Abstract
A novel proteinase, PrtR, produced by the human vaginal isolate Lactobacillus rhamnosus strain BGT10 was identified and genetically characterized. The prtR gene and flanking regions were cloned and sequenced. The deduced amino acid sequence of PrtR shares characteristics that are common for other cell envelope proteinases (CEPs) characterized to date, but in contrast to the other cell surface subtilisin-like serine proteinases, it has a smaller and somewhat different B domain and lacks the helix domain, and the anchor domain has a rare sorting signal sequence. Furthermore, PrtR lacks the insert domain, which otherwise is situated inside the catalytic serine protease domain of all CEPs, and has a different cell wall spacer (W) domain similar to that of the cell surface antigen I and II polypeptides expressed by oral and vaginal streptococci. Moreover, the PrtR W domain exhibits significant sequence homology to the consensus sequence that has been shown to be the hallmark of human intestinal mucin protein. According to its αS1- and β-casein cleavage efficacy, PrtR is an efficient proteinase at pH 6.5 and is distributed throughout all L. rhamnosus strains tested. Proteinase extracts of the BGT10 strain obtained with Ca2+-free buffer at pH 6.5 were proteolytically active. The prtR promoter-like sequence was determined, and the minimal promoter region was defined by use of prtR-gusA operon fusions. The prtR expression is Casitone dependent, emphasizing that nitrogen depletion elevates its transcription. This is in correlation with the catalytic activity of the PrtR proteinase.
Lactic acid bacteria (LAB) have multiple amino acid auxotrophies, and in order to grow in protein-rich media they depend on the expression of a complex proteolytic system. Most of the genetic and biochemical studies of the LAB proteolytic system have focused on strains used in production of fermented milk products, and these data have been extensively reviewed (28, 31, 34). Essentially, a cell wall-bound extracellular proteinase (CEP) is responsible for the breakdown of casein, the major milk protein, into oligopeptides. Oligopeptides are then transported via oligopeptide transport systems into the bacterial cell, where the intracellular peptidases hydrolyze different oligopeptides to free amino acids.
Three distinctly different genes encoding CEPs, referred to as prtP, prtB, and prtH (50), have been cloned and sequenced from dairy LAB. The prtP genes from a number of different Lactococcus lactis strains and from Lactobacillus paracasei NCDO151 have been cloned and sequenced (31). The different prtP genes encode proteinases with greater than 95% sequence identity. Distinct prt genes have been found in thermophilic lactobacilli. Genes encoding PrtB and PrtH from Lactobacillus delbrueckii subsp. bulgaricus NCDO1489 and Lactobacillus helveticus CNRZ32, respectively, have been sequenced and characterized (15, 42). In order to produce the enzymatically active PrtP, L. lactis and L. paracasei require the presence of an upstream-located and divergently transcribed prtM gene, encoding a maturation protein (31). No homologue of the prtM gene has been identified in L. delbrueckii subsp. bulgaricus and L. helveticus (15, 42). Virulent streptococci also encode cell wall-bound extracellular proteinases, e.g., the complement-inactivating C5a endopeptidase ScpA from Streptococcus pyogenes and the cell surface proteinase (Csp) from Streptococcus agalactiae (50). ScpA and Csp are the virulence factors of these pathogenic strains. These five types of CEPs from dairy LAB and virulent streptococci belong to a superfamily of subtilisin-like serine proteases, also known as subtilases (49), and they form a separate cluster in the family tree of all known subtilases (50).
Comparative studies of CEPs from diary LAB and virulent streptococci led to the prediction that a number of different functional domains exist (50). Starting from N terminus, PrtP, PrtB, PrtH, ScpA, and Csp consist of the following domains. The prepro domain (PP domain) has been shown to be important for the secretion and activation of proteinase. The catalytic serine protease domain (PR domain) comprises the sequences homologous to subtilases and an internal domain (I domain). The large A domain is present in all CEPs and is immediately preceded by the PR domain. The B domain, which is found only in the CEPs of lactococci and lactobacilli, and a helix spacer domain (H domain) present in PrtP and PrtH follow the A domain. All five types of proteinases mentioned above posses the cell wall spacer (W domain) characteristic of the surface proteins of gram-positive bacteria. The W domain of PrtP and streptococcal CEPs precedes a typical cell wall anchor (AN domain) (40), while PrtH and PrtB lack the C-terminally positioned AN domain and bind the cell wall by means of the W domain itself. The PrtH W domain is similar to a corresponding domain found in surface layer proteins (42).
The regulation of L. lactis proteinase expression has been extensively studied. It has been shown that expression of prtP is repressed in the presence of rich nitrogen sources such as casein hydrolysates (Casitone), Casamino Acids, and specific dipeptides, e.g., leucylproline and prolylleucine (16, 17, 33, 35). Regulation of the proteolytic system in lactobacilli is poorly studied, but it seems to be similar to that of L. lactis (20).
