Abstract
Specific oligonucleotide hybridization conditions were established for single-cell enumeration of uncultivated TM7 and IO25 bacteria by using clones expressing heterologous 16S rRNA. In situ analysis of human subgingival crevice specimens revealed that a greater proportion of samples from sites of chronic periodontitis than from healthy sites contained TM7 subgroup IO25. In addition, IO25 bacterial cells from periodontitis site samples were more abundant and fourfold longer than IO25 cells from healthy site samples.
Fluorescence in situ hybridization (FISH) allows for the quantification of microorganisms in mixed natural communities on a single-cell basis (1). To avoid nonspecific labeling with FISH, however, hybridization stringency conditions for single-nucleotide mismatch discrimination must be determined empirically for each oligonucleotide probe. In the stringency optimization process, one must rely on the availability of cultivated organisms closely related to the target group (10).
The diverse bacterial division TM7, found in contrasting environments ranging from peat bogs to human periodontal pockets, has no known cultivated members (3, 6, 14). The relative abundances of TM7 bacteria and members of the IO25 subgroup have recently been associated with chronic periodontitis and especially mild (presumably early-stage) periodontal disease in humans (3). Previous efforts to establish appropriate levels of hybridization for TM7 group (6) and IO25 subgroup (3) probes relied on cultivatable strains with two and four nucleotide mismatches, respectively, at the probe hybridization site, since there were no known cultivars with a single mismatch. As a result, the hybridization conditions could not be optimized for maximal target discrimination.
We present an alternative approach, cloned artificial targets for FISH (catFISH), for quantifying uncultivated prokaryotes in natural mixed communities using FISH, with single-nucleotide mismatch specificity. In place of cultivated close relatives, we used recombinant Escherichia coli strains that express cloned heterologous 16S ribosomal DNA (rDNA) with zero to four mismatches at the probe hybridization sites. In addition, specific mutations can be engineered into the cloned rDNA. Our objectives were first to establish the hybridization conditions for single-nucleotide mismatch discrimination by probes TM7-905 and IO25-136, which target the TM7 division and the TM7 subgroup IO25, respectively. Our second objective was to quantify IO25 members in dental plaque samples from healthy subgingival crevices and diseased human periodontal pockets.
FISH targets were created for validation of TM7 and IO25 probe specificity with 16S rDNAs amplified from human subgingival specimens and from cultivated strains and then expressed in E. coli (Table 1). The Ancylobacter aquaticus strain was grown in pure culture according to the American Type Culture Collection specifications. For additional details concerning methods, see http://relman.stanford.edu/supplements/catFISH.html. Recombinant pCR4-TOPO plasmid vectors (Invitrogen, Carlsbad, Calif.) containing PCR-amplified, ∼1,380-bp 16S rDNA fragments were either selected from a previous study of human subgingival plaque (clones pSBG1 and pSBG2) or prepared for this study from lyophilized cultures of Mycoplasma felifaucium (pMF1) and Porphyromonas endodontalis (pPE1) (Table 1). The PCR4-TOPO vector had two promoter regions, LacZ and T7, for expression of the plasmid insert. Plasmids were transformed into BL21Star(DE3) chemically competent cells (Invitrogen) and expressed via induction with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h before the transformants were fixed in 10% formalin. To assess the variability of results, nine of the pSBG1 clones and three of the pMF1 clones were independently tested (data not shown). In addition, one single-nucleotide mismatch mutant plasmid for probe TM7-905 (pSBG1a) and one mutant plasmid for probe IO25-136 (pSBG1b) were created from pSBG1 via the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) (Table 1). All 16S rDNA plasmid inserts were fully sequenced.
TABLE 1.
Recombinant 16S rDNA plasmids and strains used in this study
| Probe, plasmid, or strain | Sequenceg (5′-3′) |
|---|---|
| TM7-905 | CCGTCAATTCCTTTATGTTTTA |
| pSBG1a | ---------------------- |
| pSBG1ab | ---------G------------ |
| pSBG1bb | ---------------------- |
| pSBG2a | ---------------------- |
| pMF1c | ---------------A------ |
| A. aquaticusd | --------------GA------ |
| pPEe | --A-----------GA----C- |
| IO25-136 | GTCTTATCCCTCACTGCAGG |
| pSBG1 | -------------------- |
| pSBG1a | -------------------- |
| pSBG1b | -----------G-------- |
| pSBG2 | --CAA-------------C- |
| pMF1 | TCGC------AAT---AGA-f |
| A. aquaticus | AAA----T--GT--GATT--f |
| pPE | -GT-AT-----A-T-A----f |
Created through site-directed mutagenesis of pSBG1.
Created from M. felifaucium ATCC 43428.
ATCC 25396.
