Abstract
Pentose fermentation to ethanol with recombinant Saccharomyces cerevisiae is slow and has a low yield. A likely reason for this is that the catabolism of the pentoses d-xylose and l-arabinose through the corresponding fungal pathways creates an imbalance of redox cofactors. The process, although redox neutral, requires NADPH and NAD+, which have to be regenerated in separate processes. NADPH is normally generated through the oxidative part of the pentose phosphate pathway by the action of glucose-6-phosphate dehydrogenase (ZWF1). To facilitate NADPH regeneration, we expressed the recently discovered gene GDP1, which codes for a fungal NADP+-dependent d-glyceraldehyde-3-phosphate dehydrogenase (NADP-GAPDH) (EC 1.2.1.13), in an S. cerevisiae strain with the d-xylose pathway. NADPH regeneration through an NADP-GAPDH is not linked to CO2 production. The resulting strain fermented d-xylose to ethanol with a higher rate and yield than the corresponding strain without GDP1; i.e., the levels of the unwanted side products xylitol and CO2 were lowered. The oxidative part of the pentose phosphate pathway is the main natural path for NADPH regeneration. However, use of this pathway causes wasteful CO2 production and creates a redox imbalance on the path of anaerobic pentose fermentation to ethanol because it does not regenerate NAD+. The deletion of the gene ZWF1 (which codes for glucose-6-phosphate dehydrogenase), in combination with overexpression of GDP1 further stimulated d-xylose fermentation with respect to rate and yield. Through genetic engineering of the redox reactions, the yeast strain was converted from a strain that produced mainly xylitol and CO2 from d-xylose to a strain that produced mainly ethanol under anaerobic conditions.
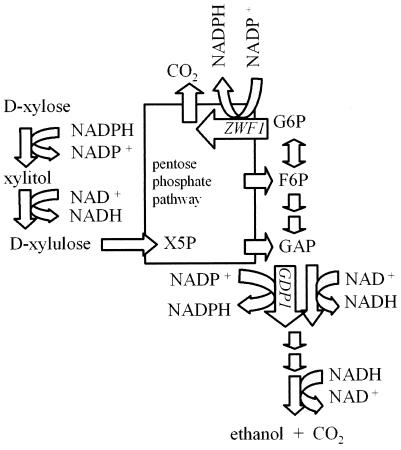
There is a fundamental distinction between NADP+/NADPH and NAD+/NADH in most biochemical pathways. Catabolic reactions are normally linked to NAD+/NADH, and anabolic reactions are normally linked to NADP+/NADPH. NAD+/NADH is the cofactor couple used by the respiratory chain and in the oxidation of glucose to ethanol or lactate through glycolysis. NADP+/NADPH is the main cofactor couple in biosynthesis. Pentose catabolism in fungi, however, is different; i.e., both cofactor couples are needed for the catabolic reactions. The catabolism of the pentose sugars d-xylose and l-arabinose requires NADPH and NAD+. d-Xylose is reduced to xylitol in a reaction that uses NADPH preferentially or exclusively. The d-xylose reductase from Pichia stipitis has a preference for NADPH but can use NADH (16, 22). All other fungal d-xylose reductases described in the literature can use only NADPH as a cofactor. Xylitol is then oxidized to d-xylulose in a strictly NAD+-dependent reaction. l-Arabinose is also converted to d-xylulose in sequential redox reactions; however, four steps are needed: two steps that require NADPH and two that require NAD+ (6, 13, 14). The conversion of l-arabinose and d-xylose to d-xylulose is redox neutral, but different redox cofactors are used. d-Xylulose then enters the pentose phosphate pathway after phosphorylation to d-xylulose-5-phosphate (Fig. 1). d-Xylulose-5-phosphate reacts in the pentose phosphate pathway to d-fructose-6-phosphate and d-glyceraldehyde-3-phosphate (GAP), which can be converted in a redox-neutral way to equimolar amounts of CO2 and ethanol. The redox reactions are catalyzed by GAP dehydrogenase (GAPDH) and alcohol dehydrogenase. Both dehydrogenases use the NAD+/NADH cofactor couple. NADPH is mainly regenerated in the oxidative part of the pentose phosphate pathway. In this pathway d-glucose-6-phosphate, which is derived from d-fructose-6-phosphate, is oxidized through d-glucose-6-phosphate dehydrogenase (G6PDH), encoded by ZWF1, and 6-phosphogluconate dehydrogenase, encoded by GND1 and GND2, thereby generating 2 mol of NADPH and 1 mol of CO2 per mol of d-glucose-6-phosphate (Fig. 1). The activities of the enzymes in the oxidative part of the pentose phosphate pathway, G6PDH and 6-phosphogluconate dehydrogenase, are increased during growth on pentose sugars in some yeast (1) and mold (26) strains, suggesting that the corresponding genes are induced under these conditions.