Lactobacilli are natural inhabitants of the human and animal intestinal and urogenital tracts. Besides their use in the production of a variety of food products, considerable evidence has implicated them in a number of potentially beneficial roles within the host (19, 44). Although the molecular bases of their probiotic properties are not very well understood, adhesion to the mucus is considered to be a prerequisite for their survival and establishment in the intestinal or urogenital tract (19). However, there are no data about CEPs of lactobacilli isolated from different ecological niches, such as the human mucosa of either the gastrointestinal or urogenital tract. In this study we identified a novel type of CEP, PrtR, from the human vaginal isolate Lactobacillus rhamnosus BGT10. The prtR gene was cloned, sequenced, and characterized. Operon fusions with a β-glucuronidase reporter gene (gusA) were used to study the medium-dependent regulation of prtR expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. L. rhamnosus BGT10, BGT16, BGT30, and BGT32 were isolated from the vagina of a healthy woman. Swabs were taken and plated on selective media specific for lactobacilli. Strains were identified by using the API 50 CH system (bioMerieux, Marcy l'Etoile, France) and L. rhamnosus-specific PCR primers (51). L. rhamnosus strains BGZL18 and BGZL20 are natural isolates from cheese and were identified by using primers specific for L. rhamnosus. Escherichia coli DH5α was grown in Luria broth medium at 37°C (46). Lactobacillus strains were generally grown in MRS (E. Merck AG, Darmstadt, Germany) at 37°C. L. lactis strains were grown in M17 broth (E. Merck AG) with 10 g of glucose per liter at 30°C. For the test of proteolytic activity, Lactobacillus cells were grown in 10% (wt/vol) pasteurized, reconstituted skim milk and on milk-citrate agar (MCA) plates containing 4.4% reconstituted skim milk, 0.8% Na3-citrate, 0.1% yeast extract, 0.5% glucose, and 1.5% (wt/vol) agar. For all proteinase production and β-glucuronidase expression studies, Lactobacillus cells were grown in chemically defined medium (CDM) for lactobacilli (glucose, 10 g/liter; Na-acetate, 6 g/liter; NH4-citrate, 1 g/liter; K2HPO4, 3 g/liter; KH2PO4, 3 g/liter; MgSO4 · 7H2O, 0.5 g/liter; MnSO4 · H2O, 0.032 g/liter; FeSO4 · 7H2O, 0.02 g/liter; para-aminobenzoic acid, 0.2 mg/liter; folic acid, 0.1 mg/liter; nicotinic acid, 1 mg/liter; pantothenic acid, 1 mg/liter; pyridoxal, 2 mg/liter; riboflavin, 1 mg/liter; biotin, 1 mg/liter; polyethylene sorbitan mono-oleate, 1 ml/liter). CDM was supplemented with Casitone (Difco Laboratories, Detroit, Mich.) at different concentrations (0.1 to 2%, wt/vol). When needed, chloramphenicol (5 μg/ml for L. lactis and 8 μg/ml for Lactobacillus casei) or ampicillin (100 μg/ml for E. coli) was added to the culture medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| L. rhamnosus | ||
| BGT10 | Human vaginal isolate | This study |
| BGT16 | Human vaginal isolate | This study |
| BGT30 | Human vaginal isolate | This study |
| BGT32 | Human vaginal isolate | This study |
| ATCC 7469 | Type strain | American Type Culture Collection |
| BGTM1 | Natural isolate from soft homemade cheese | 53 |
| BGEN1 | Natural isolate from soft homemade cheese | 27 |
| BGZL18 | Natural isolate from soft homemade cheese | This study |
| BGZL20 | Natural isolate from soft homemade cheese | This study |
| CRL576 | Type strain | CERELA Collection |
| CRL932 | Type strain | CERELA Collection |
| CRL1036 | Type strain | CERELA Collection |
| CRL1227 | Type strain | CERELA Collection |
| L. lactis MG1363 | Lac− Prt−; plasmid-free derivative of NCDO 712 | 14 |
| L. casei ATCC 393T | pLZ15− Lac− | 5 |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) supE44 λ−thi-1 gyrA96 relA1 | Laboratory collection |
| Plasmids | ||
| pGEM-T Easy | Ampr, M13ori pBR322ori, linear T-overhang vector | Promega |
| pUC18 | Ampr | 54 |
| pXS200 | Ampr, pUC18 carrying 2-kb XbaI-SphI chromosomal fragment from 3′ part of prtR gene | This study |
| pNZ273 | Cmr, promoter probe vector containing promoterless gusA gene | 43 |
| pEB640 | pNZ273 carrying T10Bam-T10Eco PCR fragment in front of the gusA gene | This study |
| pEB260 | pNZ273 carrying TR1035-T10Eco PCR fragment in front of the gusA gene | This study |
| pEB122 | pNZ273 carrying TR180-T10Eco PCR fragment in front of the gusA gene | This study |
| pEB471 | pNZ273 carrying T10Bam-TR237 PCR fragment in front of the gusA gene | This study |
| pEB99 | pNZ273 carrying TR1035-TR237 PCR fragment in front of the gusA gene | This study |
Proteinase activity assay.
Proteolytic activities of Lactobacillus strains were assayed as described previously (26). For enzyme assays, the strains were grown on MCA plates or CDM supplemented with Casitone for 48 h at 37°C prior to cell collection. Collected fresh cells (10 mg; approximate density, 1010 cells/ml) were resuspended in 100 mM K-phosphate buffer (pH 6.5). The cell suspension was mixed with the substrate αs1-, β-, or κ-casein (5 mg/ml) (Sigma, St. Louis, Mo.) dissolved in the same buffer at a 1:1 volume ratio. After incubation for 3 h at 30°C, the cells were pelleted by centrifugation, the clear supernatant fluid was taken, and samples for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were prepared.
PAGE.
Samples for analysis by SDS-PAGE were prepared by heating (100°C) for 2 min with an equal volume of 0.125 M Tris-HCl (pH 6.8) containing 4% (wt/vol) SDS, 20% (wt/vol) glycerol, and 10% (vol/vol) β-mercaptoethanol. SDS-PAGE was carried out on 15% polyacrylamide gels on vertical slab electrophoresis cells for 20 h at a 20-mA constant current, with staining with Coomassie brilliant blue G250 and destaining in a mixture of methanol (20%) and acetic acid (7%).
Influence of Ca2+ ions on proteinase activity.
To analyze the influence of Ca2+ ions on L. rhamnosus BGT10 proteinase activity, cells were grown either in MCA medium or in CDM supplemented with 0.1% Casitone and washed twice in 100 mM K-phosphate buffer (pH 6.5), and the extracts were pooled. The substrate (β-casein [5 mg/ml] in K-phosphate buffer [pH 6.5]) was mixed with crude extracts at a 1:1 volume ratio and incubated for 3 h at 30°C, and samples for SDS-PAGE were prepared.
DNA manipulation procedures.
Molecular cloning was done essentially as described by Sambrook et al. (46). Restriction enzymes, T4 DNA ligase, and deoxynucleotides were obtained from Pharmacia (Vienna, Austria) or New England Biolabs (Beverly, Mass.) and used according to the instructions of the suppliers. Large-scale isolation of E. coli plasmids for nucleotide sequence analysis was performed with the Jet Star system (Genomed GmbH, Bad Deynhausen, Germany) according to the manufacturer's instructions. Transformation of E. coli was performed as described by Miller (36). Transformation of L. lactis and plasmid isolation from L. lactis and L. casei were done as described previously (1, 22). L. casei ATCC 393T was electroporated as described by Walker et al. (55). Large-scale isolation of L. lactis plasmids for nucleotide sequence analysis was performed with the Jet Star Maxi system with a following modification: mid-log-phase cells in 200 ml of GM17 were pelleted, resuspended in 4 ml of buffer (20% sucrose, 10 mM Tris, 50 mM NaCl, 10 mM EDTA [pH 8.0]), 40 mg of lysozyme per ml, and incubated for 1 h at 55°C. After this step, isolation proceeded according to the manufacturer's instructions.
Identification of the prtR gene.
