion of limited distribution for the majority of recovered sulfate-reducing taxa and indicates that perhaps geographic scale is a better predictor of genetic similarity than habitat type. Spatially scalable patterns of genetic diversity have also been observed for denitrifying bacteria in the continental margins (44).
Extensive new sulfate reducer gene sequences were obtained from both continental margin systems and all sampling sites ranging in depth from 119 to 997 m. In contrast to this study, most recent environmental surveys of sulfate reducers have adopted the revised forward primer of Wagner et al. (50), approximately 490 nucleotides upstream of the forward primer used here. Consequently, numerous environmentally recovered National Center for Biotechnology Information database sequences were not comparable because of limited sequence overlap. It is therefore uncertain whether many of our sequences represent potentially novel phylotypes or if similar sequences have been catalogued in other systems. Nonetheless, the majority of these sequences were evolutionarily divergent from all of the authentic Bacteria and Archaea sulfate reducer species that have been formally described to date.
The G+C content of the sequenced continental margin sulfite reductase gene sequences (48 to 66% G+C; mean ± SD = 59% ± 4%) fell within the range of the five known divisions of sulfate-reducing prokaryotes (for a summary, see reference 33). However, sequences descending from node A, which accounted for 75% of the recovered sequences, were as high as or exceeded the most G+C-rich dsrAB sequences documented to date (delta subdivision of the class Proteobacteria, 47 to 62% G+C), ranging from 55 to 66% (mean ± SD = 61% ± 2%). Continental margin nosZ genes recovered by Scala and Kerkhof (43) were also examined for a comparable shift in percent G+C content, but no significant differences were found relative to all of the other nosZ database sequences (data not shown). The overwhelming predominance of G+C-rich dsrAB sequences in the continental margin sediments sampled in this study may be ecologically relevant but cannot be fully determined in the absence of pure cultures. Thus, further efforts are needed to isolate the novel sulfate-reducing bacteria revealed by dsrAB sequences, to establish their physiological and ecological functions, and to understand their potential roles in carbon dynamics in marine sediments.
Acknowledgments
X.L. and C.E.B. contributed equally to this paper.
This research was supported by the U.S. Department of Energy Office of Science as part of its Biological and Environmental Research Programs in Biotechnology Investigations—Ocean Margins program. Oak Ridge National Laboratory is managed by the University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
REFERENCES
- 1.Bagwell, C. E., and C. R. Lovell. 2000. Persistence of selected Spartina alterniflora rhizoplane diazotrophs exposed to natural and manipulated environmental variability. Appl. Environ. Microbiol. 66:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandes, J. A., and A. H. Devol. 1995. Simultaneous oxygen and nitrate respiration in coastal sediments: evidence for discrete diagenesis. J. Mar. Res. 53:771-797. [Google Scholar]
- 5.Canfield, D. E., B. B. Jørgensen, H. Fossing, R. Glud, J. Gundersen, N. B. Ramsing, B. Thamdrup, J. W. Hansen, L. P. Nielsen, and P. O. J. Hall. 1993. Pathways of organic carbon oxidation in three continental margin sediments. Mar. Geol. 113:27-40. [DOI] [PubMed] [Google Scholar]
- 6.Canfield, D. E., and D. J. Des Marais. 1991. Aerobic sulfate reduction in microbial mats. Science 251:1471-1473. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter, J. H. 1969. The Chesapeake Bay Institute technique for the Winkler dissolved oxygen method. Limnol. Oceanogr. 10:141-143. [Google Scholar]
- 8.Chang, Y.-J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. Anwar Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, J. P. 1989. Sulfate reduction and carbon oxidation rates in continental shelf sediments, an examination of off shelf carbon transport. Cont. Shelf Res. 9:223-246. [Google Scholar]
- 10.Christian, R. R., K. Bancroft, and W. J. Wiebe. 1978. Resistance of the microbial community within salt marsh soils to selected perturbations. Ecology 59:1200-1210. [Google Scholar]
- 11.Codispoti, L. A. 1995. Is the ocean losing nitrate? Nature 376:724. [Google Scholar]
- 12.Codispoti, L. A., and J. P. Christensen. 1985. Nitrification, denitrification, and nitrous oxide cycling in the eastern tropical South Pacific. Mar. Chem. 16:277-300. [Google Scholar]
- 13.Cottrell, M. T., and S. G. Cary. 1999. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 65:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 15.Degens, B. P. 1998. Microbial functional diversity can be influenced by the addition of simple organic substrates to soil. Soil Biol. Biochem. 30:1981-1988. [Google Scholar]
- 16.Degens, B. P., L. A. Schipper, G. P. Sparling, and M. Vojvodic-Vukovic. 2000. Decreases in organic C reserves in soils can reduce the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 32:189-196. [Google Scholar]
- 17.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devol, A. H., and J. P. Christensen. 1993. Benthic fluxes and nitrogen cycling in sediments of the continental margin of the eastern North Pacific. J. Mar. Res. 51:345-372. [Google Scholar]
- 19.Devol, A. H., J. J. Anderson, K. M. Kuivila, and J. W. Murray. 1984. A model for coupling sulfate reduction and methane oxidation in sediments of Saanich Inlet. Geochim. Cosmochim. Acta 48:993-1004. [Google Scholar]
- 20.Devol, A. H., L. A. Codispoti, and J. P. Christensen. 1997. Summer and winter denitrification rates in western Arctic shelf sediments. Cont. Shelf Res. 17:1029-1050. [Google Scholar]
- 21.Diling, W., and H. Cypionka. 1990. Aerobic respiration in sulphate-reducing bacteria. FEMS Microbiol. Lett. 71:123-128. [Google Scholar]
- 22.Hanson, R. B. 1977. Nitrogen fixation (acetylene reduction) in a salt marsh amended with sewage sludge and organic carbon and nitrogen compounds. Appl. Environ. Microbiol. 33:846-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartnett, H. E. 1998. Ph.D. thesis. Organic carbon input, preservation and degradation in continental margin sediments: an assessment of the role of a strong oxygen minimum zone. University of Washington, Seattle.