Created from P. endodontalis ATCC 35406.
Source: Ribosomal Database Project (9).
The sequences of oligonucleotide probes TM7-905 and IO25-136 are compared with the corresponding sequences at the probe hybridization sites in the plasmids and strain. Only the nucleotides that differ from those of the probe sequences are shown.
Formalin-fixed cells were transferred to Teflon slides with 4-mm-diameter wells (Erie Scientific, Portsmouth, N.H.) in triplicate, posttreated, and subjected to hybridization as previously described (2, 5, 11), with the following modifications. Formamide concentration in the hybridization buffer was varied from 0 to 35% in 5% increments (10). Cy3 or Cy5 fluorescently labeled oligonucleotide probes (Operon, Alameda, Calif.) Control-519 (5′CCTAGTGACGCCGTCGAC3′) (12), Bac338 (5′GCTGCCTCCCGTAGGAGT3′) (8), TM7-905 (5′GTCTTATCCCTCACTGCAGG3′) (6), and IO25-136 (5′GTCTTATCCCTCACTGCAGG3′) (3), were prepared at a final concentration of 5 ng/μl in 5 μl of hybridization buffer per slide well. Hybridizations were performed at 46°C, followed by a 15-min rinse at 48°C in wash buffer, as described by Bond et al. (2). The concentration of NaCl in the wash buffer was varied according to Lathe's equation (8). All cells were counterstained with 1.0 μM YO-PRO-1 DNA dye (Molecular Probes, Eugene, Oreg.) and observed with a laser scanning confocal microscope. Once stringency levels were established for single-mismatch discrimination, TM7-905 and IO25-136 probes were used for cell enumeration in 21 samples from human subgingival sites, 9 from clinically healthy sites and 12 from sites with chronic periodontitis (1 mild, 8 moderate, and 3 severe), as previously described (3). Healthy sites did not exhibit clinical signs of inflammation and had no loss of periodontal attachment, whereas sites with periodontitis exhibited bleeding on probing and had various amounts of loss of bone and clinical attachment as described in detail by Brinig et al. (3). Between 600 and 1,500 cells were counted per probe-YO-PRO-1 combination per sample, and results were expressed as percentages of YO-PRO-1-stained cells that were probe positive. Filaments composed of multiple segments were recorded as a single organism. All experiments beginning with IPTG induction of clones, fixation, and hybridization were repeated at least once. In addition, the morphologies (length, width, and number of segments per cell) of 116 filaments and single cells (14 from healthy sites and 102 from periodontitis sites) labeled with TM7-905-Cy5 and in some cases IO25-136-Cy3 were recorded. Approval for collection and use of clinical specimens was granted by the Stanford University Institutional Review Board for Medical Use of Human Subjects.
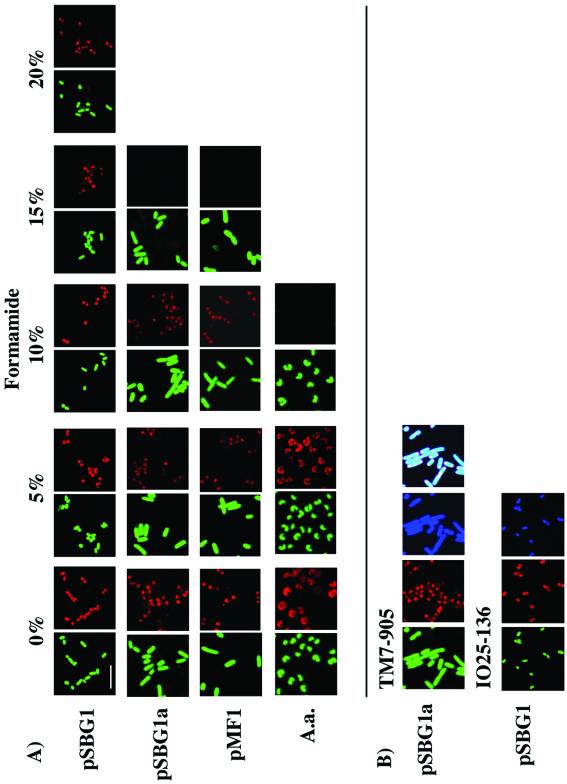
E. coli clones with plasmid inserts containing perfect identity to probe TM7-905-Cy3 (pSBG1, pSBG1b, and pSBG2) were strongly positive following hybridization in buffer containing up to 35% formamide (Fig. 1; data for ≤20% formamide are shown). Clones with a single mismatch to TM7-905 (pSBG1a and pMF1) were undetectable under conditions with ≥15% formamide (Fig. 1A). The A. aquaticus strain, with two mismatches to TM7-905, was visible only with buffer containing <10% formamide (Fig. 1A). Our A. aquaticus 16S rDNA had the same two mismatches at the probe site as the Micrococcus luteus (accession no. M38242) strain used by Hugenholtz et al. (6) to infer hybridization stringency conditions for probe TM7-905. Probe IO25-136-Cy3 also showed its strongest drop in hybridization intensity under conditions with >20% formamide with zero mismatches (pSBG1) and with 10% formamide with one mismatch (pSBG1b) and gave no detectable signal with four mismatches (pSBG2). Hence, hybridization conditions for probes TM7-905 and IO25-136 were established at 20% formamide for discrimination of targets with single-nucleotide mismatches. Positive control Bac338-Cy5-labeled cells were visible with buffer containing up to 35% formamide (Fig. 1B).