FIG. 1.
Redox cofactors in the metabolic pathway for d-xylose fermentation. d-Xylose is converted to d-xylulose through an NADPH-utilizing reductase and NAD+-utilizing dehydrogenase. d-Xylulose is then phosphorylated to the pentose phosphate intermediate d-xylulose 5-phosphate (X5P). The products of the pentose phosphate pathway are fructose-6-phosphate (F6P) and GAP. GAP is reduced through an NADP-GAPDH, encoded by GDP1, or by the endogenous NAD-GAPDH, depending on cofactor availability. In the following reactions involving an NADH requiring alcohol dehydrogenase, equimolar amounts of CO2 and ethanol are derived. A competing pathway for NADP+ is the oxidative part of the pentose phosphate pathway. Glucose-6-phosphate (G6P) is derived from fructose-6-phosphate and can enter the oxidative part of the pentose phosphate pathway through G6PDH, which is encoded by the ZWF1 gene. G6P is oxidized, thereby generating NADPH and CO2. The deletion of ZWF1 prevents this reaction.
Pentose fermentation to ethanol is desired in biotechnology when fuel ethanol is to be produced from biomass. d-Xylose and l-arabinose fermentation to equimolar amounts of ethanol and CO2 under anaerobic conditions is theoretically possible and of relevance in biotechnology. However, in practice, either the fermentation requires careful aeration or the fermentation product is mainly biomass or xylitol and CO2. The fact that the pentoses are not efficiently fermented to ethanol can be explained by the imbalance of redox cofactors. Since NADPH is regenerated mainly in the oxidative part of the pentose phosphate pathway, where the reduction of NADP+ is coupled to the generation of CO2, it has an effect on the redox balance (Fig. 1). When extra CO2 (an oxidized product) is produced in this pathway, the pentose fermentation to ethanol and CO2 is no longer redox neutral. To remove excess NADH, either xylitol is produced or aeration is required, which leads to further unwanted CO2 production or a combination of both processes. For an improved ethanol yield, it would be useful to regenerate the NADPH in a way which is not directly coupled to CO2 production and which simultaneously eliminates the production of excess NADH.
We previously described an NADP-GAPDH (EC 1.2.1.13) from Kluyveromyces lactis (23). This enzyme may function to regenerate the NADPH. It is not directly coupled to CO2 production, and furthermore, its use eliminates a stoichiometric amount of NADH production; thus, it should facilitate anaerobic pentose fermentation to ethanol (Fig. 1).
In this work we expressed the gene for NADP-GAPDH in a Saccharomyces cerevisiae strain which contains the d-xylose pathway. In addition, we deleted the gene for the competing reaction, ZWF1, in the oxidative part of the pentose phosphate pathway. We tested ethanol, CO2, and xylitol production from d-xylose under anaerobic conditions.
MATERIALS AND METHODS
Strain construction.
The cultivations were done with S. cerevisiae strains, all originating from the CEN.PK2 strain. Every strain used in this study was carrying all the genes of the d-xylose pathway (XYL1, XYL2, and XKS1). XYL1 and XYL2 are from P. stipitis and code for aldose reductase and d-xylulose reductase, respectively. XKS1 is from S. cerevisiae and codes for xylulokinase. XYL1 (under the control of the PGK1 promoter), XYL2 (under the control of the modified ADH1 promoter) (17), and XKS1 (under the control of the modified ADH1 promoter) were integrated into the chromosomes by targeted integration (15, 21).
GDP1 was amplified by PCR from K. lactis DNA with the following primers, each of which contains a BamHI restriction site (BamHI sites are underlined): AAGGATCCAAGATGCCCGATATGACAAACGAATCTTC and AAGGATCCAAGCGTCTCCTTAAACACCAGC. The PCR product was then cloned into a TOPO vector (Invitrogen), and the 1-kb BamHI fragment from the resulting vector was ligated to the BglII site of a yeast expression vector with the PGK1 promoter, p1181. The resulting plasmid, p1696, was described previously (23). Alternatively, the BamHI fragment was ligated to the BamHI site of pVT102U (24), which is a yeast expression vector with an adenine dinucleotide promoter. The resulting vector was p1731.