The strategy for isolation of the prtR gene was as follows. The first part of the prtR gene was isolated by amplification of the chromosomal DNA of L. rhamnosus strain BGT10 by PCR using degenerate primers. Two degenerate primers, prt1 and prt2r, were designed from an alignment of the conserved regions surrounding the active-site residues of the protease genes (Asp32 and His64; numbering is that of Bacillus amyloliquefaciens subtilisin BPN′). The sequences of the prt1 and prt2r primers, containing the EcoRI and BamHI sites as 5′ extensions, respectively, were as follows: prt1, 5′-CCGCGCGAATTCGAYWSNGGNRTNSAN-3′, and prt2r, 5′-CCGCGCGGATCCCCNGCNACRTGNRTNCCRTG-3′ (where Y is T or C; K is G or T; W is A or T; S is C or G; R is A or G; and N is A, C, G, or T). The primers were synthesized by Genosys. PCR amplifications were performed with a Perkin-Elmer (Norwalk, Conn.) model 9600 thermal cycler and a gradient PCR machine (Eppendorf, Hamburg, Germany) by using Taq polymerase (Pharmacia). An inverse-PCR strategy was used to identify adjacent DNA regions (46). BGT10 chromosomal DNA (1 to 4 μg) was digested with an appropriate restriction enzyme (HindIII, XbaI, or PstI), precipitated with ethanol, and resuspended in 7 μl of deionized H2O. The digested DNA was self-ligated overnight at 15°C in a 10-μl reaction mixture. Ethanol-precipitated ligation mixture was used as a template for PCR. By inverse PCR with primers prti1 (5′-ACTGAGGCGTAAGTCGTCGT-3′) and prti2 (5′-CAACACCGGGACCACGGTG-3′) we obtained 1,660-, 2,160-, and 2,620-bp DNA fragments. After sequencing, new primers were designed for a second round of inverse PCR. Novel inverse PCR fragments of 850 and 1,560 bp were obtained with the primer pairs IP5 (5′-GATAATGGCAGCCGTCATCC-3′)-prti2 and INPst345 (5′-TGTTGTATTGTTCGGTCACCAGACG-3′)-ct1 (5′-AGTCACCAAGGTCTTCCACG-3′), respectively. All PCR fragments were cloned directly into pGEM-T Easy vector (Promega, Madison, Wis.) and sequenced.
Nucleotide sequencing and sequence analyses.
Sequencing was performed with a model A.L.F. DNA sequencer (Pharmacia). The dideoxy sequencing reactions (47) were performed by using the methods recommended in the AutoRead sequencing kit manual (Pharmacia). Both DNA strands were sequenced with pUC18-specific (universal and reverse) primers and the sequence-specific oligonucleotides used for primer walking. Additional sequence data were obtained from sequencing an XbaI-SphI chromosomal fragment (2.082 kb of the prtR 3′ region) cloned onto pUC18, creating plasmid pXS200. The Clone5 software package was used for assembling and analyzing DNA sequences. Protein secondary structure prediction was performed by the methods of Garnier et al. (13), Chou and Fasman (6), and Rost and Sander (45). Hydrophobicity analyses were performed by the method of Kyte and Doolittle (32). Signal peptide prediction was made with the SignalP server (41). Protein homology searches were carried out with the SWISS-PROT database with the EMBL BLAST and EMBL FASTA servers. All reported DNA sequence data were confirmed by sequencing both DNA strands from at least two independent cloned PCR products.
Southern blot analyses.
Chromosomal DNA from L. rhamnosus was isolated by the procedure described by Hopwood et al. (23). Different probes were used to analyze the potential presence of known prt genes in L. rhamnosus BGT10. PCR amplification with primers JP22 and JP23 (42) and PRTB10 and PRTB20 (11) was used to synthesize the probes from the catalytic domains of prtH and prtB, respectively. Probes Q1 and Q6 from L. lactis Wg2 (26) were used to analyze the potential presence of the prtM and prtP genes, respectively. All probes were labeled by using the BioNick labeling system (GibcoBRL Life Technologies, Gaithersburg, Md.). The hybridizations were performed with the PhotoGene nucleic acid detection system (BRL). Hybridizations were carried out at 45°C.
For detection of the prtR DNA fragment on plasmid pXS200, a XbaI-KpnI probe comprising the nucleotide sequence of the prtR gene from nucleotide 2892 to 3334 was used. Hybridization was carried out at 65°C.
A 1-kb internal fragment from the catalytic domain of prtR was used as a probe to determine the distribution of prtR among various L. rhamnosus strains. The probe was made by PCR amplification with primers prti2 and IP6RXba (5′-TCTGATCGTGGACGGTGTTGC-3′). Hybridizations were carried out at 65°C.
Primer extension analysis.
RNA was isolated from exponentially growing L. rhamnosus BGT10 and L. casei ATCC 393T cells carrying pEB640 and pEB471 by the Macaloid method described by Kuipers et al. (29). Primer extension reactions were performed by annealing 20 ng of oligonucleotides specific for the 5′ end of prtR (IP5) or the gusA reporter gene (GUS-AS) (52) to 40 μg of RNA as described previously (29). The radioactive products of the reactions were run on a denaturing sequencing gel next to the DNA sequence (47) generated with the same oligonucleotide primers.
Construction of prtR-gusA operon fusions.
To make operon (transcriptional) fusions with the promoterless β-glucuronidase gene, different fragments of the prtR regulatory region were amplified by PCR with Taq polymerase (Pharmacia). Forward primers T10Bam (5′-CGCGGATCCGAAACTGTCTAATCAGTACC-3′), TR1035 (5′-CGCGGATCCTTTGGCGCTTGACACAGGCG-3′), and TR180 (5′-CGCGGATCCGCGGTTATTAAGAAGAAATG-3′) (with a BamHI extension at the 5′ end) and reverse primers T10Eco (5′-CCGGAATTCACTAAACTCGTCCCAAATGT-3′) and TR237 (5′-CCGGAATTCCCAAAATACCCAAAGAAATCACA-3′) (with an EcoRI extension at the 5′ end) were used. The primers were synthesized by Genosys. The products T10Bam-T10Eco (640 bp), TR1035-T10Eco (260 bp), TR180-T10Eco (122 bp), T10Bam-TR237 (471 bp), and TR1035-TR237 (99 bp) were digested with BamHI and EcoRI restriction enzymes and ligated with vector pNZ273 (52) in order to construct plasmids pEB640, pEB260, pEB122, pEB471, and pEB99, respectively. L. lactis MG1363 was used as an intermediate host for transformations with the ligation mixtures. Transformants containing constructs were selected by PCR with the same primers used for operon fusion constructions. The constructs obtained were used to transform L. casei ATCC 393T.
GusA enzyme assay.