- 24.Hartnett, H. E., and A. H. Devol. 2003. The role of a strong oxygen deficient zone in the preservation and degradation of organic matter: a carbon budget for the continental margins of NW Mexico and Washington State. Geochim. Cosmochim. Acta 67:247-264. [Google Scholar]
- 25.Hedges, J. I., F. S. Hu, A. H. Devol, H. E. Hartnett, E. Tsamakis, and R. G. Keil. 1999. Sedimentary organic matter preservation: a test for selective degradation under oxic conditions. Am. J. Sci. 299:529-555. [Google Scholar]
- 26.Hedges, J. I., J. A. Baldock, Y. Gelinas, C. Lee, M. Peterson, and S. G. Wakeham. 2001. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 409:801-804. [DOI] [PubMed] [Google Scholar]
- 27.Hedges, J. I., and J. H. Stern. 1984. Carbon and nitrogen determinations of carbonate-containing solids. Limnol. Oceanogr. 29:657-663. [Google Scholar]
- 28.Hines, M. E., R. S. Evans, B. R. S. Genthner, S. G. Willis, S. Friedman, J. N. Rooney-Varga, and R. Devereux. 1999. Molecular phylogenetic and biochemical studies of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 65:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea bed—the role of sulfate reduction. Nature 296:643-645. [Google Scholar]
- 30.Jørgensen, B. B. 1983. Processes at the sediment-water interface, p. 477-509. In B. Bolin and R. B. Cook (ed.), The major biogeochemical cycles and their interactions, vol 21. Wiley, New York, N.Y.
- 31.Jørgensen, B. B., and F. Bak. 1991. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karkhoff-Schweizer, R. R., D. P. W. Huber, and G. Voordouw. 1995. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen, E., S. I. Ahmed, and A. H. Devol. 1995. Aerobic and anaerobic decomposition of organic matter in marine sediments: which is faster? Limnol. Oceanogr. 40:1430-1437. [Google Scholar]
- 35.Li, J., K. J. Purdy, S. Takii, and H. Hayashi. 1999. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in a freshwater lake sediment. FEMS Microbiol. Ecol. 28:31-39. [Google Scholar]
- 36.Marschall, C., P. Frenzel, and H. Cypionka. 1993. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 159:168-173. [Google Scholar]
- 37.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naqvi, S. W. A., D. A. Jayakumar, P. V. Narvekar, H. Naik, V. V. S. S. Sarma, W. D'Souza, S. Joseph, and M. D. George. 2000. Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408:346-349. [DOI] [PubMed] [Google Scholar]
- 40.Premuzic, E. T., C. M. Benkovitz, J. S. Gaffney, and J. J. Walsh. 1982. The nature and distribution of organic matter in the surface sediments of world oceans and seas. Org. Geochem. 4:63-77. [Google Scholar]
- 41.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S RNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooney-Varga, J. N., R. Devereux, R. S. Evans, and M. E. Hines. 1997. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 63:3895-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skoog, A., and R. Benner. 1997. Aldoses in various size fractions of marine organic matter: implications for carbon cycling. Limnol. Oceanogr. 42:1803-1813. [Google Scholar]
- 46.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analysis. FEMS Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 47.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 48.Thamdrup, B., and D. E. Canfield. 1996. Pathways of carbon oxidation in continental margin sediments off central Chile. Limnol. Oceanogr. 41:1629-1650. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakeham, S. G., C. Lee, J. I. Hedges, P. J. Hernes, and M. L. Peterson. 1997. Molecular indicators of diagenetic status in marine organic matter. Geochim. Cosmochim. Acta 61:5363-5369. [Google Scholar]
- 52.Walsh, J. J. 1991. Importance of continental margins in the marine biogeochemical cycling of carbon and nitrogen. Nature 350:53-55. [Google Scholar]
- 53.Whittaker, R. H. 1975. Communities and ecosystems, 2nd ed. Macmillan Publishing Co., Inc., New York, N.Y.
- 54.Widdel, F. 1992. The genus Desulfotomaculum, p. 1792-1799. In A. Balows, H. G. Trüper, M. Dworkin, W. Hardin, and K.-H. Schleifer (ed.), The procaryotes, 2nd ed. Springer Verlag, New York, N.Y.
- 55.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer Verlag, New York, N.Y.
- 56.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, J.-Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]