The ZWF1 gene coding for the G6PDH was obtained by PCR using S. cerevisiae genomic DNA as a template. The specific primers GCTATCGGATCCAAGCTTAGGCAAGATGAGTGAAGGTT and GCTATCGGATCCAAGCTTAGTGACTTAGCCGATAAATG were used. Both primers had BamHI and HindIII sites to facilitate the cloning. The restriction sites are underlined. The ZWF1 fragment obtained by the PCR was digested with BamHI and ligated into the pBluescript SK plasmid (Stratagene). The resulting plasmid was digested with BglII to remove a 1,063-bp fragment from the middle of the ZWF1 gene. The digested vector was blunted with mung bean nuclease. The HIS3 marker gene was obtained from the pRS423 plasmid (7) by BsmBI and DraIII digestion. The 1,591-bp fragment containing the HIS3 gene was blunted with mung bean nuclease and ligated into the BglII-digested and -blunted vector. From the resulting plasmid, the ZWF1 deletion cassette was released with BamHI and transformed into the S. cerevisiae strain with the d-xylose pathway as described above. The deletion of the ZWF1 gene was confirmed by PCR analysis, by Southern blot analysis, and by assaying the G6PDH activity in the yeast extract. The resulting strain was then transformed with a plasmid containing the GDP1 gene as described above or with the corresponding empty plasmid. All the strains and their genetic modifications are listed in the Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| CEN.PK2 | MATαleu2-3/112 ura3-52 trp1-289 his3Δ1 MAL2-8c SUC2 | 3 |
| H2217 | CEN.PK2 ura3::XYL1 XYL2 his3::XKS1 kanMX | 14 |
| H2750 | H2217(p1181) | 23 |
| H2757 | H2217(p1696) | 23 |
| H2674 | H2217(pVT120U) | This study |
| H2673 | H2217(p1731) | This study |
| H2723 | H2217 zwf1::HIS3 (p1181) | This study |
| H2684 | H2217 zwf1::HIS3 (p1696) | This study |
| Plasmids | ||
| p1181 | URA3 | 23 |
| p1696 | URA3 GDP1 | 23 |
| pVT102U | URA3 | 24 |
| p1731 | URA3 GDP1 | This study |
Enzyme activity assays.
The cell extracts for the G6PDH enzyme activity measurement were prepared by disrupting the yeast cells in 10 mM Na-phosphate (pH 7.0) buffer with glass beads. The protease inhibitors phenylmethylsulfonyl fluoride (final concentration, 1 mM) and pepstatin A (0.01 mg/ml) were added into the extraction buffer. The activity was measured with a Cobas Mira automated spectrophotometric analyzer (Roche). The activity was measured in buffer containing 10 mM Na-phosphate (pH 7.0) and 1 mM NADP+. d-Glucose-6-phosphate, which was used as start reagent, was added to a final concentration of 10 mM.
The NADP-GAPDH activity was measured as described by Verho et al. (23).
Fermentation conditions.
The cultivations were batch cultivations and performed in Biostat CT fermentors (B. Braun Biotech International, Melsungen, Germany). Each strain was tested in a single experiment. The volume was 1 liter during the first 2 days, and the medium was synthetic complete medium (19) lacking uracil with 3% glucose as a carbon source. After 48 h, the biomass was around 3 g/liter, the glucose was consumed, and the ethanol concentration was between 0.5 and 1 g/liter. Then 0.5 liter of synthetic medium with 15% d-xylose was added to give a final d-xylose concentration of 5%. The temperature was 30°C, and the agitation speed was 300 rpm. During cultivation on d-glucose, the medium was sparged with air at a flow rate of 1.0 liter/min and the pH was adjusted to 5.0 with 2 M potassium hydroxide. When the d-xylose was added, the airflow was changed to nitrogen at 0.1 liter/min to have anaerobic conditions. The outlet gas was analyzed for CO2 by online mass spectroscopy. Samples of the culture were taken at different time intervals. The dry biomass was measured from 10 ml of cell suspension. The suspension was centrifuged, and the cell pellet was washed twice with water and dried overnight at 95°C. The first supernatant was analyzed by high-performance liquid chromatography for ethanol, d-xylose, xylitol, and other components.
Metabolite analysis.