The activity of the β-glucuronidase (GusA) enzyme was measured as described previously (30). Protein concentrations were determined as described by Bradford (2), using the Bio-Rad protein assay.
Nucleotide sequence accession number.
The EMBL accession number for the prtR nucleotide sequence reported in this paper is AJ496666.
RESULTS
Identification of human vaginal L. rhamnosus strains.
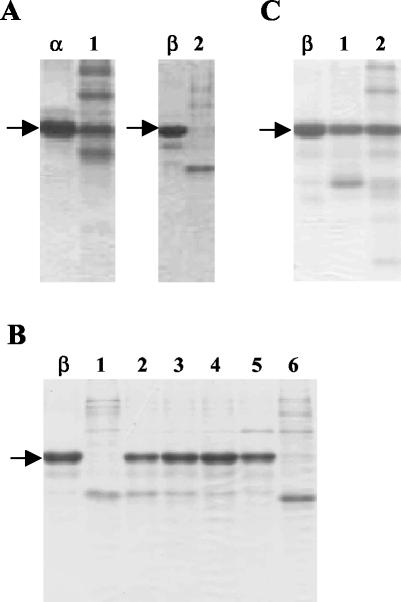
Human vaginal isolates of Lactobacillus species were identified and screened for their proteolytic activities (see Materials and Methods). Among them, we identified isolates of L. rhamnosus by using specific PCR primers (51), and the individual strains were named BGT10, BGT16, BGT30, and BGT32. The proteolytic activities of BGT10, -16, -30, and -32 whole cells grown on MCA were monitored by the efficiency of hydrolysis of different caseins. All strains degraded the αS1- and β-caseins at pH 6.5 (Fig. 1A [data shown only for BGT10]), while the κ-casein was not hydrolyzed (data not shown).
FIG. 1.
Proteolytic activity of L. rhamnosus BGT10. (A) Caseins hydrolyzed by whole cells collected from MCA plates. Lanes: α, αS1-casein substrate; 1, hydrolyzed αS1-casein; β, β-casein substrate; 2, hydrolyzed β-casein. (B) Nutrient-dependent proteolytic activity of L. rhamnosus BGT10. Lanes: β, β-casein substrate; 1 to 4, β-casein hydrolyzed by whole cells collected from CDM plates containing 0.1, 0.5, 1, and 2% Casitone, respectively; 5, whole cells collected from MRS plates; 6, whole cells collected from MCA plates. (C) Hydrolysis of β-casein by the proteinase extracts. Lanes: β, β-casein substrate; 1, β-casein hydrolyzed by extract obtained from cells grown on CDM plates containing 0.1% Casitone; 2, β-casein hydrolyzed by extract obtained from the cells grown on MCA plates. Arrows indicate α- or β-casein.
Proteolytic activity of L. rhamnosus BGT10.
The proteolytic activity of whole cells of L. rhamnosus BGT10 was determined by monitoring β-casein hydrolysis. For this purpose, cells were grown on either CDM containing Casitone, peptide-rich growth medium (MRS), or MCA plates. The greatest β-casein hydrolysis was obtained at pH 6.5 when the cells were grown on CDM plates supplemented with 0.1% Casitone (Fig. 1B, lane 1) or on MCA plates (Fig. 1B, lane 6). Lower proteolytic activities of the BGT10 proteinase were detected when the cells were grown on CDM plates supplemented with the higher concentrations of Casitone (0.5 and 1%). Hydrolysis of β-casein was negligible when the cells were grown on MRS or CDM plates containing 2% Casitone (Fig. 1B).
The minimal lysis treatment is the most widely used procedure for releasing the proteinase from the cell envelope of LAB, by washing the cells with a Ca2+-free buffer (37). Active proteinase extracts at pH 6.5 were obtained by washing the L. rhamnosus BGT10 cells in Ca2+-free K-phosphate buffer (Fig. 1C). When cells were propagated on CDM plates supplemented with 0.1% Casitone, they showed growth in mass as a layer, while cells on MCA plates grew in the agar. Consequently, the extract of cells grown on CDM plus 0.1% Casitone showed stronger proteolytic activities than the extract obtained from the cells grown on MCA medium (Fig. 1C).
Finally, L. rhamnosus BGT10 coagulated milk and reduced the pH to 4.8 after 27 h of incubation at 37°C, using a 2% inoculum.
Identification and sequencing of a cell wall-bound extracellular proteinase gene, prtR, from L. rhamnosus BGT10.
In order to define the prt locus in human vaginal isolates of L. rhamnosus, we initially used probes deduced from the previously characterized CEP genes prtP, prtH, and prtB, as well as the PrtP maturation protein gene, prtM, in Southern blot analyses. Chromosomal DNA isolated from L. rhamnosus strains, including strain BGT10, did not hybridize either with probes encoding the catalytic regions of PrtP, PrtB, and PrtH or with a probe characteristic of the prtM gene (data not shown).
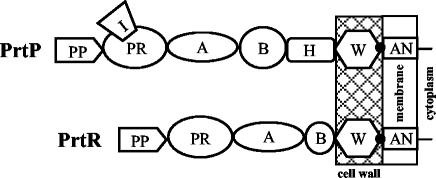
Subsequently, PCR with degenerate primers was used to identify and clone part of the catalytic domain of the CEP gene from L. rhamnosus BGT10 (see Materials and Methods). A single 214-bp product (prt1-prt2r fragment) was obtained (data not shown) and sequenced. New primers were designed based on the sequence of this product, and inverse PCR was used to identify adjacent DNA regions. Additional sequence data were obtained from sequencing a 2.1-kb XbaI-SphI chromosomal fragment (pXS200). In total, we sequenced 5.1 kb of double-stranded DNA containing a 4,443-nucleotide putative proteinase gene named prtR (R for L. rhamnosus). The encoded modular protein PrtR was 1,480 amino acids with a deduced molecular mass of 154 kDa and showed significant amino acid sequence similarity to most functional domains of previously characterized CEPs (see below) (Fig. 2).
FIG. 2.
Schematic representation of PrtP as a prototype for the subtilases and the PrtR of L. rhamnosus strain BGT10, showing different domains according to the model of Siezen (50). PP, prepro domain; PR, protease domain; I, insert domain; A, A domain; B, B domain; H, helix domain; W, cell wall spacer domain; AN, anchor domain; black dot, sorting signal.
Domain structure of PrtR and comparison with other CEPs.