FIG. 1.
(A) FISH stringency tests with clones pSBG1, pSBG1a, pMF1, and A. aquaticus and probe TM7-905 at 0 to 20% formamide concentrations. All green panels refer to the use of the nonspecific YO-PRO-1 DNA stain, and red panels refer to the use of the Cy3-labeled TM7-905-specific probe. (B) Staining with TM7-905-Cy3 (10% formamide; clone pSBG1a) or IO25-136-Cy3 (20% formamide; pSBG1), demonstrating, in addition to YO-PRO-1 (green) and probe-Cy3 (red) labels, the positive-control Bac338-Cy5 (blue) probe. The last TM7-905 panel shows colocalization of the three stains with heterologous 16S rRNA at the cell poles. Scale bar = 5 μm for all panels.
Interestingly, TM7-905 and IO25-136 probe fluorescence was strongest at bacterial cell poles (Fig. 1B). Such “segregation” of the 16S rRNA fluorescence signal may be linked to plasmid segregation in E. coli (4, 7). In contrast, Bac338 fluorescence was homogeneously spread throughout the cell; Bac338 recognizes both the heterologous plasmid-encoded and native E. coli 16S rRNA (Fig. 1B). IPTG induction was required for a detectable fluorescence signal. Only expression under the control of the T7 plasmid promoter resulted in cell detection. Likewise, cells were not detectable when labeled with the Control-519 probe or when no probe was used (data not shown). Clones with four mismatches to either the TM7-905 (pPE1) or the IO25-136 (pSBG2) probe were nondetectable, even under conditions with 0% formamide (data not shown).
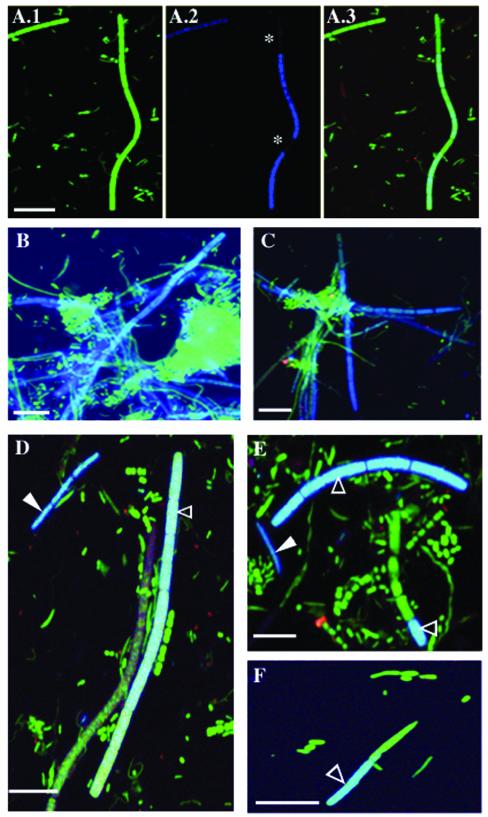
FISH analysis of the microbiota in the human subgingival sites revealed cells that hybridized with the TM7-905 probe in all 12 periodontitis samples (Fig. 2A to E), compared to 44% (4 of 9) of healthy samples. IO25-136-labeled cells were detected in 83% (10 of 12) of the disease samples (Fig. 2D to E) compared to 55% (5 of 9) of the healthy samples (Fig. 2F). The relative abundance of TM7-905-labeled cells was nearly three times higher in disease samples (mean ± standard error [SE], 1.90% ± 0.36%) than in healthy samples (0.67% ± 0.47%), and the difference was statistically significant (P = 0.0492; unpaired t test). IO25-136-labeled cells were 2.5 times more abundant in disease (0.84% ± 0.61%) than in healthy (0.34% ± 0.62%) samples, but this difference was not significant (P = 0.778; unpaired t test).
FIG. 2.