Levels of d-xylose, xylitol, glycerol, acetate, and ethanol were analyzed by high-performance liquid chromatography. The detection unit was a Waters 410 refractive-index detector, and the column was an Aminex HPX-87H column (Bio-Rad Laboratories), which was maintained at 35°C. The eluent consisted of 5 mM H2SO4 at a constant flow rate of 0.3 ml min−1. CO2 was analyzed with a model QMG 421C quadrupole mass spectrometer (Pfeiffer Balzers, Balzers, Liechtenstein). Xylitol was quantified enzymatically using a commercial kit (Roche) where indicated below.
Shake flask cultures with mixed sugars.
Mixtures of d-glucose and uniformly 14C-labeled d-xylose were fermented in anaerobic shake flasks. The initial biomass was adjusted to 0.365 g (dry mass)/liter. Yeast suspension (100 ml) in a mixture of 2.4% d-glucose and 2.4% 14C-labeled d-xylose was stirred with a magnetic stirrer at 30°C in a 100-ml Erlenmeyer flask with a water lock to maintain anaerobiosis. Fermentation was 75 h. After the fermentation, the biomass was determined and the xylitol concentration was measured enzymatically. The ethanol was distilled from 50 ml of medium, and the distillate was filled up with distilled water to a volume of 50 ml. The total ethanol was then determined by measuring the density of the distillate with an Anton-Paar DMA58 densitometer. The ethanol derived from d-xylose was estimated from the radioactivity of the distillate. In a separate experiment, the fermentation was performed in the absence of d-xylose and the ethanol produced from glucose was estimated by densitometry. The ethanol from d-xylose was then calculated from the difference in the levels of ethanol with and without d-xylose. Each strain was tested in four independent experiments.
RESULTS
Anaerobic shake flask cultivations with d-glucose and d-xylose.
A mixture of 2.4% d-glucose and 2.4% uniformly 14C-labeled d-xylose were fermented in shake flasks under anaerobic conditions. The initial biomass was in all cases 0.365 g/liter. The fermentation time was 75 h. After this time, all of the d-glucose and part of the d-xylose were consumed. The biomass after this time and the ethanol and xylitol concentrations are summarized in Table 2. The xylitol concentration was measured enzymatically. The amount of ethanol was calculated from the radioactivity of the distilled ethanol. In separate experiments the total ethanol was measured after fermentation in the presence or in the absence of d-xylose by densitometry. The difference in the ethanol concentrations between these two sets of fermentations is due to the ethanol derived from d-xylose. The values obtained for the ethanol from d-xylose in these experiments were in agreement with the values obtained by measuring the incorporated radiolabel; however, measuring the incorporation of radiolabel is more precise. The overexpression of GDP1 led to an increased ethanol yield, while the deletion of zwf1 had a detrimental effect. The largest increase in the ethanol yield was observed when the two genetic modifications, overexpression of GDP1 and deletion of zwf1, were combined. The xylitol production was reduced in the Δzwf1 strains. Relative to levels in the control, the yield of ethanol production from d-xylose in the strain with the two genetic modifications increased by about 50% while at the same time the xylitol production decreased by a similar percentage.
TABLE 2.
Results of anaerobic shake flask cultivations with d-glucose and d-xylose
| Strain | Final dry mass (g/liter) | Final xylitol concn (mM) | Concn of ethanol derived from 14C-labeled d-xylose (mM) |
|---|---|---|---|
| H2750 (control) | 2.15 ± 0.07 | 33 ± 1 | 29 ± 2 |
| H2757 (GDP1) | 2.25 ± 0.05 | 37 ± 2 | 31 ± 2 |
| H2723 (Δzwf1) | 1.95 ± 0.15 | 9 ± 0.5 | 25 ± 1 |
| H2684 (GDP1 Δzwf1) | 2.1 ± 0.1 | 17 ± 0.8 | 44 ± 3 |
Anaerobic fermenter cultivations.