Analysis of the deduced PrtR sequence revealed the existence of a domain structure characteristic of CEPs from lactococci, lactobacilli, and virulent streptococci (Fig. 2). The PrtR has a putative 33-amino-acid N-terminal secretion signal sequence with a predicted cleavage site between positions A33 and D34. Adjacent to the signal peptide is a putative pro sequence of 147 amino acids that does not share significant amino acid homology with pro sequences of other CEPs. An obvious consensus sequence for autoproteolytic cleavage is not present. The potential length of the PrtR propeptide (147 amino acids), the alignment of the N-terminal part of the subsequent catalytic domain with other CEPs, and the position of Pro before and Asn after the putative cleavage site found in PrtR (e.g., in Csp, SKETPQ↓N) suggest that the autoproteolytic site for the PrtR propeptide could be between A147 and N148. Therefore, the PrtR might be synthesized as a prepro enzyme. Such processing would result in a mature PrtR of 1,300 amino acids.
The N-terminal region of the mature PrtR proteinase (370 amino acids) has all of the characteristics of the CEP catalytic domain. This region has similarity to the subtilisin-like serine proteinases (subtilases); therefore, PrtR should be classified within this family. The subtilase family of proteins are characterized by a catalytic triad, Asp-His-Ser (D-H-S) (49). The putative catalytic residues of PrtR are at positions D29, H93, and S298. The catalytic sites, as well as adjacent residues such as the oxyanion hole residue N200 of PrtR, are very well conserved among CEPs and other members of the subtilase family (49). The catalytic domain of PrtR has 38.6% identity with the same domain of PrtB. PrtR does not contain the I domain (Fig. 2), which is otherwise present in PrtP, PrtH, PrtB, streptococcal CEPs, B. subtilis Vpr protease, and plant subtilases.
The putative A domain of PrtR (415 amino acids) is located after the catalytic domain. Eight out of nine segments of the A domain from PrtR have high sequence identity with suggested conserved sequences derived from PrtP, PrtH, PrtB, Csp, and ScpA (50) (Fig. 3A). The fourth segment, VRVLDKE, is different from that in other characterized CEPs, but it retains the β-sheet secondary structure characteristic of all conserved sequences. The sequence predicted to be a B domain (348 amino acids) of PrtR follows the A domain, and it is approximately 100 residues shorter than the B domains (500 amino acids) of PrtP, PrtH, and PrtB (50) (Fig. 2). The PrtR B domain has a high level of predicted β-sheet secondary structures and, similarly to PrtP, a high level of Thr residues (14%). The overall sequence identity between the B domains of previously characterized CEPs and PrtR is not significant, but there are some similarities with the proposed conserved segments found in PrtP (Fig. 3B). Additionally, the 100% identical sequence TEPAKTVT at the end of this domain is present in PrtP from L. paracasei NCDO151 and L. lactis Wg2 and PrtR from L. rhamnosus BGT10 (Fig. 3B). PrtR entirely lacks the H domain (Fig. 2).
FIG. 3.
The A, B, and W domains of PrtR. (A) Alignment of the A-domain segments of L. rhamnosus PrtR with conserved residues from PrtP (L. lactis), PrtH (L. helveticus), PrtB (L. delbrueckii subsp. bulgaricus), Csp (S. agalactiae), and ScpA (S. pyogenes). Consensus residues were derived from an alignment of known CEPs; uppercase indicates identical residues, and lowercase indicates highly conserved residues (50). (B) Comparison of B-domain segments of PrtR with conserved sequences residues of PrtP and consensus residues derived from known CEPs. Consensus residues (Con.) are as described for panel A. Identical amino acids are shaded in black; homologous residues are shaded in grey. (C) Alignment of the W domain of PrtR from L. rhamnosus strain BGT10 and the C-terminal part of SspB from S. gordonii (8). Identical amino acids are shaded in black; homologous residues are shaded in grey.(D) Alignment of the W domain of PrtR from L. rhamnosus strain BGT10 and a fragment of the human intestinal mucin (IMuc) (18). Identical amino acids are shaded in black; homologous residues are shaded in grey.
A 133-amino-acid fragment of the PrtR C-terminal region is predicted to be the cell wall spacer (W domain). Like the W domains of PrtP and ScpA, this domain of PrtR is quite hydrophilic; is Thr, Pro, and Lys rich (47%); and lacks some hydrophobic and aromatic residues (C and W). The region comprising 100 amino acids of the PrtR W domain exhibits 35% identity to the C terminus of the Streptococcus gordonii surface proteins SspA and SspB (amino acid residues 1365 to 1464 and 1432 to 1531, respectively) (9) (Fig. 3C). Moreover, the same 100 amino acids of the cell wall spacer domain of PrtR has Thr- and Pro-rich imperfect repeats homologous to consensus sequences of human intestinal mucin (22) (Fig. 3D).
The W domain of PrtR precedes a putative cell wall anchor (Fig. 2). The C-terminal sequence of PrtR has most characteristics of the AN domain but differs in the sorting signal. Instead of typical sorting signal (LPxTG), the sequence MPQAG is present. This unusual sequence is followed by a hydrophobic putative membrane-spanning α-helix and a small, four-residue-long charged tail.
Distribution of prtR within L. rhamnosus strains.
Southern blot analysis was used to determine the distribution of prtR among various strains of L. rhamnosus (BGT10, BGT16, BGT30, BGT32, BGTM1, BGEN1, BGZL18, BGZL20, ATCC 7469, CRL576, CRL932, CRL1036, and CRL1227). All L. rhamnosus strains tested exhibit cell surface proteinase activity, as shown by the hydrolysis of β-casein (data not shown). Under high-stringency condition (65°C), the probe from the catalytic domain of L. rhamnosus BGT10 prtR hybridized to a 2.3-kb XbaI DNA fragment in all tested strains. This band is specific for the prtR gene, since this region lacks the sequence that encodes the I domain (data not shown).
Determination of prtR minimal promoter region and Casitone-dependent regulation of prtR expression.