FISH on dental plaque samples from human subgingival sites of moderate chronic periodontitis (A to E) and from healthy sites (F). (A.1 to A.3) Staining with YO-PRO-1 (green) general stain (A.1) and TM7-905-Cy5 (blue) (A.2) and colocalization of these two stains (A.3). Asterisks, two segments of the TM7 filament that are not labeled by the probe. In all other panels, YO-PRO-1 and probe signals are merged as in panel A.3. (B and C) TM7-905 probe and YO-PRO-1-labeled cells (blue-green); (D to F) TM7-905 probe-labeled cells (solid arrowhead, dark blue) plus IO25-136 probe-labeled cells (open arrowhead, light blue) and cells labeled with YO-PRO-1 alone (green). Scale bar = 5 μm in all panels.
The morphology of TM7-905-labeled cells varied from cocci with dimensions of 1.0 by 0.45 μm to filaments 3.0 to 75.0 μm in length and 0.6 to 1.0 μm in width, with an average (± SE) of 3.3 ± 0.22 segments per filament. Not all segments of each TM7 filament were labeled by either the TM7-905 probe (Fig. 2A.2, E, and F) or the other probes used in this study (data not shown), suggesting that these unlabeled segments may be metabolically inactive (13, 16). Average TM7 filament length did not differ significantly between samples from healthy (mean ± SE, 13.61 ± 5.73 μm) and disease sites (16.0 ± 1.8 μm). IO25-136-labeled filaments, however, were more than fourfold longer in disease samples (21.0 ± 2.2 μm; Fig. 2D) than in healthy samples (5.0 ± 1.0 μm; Fig. 2F) (P = 0.0003; unpaired t test) and often coassociated in clusters (Fig. 2B to E).
Overall, these data provide further evidence for a possible role of TM7 division members, and IO25 bacteria in particular, in chronic periodontitis and support earlier conclusions to this effect based on real-time quantitative PCR (3). Even though members of the uncultivated TM7 division exist in relatively low abundance in the human subgingival crevice, the differences in abundance between healthy sites and those with various degrees of disease suggest that they are capable of surviving and growing under a wide range of conditions (high individual plasticity or high group diversity) and may be involved in the development of chronic periodontitis. Time course studies looking at the progression of the microflora at individual subgingival sites could help answer these questions. FISH data pertaining to IO25 also suggest that these filaments, as well as other TM7 members, may be involved in the formation of a scaffold or biofilm, which could support the development of a disease-associated microbial community (Fig. 2B and C). In addition, longer IO25 filaments in disease sites might reflect higher growth rates. Although the longer filaments might correspond to species or strains that are different from those that form shorter filaments, our previous study found no differences in sequence diversity that might support this alternative explanation.
Recently, a clone-based FISH approach for establishment of hybridization stringency conditions was used to screen clone libraries (15). Although our approach was similar, we focused on the challenges associated with the study of uncultivated microbial community members. In theory, catFISH can be applied for use with any 16S rDNA molecule, whether derived from a naturally occurring organism or generated with in vitro recombinant methods. catFISH may be the preferred approach for testing FISH probes in situ when target microorganisms are fastidious or are pathogenic and is certainly the only option when the target is an uncultivated organism.
Acknowledgments
This work was supported in part by NIH postdoctoral training grant 5 T32 AI07328 (Stanford Department of Microbiology and Immunology) and by NIH grant R01 DE13541 and an Ellison Medical Foundation Senior Scholar Award (D.A.R.).
We thank Muna Khan at UCSF Dental School and Paul Lepp, Mary A. Brinig, Elisabeth Bik (who coined the acronym catFISH), Danielle Peres, Christine Dieterich, Caroline Heckman, and Jennifer Maynard at Stanford University for their assistance. Special thanks to Ji-Yun Kim for advice regarding site-directed mutagenesis.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond, P. L., R. Erhart, M. Wagner, J. Keller, and L. L. Blackall. 1999. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl. Environ. Microbiol. 65:4077-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinig, M. M., P. W. Lepp, C. C. Ouverney, G. C. Armitage, and D. A. Relman. 2003. Prevalence and disease association of the uncultivated bacterial division TM7 in human subgingival plaque. Appl. Environ. Microbiol. 69:1687-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 5.Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29-42. [DOI] [PubMed] [Google Scholar]
- 6.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO. J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lathe, R. 1985. Synthetic oligonucleotide probes deduced from amino acid sequence data: theoretical and practical considerations. J. Mol. Biol. 183:1-12. [DOI] [PubMed] [Google Scholar]
- 9.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 11.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouverney, C. C., and J. A. Fuhrman. 1997. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl. Environ. Microbiol. 63:2735-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 16.Zweifel, U. L., and A. Hagström. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts). Appl. Environ. Microbiol. 61:2180-2185. [DOI] [PMC free article] [PubMed]