To confirm the results from the shake flasks and to obtain additional information, e.g., about the CO2 production, we performed fermenter cultivations. All strains were first grown aerobically with glucose as a carbon source until most of the produced ethanol was utilized. This process took 48 h, and the residual ethanol was between 0.5 and 1 g/liter. The biomass at this stage varied between 2 and 4 g (dry mass) per liter. d-Xylose medium was added, and by changing the gas from air to nitrogen, the fermentation was switched to anaerobiosis. Anaerobic d-xylose fermentations were then monitored for about 120 h, and the results are summarized in Fig. 2 and 3 and Table 3. During this time the biomass increased by 10 to 20%. In earlier similar experiments the cells were pregrown on d-glucose and then harvested and washed before they were mixed with the d-xylose medium and anaerobic conditions were applied. Under these earlier conditions the biomass decreased by about 50% during 120 h of anaerobic d-xylose fermentation (21). When the washing was omitted, the biomass did not decrease but on the contrary increased. Drawbacks are that the initial biomass for the d-xylose fermentation is not adjusted and there is some initial ethanol, which has to be accounted for.
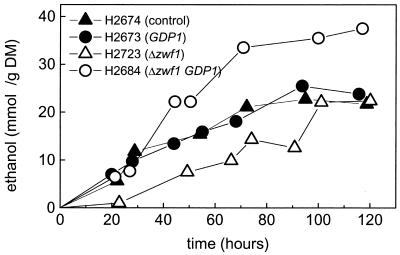
FIG. 2.
Ethanol production in the fermenter cultivations. Pure d-xylose is fermented under anaerobic conditions. The ethanol production is normalized to the biomass. The average biomass is indicated in Table 3. DM, dry mass.
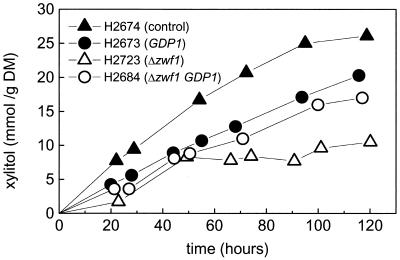
FIG. 3.
Xylitol production in the fermenter cultivations. Conditions are as described for Fig. 2 and Table 3. DM, dry mass.
TABLE 3.
Results of anaerobic fermenter cultivations
| Strain | Concn (mM) (%) of:
|
CO2/ethanol ratio (mol/mol) | % Carbon recovery | Dry mass (g/liter) | |||
|---|---|---|---|---|---|---|---|
| d-Xylose used | Ethanol | Xylitol | CO2 | ||||
| H2674 (control) | 200 (100) | 90 (18) | 110 (53) | 225 (22) | 2.5 | 93 | 3.8 |
| H2673 (GDP1) | 135 (100) | 76 (23) | 65 (48) | 150 (22) | 2.0 | 93 | 3.0 |
| H2723 (Δzwf1) | 175 (100) | 100 (24) | 50 (28) | 160 (18) | 1.6 | 70 | 4.2 |
| H2684 (GDP1 Δzwf1) | 98 (100) | 100 (41) | 34 (34) | 125 (25) | 1.3 | 100 | 1.9 |
The main fermentation products of anaerobic d-xylose fermentation were ethanol, xylitol, and CO2. The levels of ethanol and xylitol production are shown in Fig. 2 and 3. In the figures the productivities are normalized to the biomass. The average biomasses of the different experiments are listed in Table 3. The ethanol production rate is about 0.2 mmol g (dry mass)−1 h−1 for all strains except for strain H2684 (GDP1 Δzwf1), where it is about 0.4 mmol g (dry mass)−1 h−1. The expression of GDP1 had no clear effect on the rate of ethanol production; however, the rate of xylitol production was decreased. The simultaneous overexpression of GDP1 and the deletion of zwf1 increased the rate and yield of ethanol production and decreased xylitol production (Fig. 2 and 3). The specific xylose utilization rates were similar in all strains (not shown). In Table 3 the molar yields of ethanol, xylitol, and CO2 are summarized. The yields were calculated after 120 h of fermentation. The control strain produced more xylitol than ethanol; i.e., the molar ratio was below 1. This situation was reversed when GDP1 was overexpressed or zwf1 was deleted. The biggest effect was seen when both genetic modifications were combined. In this case about three times more ethanol than xylitol was produced. The increased ethanol yield was also reflected by a decreased CO2/ethanol ratio. In the control strain, about 2.5 mol of CO2 per ethanol is produced, while in the strain with both genetic modifications, the ratio is about 1.3.