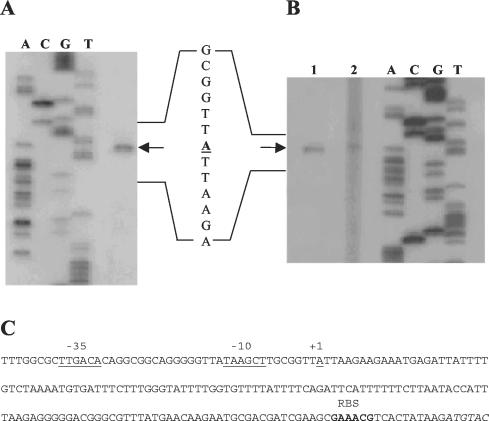
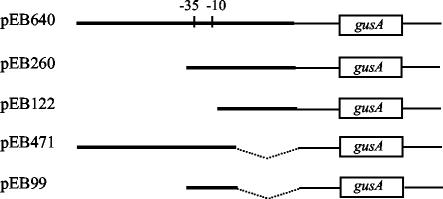
Total RNA was isolated from log-phase L. rhamnosus BGT10 cells grown in MRS medium. The prtR transcription start was identified by primer extension and was located 154 nucleotides upstream of the proposed prtR ATG start codon (Fig. 4). The same transcription start point was identified for RNA isolated from L. casei ATCC 393T harboring pEB471 or pEB640 (Fig. 4A and B). A putative promoter-like sequence with the configuration TTGACA-N17-TAAGCT was found (Fig. 4C). We designed an experiment to determine the minimal promoter region and to analyze the regulation of the prtR gene. Different parts of the prtR regulatory region were used to construct operon fusions with a β-glucuronidase reporter gene (gusA) (Fig. 5). These fusions were found to be inactive in L. lactis. Therefore, L. casei ATCC 393T was used as a host to accommodate the prtR-gusA operon fusions. Deletion analysis of the prtR regulatory region was performed by using constructs pEB640, pEB260, pEB122, pEB471, and pEB99. Comparison of the GusA activities of these operon fusions revealed that only construct pEB122 lacks an active promoter (Table 2). The results showed that the smallest construct, pEB99, is active (Table 2) and carries the minimal promoter region of the prtR gene situated on a DNA fragment with the 5′ end 44 bp upstream and the 3′-end 54 bp downstream from the prtR transcription start point (Fig. 4 and 5).
FIG. 4.
Determination of the prtR transcription start point. (A) Primer extension analysis of the prtR promoter was performed with RNA from L. casei ATCC 393T cells harboring pEB471. (B) L. casei ATCC 393T cells harboring pEB640 (lane 1) and L. rhamnosus BGT10 (lane 2). The sequencing reactions (A, C, G, and T) were performed with the same primers as the corresponding primer extension reactions. The primer extension products are indicated with an arrow. (C) Interpretation of the primer extension results. The putative −35 and −10 prtR promoter haxamers (underlined), the transcription start point (+1, underlined), and the putative ribosome binding site (RBS) (boldface) are shown. The prtR coding sequence is in italic.
FIG. 5.
Schematic representation of different prtR-gusA operon fusions. The putative prtR promoter position (−35 and −10 hexanucleotides) is marked. Thick lines represent different fragments of the prtR regulatory region, and dashed lines represent deleted regions.
TABLE 2.
Expression of the prtR-gusA fusions in CDM supplemented with casitone and in MRS broth
| Transformant | β-Glucuronidase activity (mean ± SD)a in:
|
Induction ratio (0.1%/2% Casitone) | ||||
|---|---|---|---|---|---|---|
| CDM supplemented with Casitone at:
|
MRS | |||||
| 0.1% | 0.5% | 1% | 2% | |||
| ATCC 393T/pEB640 | 93 ± 44 | 31 ± 23 | 8 ± 6 | 10 ± 5 | 3 ± 2 | 9.30 |
| ATCC 393T/pEB260 | 181 ± 86 | 50 ± 13 | 20 ± 8 | 14 ± 5 | 3 ± 1 | 12.93 |
| ATCC 393T/pEB471 | 121 ± 51 | 54 ± 24 | 45 ± 34 | 20 ± 13 | 15 ± 7 | 6.05 |
| ATCC 393T/pEB99 | 337 ± 167 | 129 ± 57 | 75 ± 10 | 50 ± 14 | 12 ± 2 | 6.74 |
| ATCC 393T/pEB122 | 0 | 0 | 0 | 0 | 0 | |
Expressed as nanomoles per minute per milligram of protein. Values are means and standard deviations from three independent determinations.
Since the proteolytic activity of whole cells of L. rhamnosus strain BGT10 was Casitone dependent, we decided to study the expression of prtR under different growth conditions. L. casei ATCC 393T harboring either pEB640, pEB260, pEB471, or pEB99 was used to study the regulation of prtR expression. For that purpose, cells were grown in CDM containing 0.1, 0.5, 1, and 2% Casitone or in MRS broth. The results of GusA assays strongly indicated that the prtR promoter is regulated by Casitone (Table 2). GusA activities gradually decreased with increasing concentration of Casitone in the CDM. The minimal activity was achieved in the nitrogen-rich MRS medium. Note that the induction rate is the highest in constructs containing the 3′ regulatory region positioned downstream of the prtR promoter-like sequence. Maximal expression of the prtR-gusA operon fusion was obtained with construct pEB99, which lacks both 5′ and 3′ regions, upstream and downstream of the prtR promoter-like sequence, respectively (Table 2).
DISCUSSION
The prtR gene of the human isolate L. rhamnosus BGT10 encodes a novel proteinase. Characterization of this CEP could be relevant for understanding the functionality of CEPs and their domains or of cell surface proteins carrying similar functional domains. Since L. rhamnosus is already available commercially as a probiotic, further analyses will enable estimation of the possible application of BGT10 as a starter in the food industry and in dairy products.
L. rhamnosus BGT10 exhibits proteolytic activity and degrades αS1- and β-caseins. The efficiency of proteolysis was optimal at pH 6.5 when the bacteria were grown in peptide-depleted medium such as MCA or CDM supplemented with 0.1% Casitone. The 4,443-bp prtR gene of BGT10 encodes a 154-kDa modular protein with functional domains resembling the CEPs previously identified in dairy LAB and streptococci, revealing the main features characteristic of the members of the superfamily of subtilisin-like serine proteases, subtilases (50). The PrtR N-terminal PP domain carries a putative sec-dependent translocation signal sequence (10) and a pro sequence. The catalytic PrtR serine protease PR domain exhibits homology to subtilases. A striking feature of the PrtR PR domain is a complete lack of the I domain, which is otherwise present in all CEPs of LAB (50). Usually, the I domain is 134 to 156 residues long and modulates the activity and specificity of the PR domain itself (3, 50). Deletion of the PrtP I domain reduces the proteolytic activity for caseins (3). It is possible that the lack of the I domain in PrtR accounts for the low efficiency of proteolytic degradation of caseins in milk, where they are not directly accessible.