DISCUSSION
A strain of S. cerevisiae expressing the fungal d-xylose pathway can ferment d-xylose to ethanol but only at a low rate and yield. To improve the rate and yield of d-xylose fermentation to ethanol, an NADP+-utilizing GAPDH (GDP1) was expressed and the gene for G6PDH (ZWF1) was deleted in order to shut down the oxidative part of the pentose phosphate pathway. As a result of these genetic modifications, the strain was converted from a mainly xylitol- and CO2-producing strain to a mainly ethanol-producing strain.
d-Xylose fermentation to equimolar amounts of CO2 and ethanol is redox neutral; however, in this pathway NADPH is utilized, which has to be regenerated elsewhere. The expression of an NADP-GAPDH provides such a mechanism for the regeneration of NADPH within the pathway. For the conversion of 3 mol of d-xylose to d-xylulose, 3 mol of NADPH and NAD+ have to be regenerated. Three moles of d-xylulose can lead to up to 5 mol of GAP; i.e., 3 mol of GAP must go through the NADP-GAPDH and 2 mol must go through the NAD-GAPDH, leaving 5 mol of NADH for the alcohol dehydrogenase reaction to produce equimolar amounts of ethanol and CO2. The simultaneous presence of NAD- and NADP-GAPDH activities allows the cell to automatically adjust the flux through these enzymes according to the cofactor requirement.
The main path for the regeneration of NADPH is the oxidative part of the pentose phosphate pathway. This path, in which the regeneration is furthermore coupled to CO2 production, competes with the NADP-GAPDH. To block this part of the pathway, we deleted the gene for the G6PDH, ZWF1. A haploid strain of S. cerevisiae has only one gene coding for such an enzyme, but the deletion of this enzyme has no major detrimental effects except that the sensitivity to oxidizing agents is elevated (12). The combination of overexpression of GDP1 and deletion of zwf1 had a bigger effect than expression of GDP1 alone, indicating that the oxidative part of the pentose phosphate pathway was indeed a pathway competing for NADP+, and by closing this pathway more NADP+ was forced through the NADP-GAPDH.
Since NADPH regeneration through NAPD-GAPDH is not coupled to CO2 production, anaerobic d-xylose fermentation by only this route would yield equimolar amounts of CO2 and ethanol; i.e., the CO2/ethanol ratio is 1. A higher CO2/ethanol ratio (under anaerobic conditions) is an indication that other routes for NADPH regeneration, which are coupled to CO2 production, are used. In the control strain H2674, this CO2/ethanol ratio was 2.5 (Table 3), indicating that NADPH regeneration is coupled mainly to CO2 production. Through expression of GDP1 in combination with deletion of zwf1, this ratio was lowered to about 1.3, indicating that the NADPH regeneration was redirected to a non-CO2-producing reaction.
A high CO2/ethanol ratio was correlated with a low ethanol/xylitol ratio. When the NADPH is regenerated in a reaction, which is directly coupled to CO2 production, d-xylose fermentation to equimolar amounts of CO2 and ethanol is no longer redox neutral; i.e., the reaction will not occur. In the control strain H2674, the CO2/ethanol ratio is about 2.5. One mole of CO2 per mole of ethanol is produced in the pyruvate decarboxylase reaction; the rest is produced in other reactions, such as the oxidative reaction of the pentose phosphate pathway, where through CO2 production the redox balance is shifted so that more of a reduced product, i.e., xylitol, is formed. In strain H2684, with overexpression of GDP1 and deletion of zwf1, the CO2/ethanol ratio is 1.3; i.e., much less NADPH is regenerated in reactions coupled to CO2 production, which is reflected by less xylitol formation or a higher ethanol/xylitol ratio than that of the control strain (Table 3).
In the strain with a zwf1 deletion, H2723, the CO2/ethanol ratio was lower than in the control strain. It is not clear whether in this strain the NADPH is regenerated in an alternative way or whether the P. stipitis-derived d-xylose reductase, which can use NADH, now uses it as its major coenzyme. Alternative ways to regenerate NADPH, which are not directly coupled to CO2 production, are transhydrogenases or transhydrogenase cycles such as the d-mannitol cycle (20), a cycle with glutamate dehydrogenases with different cofactor specificities (4), or a cycle around the malic enzyme (2). However there is no experimental support for the existence of natural transhydrogenase activities or transhydrogenase cycles in yeasts or molds (5, 20).
The effect of the zwf1 deletion alone on anaerobic d-xylose fermentation in S. cerevisiae was previously described by Jeppsson et al. (8). Those authors tested a d-glucose and d-xylose cofermentation in continuous culture, which is difficult to compare with our batch fermentations; however, those authors also observed a lowered xylitol production.