The putative A domain of PrtR, which is predicted to regulate the proteolytic activity and/or specificity of the PR domain of other CEPs, shares all sequence and structural characteristics seen previously (48, 50, 53). It seems that this domain is not dispensable in CEPs and is very important for the proteolytic activity. The A domain of PrtR precedes a B domain found only in PrtP, PrtH, and PrtB (50). This domain, which is predicted to provide stability and/or protection against autoproteolysis (4, 9), is much smaller in PrtR and has 3 out of 12 segments homologous to the PrtP B domain. On the very C-terminal end of the B domain, PrtR and PrtP share a motif with 100% sequence identity, TEPAKTVT. No exact function can be assigned to this sequence at this point. However, since PrtR as well as PrtP (26, 37, 39) could be liberated from the cell wall by washing the cells in Ca2+-free buffer, we can speculate that this sequence in these CEPs could have some role in autoproteolytic cleavage.
Like PrtB and ScpA, PrtR does not have the H domain present in the protein structure, and hence the W domain is situated immediately after the B domain. The putative cell wall spacer W domain of PrtR has similarities to the corresponding domains in PrtP and ScpA. In addition, we found that 100 amino acids out of 133 present in this domain show a high sequence identity to C-terminal regions of two cell surface antigen I and II polypeptides, SspA and SspB, expressed by oral and vaginal commensal S. gordonii strains (7, 8, 24). These polypeptides are multifunctional cell surface adhesins involved in colonization of oral tissues and adhesion of these bacteria to human and bacterial receptors (8). More precisely, the C-terminal regions of these proteins provide adhesion to immobilized glycoproteins (24). The same region of the PrtR W domain contains the imperfect repeats of threonine and proline that are homologous to the consensus sequence characteristic of the human intestinal mucin (18, 38). The vaginal tract is colonized by a complex resident microflora with a potential protective role. Lactobacillus is the most prominent member of the vaginal microflora, and it has been suggested that this bacterium helps to prevent pathogens from colonizing the vagina by keeping the pH low and by forming a biofilm (44). Mucus is a mixture of protein mucin and carbohydrates; it covers the mucosal membranes of the urogenital tract, including the vagina, and protects from colonization by bacterial pathogens as well. It has been shown that two or more repeats of either the L. lactis BGMN1-5 or L. lactis SK11 proteinase W domain are involved in enhanced sedimentation of bacteria and facilitate adhesion and interaction with human cancer cells of the small intestine (12). Taken together, these facts and predictions imply some direct and accessory roles that could be assigned for the W domain of the human vaginal isolate CEP, PrtR. The primary role should be to link the proteinase to the cell wall and to localize this CEP on the cell surface of bacteria. The accessory role might be to facilitate the adhesion of L. rhamnosus to vaginal mucosa and/or involvement in biofilm formation. The potential application of PrtR, the W domain itself, or specific peptides extracted from the PrtR W domain could be to compete with bacterial pathogens and protect the human vaginal or intestinal mucosa from their adhesion and colonization. Human Lactobacillus isolates carrying CEPs such as PrtR might have potential as probiotics. It would be interesting to investigate possible W domain resistance of such proteinases or extracted peptides to the enzyme mucinase, the virulence factor produced extensively by some pathogens (e.g., Vibrio cholerae) during colonization of mucosa in different tracts.
The C terminus of PrtR resembles the AN domain that is found and conserved in many surface proteins (40). The AN domain of PrtR has an unusual sorting signal, MPQAG instead of LPxTG. Unlike the other CEPs, at position 4 in the sorting signal PrtR carries the amino acid alanine instead of threonine, which has been shown to be important for peptide cleavage and cell wall binding (40). Hence, the PrtR cell wall sorting signal is more similar to those found in the Listeria monocytogenes proteins internalin C2 and internalin D and the Peptostreptococcus magnus proteins L and PAB (40). Additionally the PrtR sorting signal carries an even more rare but neutral change at position 1, methionine instead of leucine. It would be interesting to test and analyze the AN domains and sorting signals of other L. rhamnosus cell surface proteins and to determine whether this MPQAG sequence is a signal specific for this species.
The sequencing of prtR flanking regions as well as the Southern blot analyses revealed that no homologue of the prtM gene could be identified in L. rhamnosus BGT10, BGT16, BGT30, or BGT32. PrtM is a maturation protein that is required for the activation of PrtP-like proteinases and is not found in strains encoding the other CEPs, such as PrtH, PrtB, Csp and ScpA (50). This does not exclude the presence of a PrtM analogue protein in L. rhamnosus strains.
The growth of L. rhamnosus strains depends on proteinase activity, since their niches are rich in proteins but depleted of free amino acids regardless of their origin. All 14 L. rhamnosus strains that we tested, including some human vaginal isolates, some isolated from fermented milk products (e.g., cheese), and some representing strains from different collections, were proteolytically active and carried the prtR gene, suggesting that this proteinase gene could be characteristic of the species L. rhamnosus.
The prtR promoter-like sequence is similar to promoters found in lactococci and lactobacilli (25). A long distance between the transcription start and the start of the gene is characteristic of the proteinase genes of LAB (15, 21, 42). A minimal prtR promoter region determined by deletion analysis of the regulatory region is 99 nucleotides long and comprises the identified putative prtR promoter-like sequence (pEB99), implying that this may be the active promoter. prtR expression is decreased in peptide-rich medium, while nitrogen depletion (0.1% Casitone) elevates the activity of the prtR promoter. The results of deletion analysis experiments showed that both 3′ and 5′ prtR promoter flanking sequences might contribute to the Casitone-dependent regulation. These results and the patterns of prtR expression are in correlation with the proteolytic activities obtained for whole cells of L. rhamnosus BGT10. This marked decrease of proteolytic activity with increasing concentrations of Casitone is also in accordance with the results obtained with L. lactis PrtP and L. helveticus PrtH (16, 20, 33, 35). Therefore, we assume that when nutrition-dependent negative regulation is lifted, expression is up-regulated and the proteolytic activity of PrtR is elevated.
Acknowledgments
We thank Raúl R. Raya for sharing the strains of the CERELA collection and Gaspar Pérez-Martínez for kindly providing L. casei strain ATCC 393T.
This work was supported by MSTDRS grant 1442 and by an FEMS Young Scientist grant awarded to I.P.