In biotechnological applications, it is often a mixture of d-glucose and d-xylose and not pure d-xylose which has to be fermented. Under our conditions, all the d-glucose and parts of the d-xylose were fermented. By using radiolabeled d-xylose, we were able to distinguish between the ethanol produced from d-xylose and that produced from d-glucose. In these experiments the overexpression of GDP1 increased the ethanol yield. The yield was further improved by the combination of deleting zwf1 and overexpressing GDP1. The zwf1 deletion alone led to a lower ethanol production.
We overexpressed the genes of the d-xylose pathway from P. stipitis because P. stipitis is one of the best d-xylose-fermenting yeasts. One reason for this might be that the P. stipitis d-xylose reductase can use NADH as a cofactor; however, it has a preference for NADPH (16, 22). In a strain where NADPH is efficiently regenerated, the ability of the P. stipitis xylose reductase to use NADH as a cofactor would not be an advantage. This fact is of relevance since the P. stipitis xylose reductase has a relatively low affinity for d-xylose. Other d-xylose reductases which are strictly NADPH dependent, such as the d-xylose reductase from S. cerevisiae, have a higher affinity for d-xylose (Kms, 27.9 mM for S. cerevisiae and 42 mM for P. stipitis) (11, 22). For an accelerated fermentation, especially of low concentrations of d-xylose, it might be of advantage to use a d-xylose reductase with a higher affinity for d-xylose.
The ethanol production rate was about 0.2 and 0.4 mmol per g (dry mass) and h in the control strain H2674 and in strain H2684 (GDP1 Δzwf1), respectively (Fig. 1). The volumetric productivities were not compared since the biomasses were different in the different fermentations. Through our modifications, which affected the redox reactions, the rate of ethanol production was not greatly improved. The specific ethanol production rates under anaerobic conditions from d-xylose are still about 2 orders of magnitude lower than the corresponding rate on d-glucose. There are several possible reasons for the low fermentation rate on d-xylose. One is d-xylose transport. Kötter and Ciriacy estimated the initial d-xylose uptake rate under comparable conditions to be 100 nmol mg (fresh weight)−1 min−1, corresponding to about 20 mmol g (dry mass)−1 h−1 (10). The d-xylose utilization during fermentation was about 0.4 mmol g (dry mass)−1 h−1; i.e., the d-xylose transport capacity seems not to limit the d-xylose fermentation rate, at least not at high d-xylose concentrations. At a low d-xylose concentration it can be different. For d-xylose transport, a high-affinity system with a Km of 190 mM and a low-affinity system with a Km of 1.5 M were reported for S. cerevisiae (10).
Other factors limiting the rate of fermentation might be a nonoptimal xylulokinase activity (9) and a low capacity of reactions of the pentose phosphate pathway, such as a limiting activity of transaldolase (18, 25). Also, NADPH regeneration can still be limiting. Under our enzyme assay conditions, i.e., in the reverse reaction, the NADP-GAPDH activity of this enzyme was 0.3 nkat/mg of protein in the recombinant yeast, which compares to an endogenous NAD-GAPDH activity of 15 nkat/mg under similar conditions (23). Three-tenths nanokatal per milligram corresponds to 2.7 mmol g (dry mass)−1 h−1, assuming that 40% of the dry weight is extractable protein. The NADP-GAPDH activity was measured in the reverse direction, the thermodynamically favorable direction, with 1,3-bisphosphoglycerate as a substrate. The velocity in the forward direction with GAP as a substrate might be slower under in vivo conditions, and despite the improvements seen in this work, this step might indeed be rate limiting.
Although the rate of ethanol production was not greatly improved, there was a significant effect on the ethanol yield. Through redox engineering we were able to modify a strain that produced mainly xylitol and CO2 from d-xylose to a strain that produced mainly ethanol.
Acknowledgments
This work was supported by the Sustainable Use of Natural Resources (SUNARE) program of the Academy of Finland and the VTT Industrial Biotechnology research program (Academy of Finland; Finnish Centre of Excellence program, 2000 to 2005, project no. 64330).
We thank Eila Leino for technical help with the fermentations.