REFERENCES
- 1.Bojovic, B., G. Djordjevic, and L. Topisirovic. 1991. Improved vector for promoter screening in lactococci. Appl. Environ. Microbiol. 57:385-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 12:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bruinenberg, P. G., P. Doesburg, A. C. Alting, F. A. Exterkate, W. M. de Vos, and R. J. Siezen. 1994. Evidence for a large dispensable segment in the subtilisin-like catalytic domain of the Lactococcus lactis cell-envelope proteinase. Protein Eng. 7:991-996. [DOI] [PubMed] [Google Scholar]
- 4.Bruinenberg, P. G., W. M. De Vos, and R. J. Siezen. 2000. Deletion of various carboxy-terminal domains of Lactococcus lactis SK11 proteinase: effects on activity, specificity, and stability of the truncated enzyme. Appl. Environ. Microbiol. 66:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassy, B. M., and L. Flickinger. 1987. Transformation of Lactobacillus casei by electroporation. FEMS Microbiol. Lett. 44:173-177. [Google Scholar]
- 6.Chou, P. Y., and G. D. Fasman. 1978. Empirical predictions of protein conformation. Annu. Rev. Biochem. 47:251-276. [DOI] [PubMed] [Google Scholar]
- 7.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 8.Demuth, D. R., and D. C. Irvine. 2002. Structural and functional variation within the alanine-rich repetitive domain of streptococcal antigen I/II. Infect. Immun. 70:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exterkate, F. A., and A. C. Alting. 1999. The role of calcium in the activity and stability of the Lactococcus lactis cell envelope proteinase. Appl. Environ. Microbiol. 65:1390-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekkes, P., and A. J. M. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fira, D., M. Kojic, A. Banina, I. Spasojevic, I. Strahinic, and L. Topisisrovic. 2001. Characterization of cell envelope-associated proteinases of thermophilic lactobacilli. J. Appl. Microbiol. 90:123-130. [DOI] [PubMed] [Google Scholar]
- 12.Gajic, O. 2003. Relationships between MDR proteins, bacteriocin production and proteolysis in Lactococcus lactis. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 13.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97-120. [DOI] [PubMed] [Google Scholar]
- 14.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, C., D. Atlan, B. Blanc, R. Portalier, G. J. Germond, L. Lapierre, and B. Mollet. 1996. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbrueckii subsp. bulgaricus. J. Bacteriol. 178:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:227-1239. [DOI] [PubMed] [Google Scholar]
- 18.Gum, J. R., J. C. Byrd, J. W. Hicks, N. W. Toribara, D. T. Lamport, and Y. S. Kim. 1989. Molecular cloning of human intestinal mucin cDNAs. Sequence analysis and evidence for genetic polymorphism. J. Biol. Chem. 264:6480-6487. [PubMed] [Google Scholar]
- 19.Havenaar, R., and J. H. J. Huis in ‘t Veld. 1992. Probiotics: a general view, p. 151-170. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. I. The lactic acid bacteria in health and disease. Elsevier Applied Science, Amsterdam, The Netherlands.
- 20.Hebert, E. M., R. R. Raya, and G. S. De Giori. 2000. Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. Appl. Environ. Microbiol. 66:5316-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holck, A., and H. Naes. 1992. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO151. J. Gen. Microbiol. 138:3119-3123. [DOI] [PubMed] [Google Scholar]
- 22.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopwood, D. A., J. M. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, J. D. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 24.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, P. R., and K. Hammer. 1997. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojic, M., D. Fira, A. Banina, and L. Topisirovic. 1991. Characterization of the cell wall-bound proteinase of Lactobacillus casei HN14. Appl. Environ. Microbiol. 57:1753-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojic, M., D. Fira, B. Bojovic, A. Banina, and L. Topisirovic. 1995. Comparative study on cell-envelope associated proteinases in natural isolates of mesophilic lactobacilli. J. Appl. Bacteriol. 79:61-68. [Google Scholar]
- 28.Kok, J., and W. M. de Vos. 1994. The proteolytic system of lactic acid bacteria, p. 169-210. In M. Gasson and de W. M. de Vos (ed.), Genetics and bio/technology of lactic acid bacteria. Blackie Academic and Professional, London, England.
- 29.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers, O. P., M. M. Beerthuyzen, P. G. G. A. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 31.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 32.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 33.Marugg, J. D., W. Meijer, R. van Kranenburg, P. Laverman, P. G. Bruinenberg, and W. M. de Vos. 1995. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J. Bacteriol. 177:2982-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mierau, I., E. R. S. Kunji, G. Venema, and J. Kok. 1997. Casein and peptide degradation in lactic acid bacteria. Biotechnol. Genet. Eng. Rev. 14:279-301. [DOI] [PubMed] [Google Scholar]
- 35.Miladinov, N., O. P. Kuipers, and L. Topisirovic. 2001. Casitone-mediated expression of the prtP and prtM genes in Lactococcus lactis subsp. lactis BGIS29. Arch. Microbiol. 177:54-61. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Mills, O. E., and T. D. Thomas. 1981. Nitrogen sources for growth of lactic streptococci in milk. N. Z. J. Dairy Sci. Technol. 16:43-55. [Google Scholar]
- 38.Moniaux, N., F. Escande, N. Porchet, J. P Aubert, and S. K. Batra. 2001. Structural organization and classification of the human mucin genes. Front. Biosci. 6:1192-1206. [DOI] [PubMed] [Google Scholar]
- 39.Naes, H., and J. Nissen-Meyer. 1992. Purification and N-terminal amino acid determination of the cell wall bound proteinase from Lactobacillus paracasei subsp. paracasei. J. Gen. Microbiol. 138:313-318. [DOI] [PubMed] [Google Scholar]
- 40.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of procaryotic and eucaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 42.Pederson, J. A., G. M. Mileski, B. C. Weimer, and J. L. Steele. 1999. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 45.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siezen, R. J., P. G. Bruinenberg, P. Vos, I. J. van Alen-Boerrigter, M. Nijhuis, A. C. Alting, F. A. Exterkate, and W. M. de Vos. 1993. Engineering of the substrate binding region of the subtilisin-like, cell-envelope proteinase of Lactococcus lactis. Protein Eng. 4:719-737. [DOI] [PubMed] [Google Scholar]
- 49.Siezen, R. J., and J. A. M. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siezen, R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Leeuwenhoek 76:139-155. [PubMed] [Google Scholar]
- 51.Tilsala-Timisjarvi, A., and T. Altossava. 1997. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbiol. 35:49-56. [DOI] [PubMed] [Google Scholar]
- 52.Vos, P., I. J. Boerrigter, G. Buist, A. J. Haandrikman, M. Nijhuis, M. B. de Reuver, R. J. Siezen, G. Venema, W. M. de Vos, and J. Kok. 1991. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 4:479-484. [DOI] [PubMed] [Google Scholar]
- 53.Vukasinovic, M., D. Fira, and L. Topisirovic. 2001. Characteristics of natural isolates of lactic acid bacteria selected for semi-hard cheese on the trapist type. Acta Vet. 51:53-66. [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Walker, D. C., K. Aoyama, and T. R. Klaenhammer. 1996. Electrotransformation of Lactobacillus acidophilus A1. FEMS Microbiol. Lett. 138:233-237. [DOI] [PubMed] [Google Scholar]