REFERENCES
- 1.Alexander, M. A., V. W. Yang, and T. W. Jeffries. 1988. Levels of pentose phosphate pathway enzymes from Candida shehatae grown in continuous culture. Appl. Microbiol. Biotechnol. 29:282-288. [Google Scholar]
- 2.Aristidou, A., P. Richard, L. Ruohonen, M. Toivari, J. Londesborough, and M. Penttilä. 1999. Redox balance in fermenting yeast. Monogr. Eur. Brewery Convent. 28:161-169. [Google Scholar]
- 3.Boles, E., H. W. Gohlmann, and F. K. Zimmermann. 1996. Cloning of a second gene encoding 5-phosphofructo-2-kinase in yeast, and characterization of mutant strains without fructose-2, 6-bisphosphate. Mol. Microbiol. 20:65-76. [DOI] [PubMed] [Google Scholar]
- 4.Boles, E., W. Lehnert, and F. K. Zimmermann. 1993. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 217:469-477. [DOI] [PubMed] [Google Scholar]
- 5.Bruinenberg, P. M., R. Jonker, J. P. van Dijken, and W. A. Scheffers. 1985. Utilization of formate as an additional energy source by glucose-limited chemostat cultures of Candida utilis CBS621 and Saccharomyces cerevisiae CBS 8066. Evidence for the absence of transhydrogenase activity in yeasts. Arch. Microbiol. 142:302-306. [Google Scholar]
- 6.Chiang, C., and S. G. Knight. 1960. A new pathway of pentose metabolism. Biochem. Biophys. Res. Commun. 3:554-559. [DOI] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Jeppsson, M., B. Johansson, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, Y. S., H. Ni, J. M. Laplaza, and T. W. Jeffries. 2003. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate d-xylulokinase activity. Appl. Environ. Microbiol. 69:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 11.Kuhn, A., C. van Zyl, A. van Tonder, and B. A. Prior. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogae, I., and M. Johnston. 1990. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene 96:161-169. [DOI] [PubMed] [Google Scholar]
- 13.Richard, P., J. Londesborough, M. Putkonen, N. Kalkkinen, and M. Penttilä. 2001. Cloning and expression of a fungal l-arabinitol 4-dehydrogenase gene. J. Biol. Chem. 276:40631-40637. [DOI] [PubMed] [Google Scholar]
- 14.Richard, P., M. Putkonen, R. Väänänen, J. Londesborough, and M. Penttilä. 2002. The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41:6432-6437. [DOI] [PubMed] [Google Scholar]
- 15.Richard, P., M. H. Toivari, and M. Penttilä. 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190:39-43. [DOI] [PubMed] [Google Scholar]
- 16.Rizzi, M., P. Erleman, N.-A. Bui-Tanh, and H. Dellweg. 1988. Xylose fermentation by yeasts. 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl. Biochem. Biotechnol. 29:148-154. [Google Scholar]
- 17.Ruohonen, L., M. K. Aalto, and S. Keränen. 1995. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J. Biotechnol. 39:193-203. [DOI] [PubMed] [Google Scholar]
- 18.Senac, T., and B. Hahn-Hägerdal. 1991. Effects of increased transaldolase activity on d-xylulose and d-glucose metabolism in Saccharomyces cerevisiae cell extracts. Appl. Environ. Microbiol. 57:1701-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman, F., G. Fink, and J. B. Hicks. 1983. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Singh, M., N. S. Scrutton, and M. C. Scrutton. 1988. NADPH generation in Aspergillus nidulans: is the mannitol cycle involved? J. Gen. Microbiol. 134:643-654. [DOI] [PubMed] [Google Scholar]
- 21.Toivari, M. H., A. Aristidou, L. Ruohonen, and M. Penttilä. 2001. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab. Eng. 3:236-249. [DOI] [PubMed] [Google Scholar]
- 22.Verduyn, C., R. Van Kleef, J. Frank, H. Schreuder, J. P. Van Dijken, and W. A. Scheffers. 1985. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem. J. 226:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verho, R., P. Richard, P. H. Jonson, L. Sundqvist, J. Londesborough, and M. Penttilä. 2002. Identification of the first fungal NADP-GAPDH from Kluyveromyces lactis. Biochemistry 41:13833-13838. [DOI] [PubMed] [Google Scholar]
- 24.Vernet, T., D. Dignard, and D. Y. Thomas. 1987. A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52:225-233. [DOI] [PubMed] [Google Scholar]
- 25.Walfridsson, M., J. Hallborn, M. Penttilä, S. Keränen, and B. Hahn-Hägerdal. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61:4184-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witteveen, C. F. B., R. Busink, P. van de Vondervoort, C. Dijkema, K. Swart, and J. Visser. 1989. l-Arabinose and d-xylose catabolism in Aspergillus niger. J. Gen. Microbiol. 135:2163-2171. [Google Scholar]