Abstract
An epoxyalkane:coenzyme M (CoM) transferase (EaCoMT) enzyme was recently found to be active in the aerobic vinyl chloride (VC) and ethene assimilation pathways of Mycobacterium strain JS60. In the present study, EaCoMT activity and genes were investigated in 10 different mycobacteria isolated on VC or ethene from diverse environmental samples. In all cases, epoxyethane metabolism in cell extracts was dependent on CoM, with average specific activities of EaCoMT between 380 and 2,910 nmol/min/mg of protein. PCR with primers based on conserved regions of EaCoMT genes from Mycobacterium strain JS60 and the propene oxidizers Xanthobacter strain Py2 and Rhodococcus strain B-276 yielded fragments (834 bp) of EaCoMT genes from all of the VC- and ethene-assimilating isolates. The Mycobacterium EaCoMT genes form a distinct cluster and are more closely related to the EaCoMT of Rhodococcus strain B-276 than that of Xanthobacter strain Py2. The incongruence of the EaCoMT and 16S rRNA gene trees and the fact that isolates from geographically distant locations possessed almost identical EaCoMT genes suggest that lateral transfer of EaCoMT among the Mycobacterium strains has occurred. Pulsed-field gel electrophoresis revealed large linear plasmids (110 to 330 kb) in all of the VC-degrading strains. In Southern blotting experiments, the strain JS60 EaCoMT gene hybridized to many of the plasmids. The CoM-mediated pathway of epoxide metabolism appears to be universal in alkene-assimilating mycobacteria, possibly because of plasmid-mediated lateral gene transfer.
Aerobic bacteria that grow on ethene and vinyl chloride (VC) are widely distributed in the environment and have attracted interest because of their potential applications in bioremediation and biocatalysis (5, 6, 11, 12, 32, 33). The first step in ethene and VC assimilation is known to be a monooxygenase reaction yielding epoxyethane from ethene (5, 7) and chlorooxirane from VC (12, 33), but the downstream pathways are not well understood. Our recent work (5a) has revealed that an epoxyalkane:coenzyme M (CoM) transferase (EaCoMT) enzyme is involved in epoxyethane and chlorooxirane metabolism in Mycobacterium strain JS60, a strain isolated from contaminated groundwater by enrichment on VC as the sole carbon source (5). The EaCoMT reaction is a key step that channels the initial intermediates into central metabolic pathways and also guards against the accumulation of highly toxic and reactive epoxides in the cytoplasm.
EaCoMT enzymes have previously been found only in the propene-oxidizing bacteria Xanthobacter strain Py2 and Rhodococcus strain B-276. In such strains, EaCoMT is part of an unusual epoxide carboxylase enzyme complex consisting of EaCoMT, two stereoselective dehydrogenases, and an oxidoreductase/carboxylase (1, 2, 9, 16). In addition to unique biochemical reactions, the propene assimilation pathway is also distinguished by unusual genetic elements. In both strains Py2 and B-276, the propene monooxygenase genes are carried on linear megaplasmids, and in strain Py2, the epoxide carboxylase system and CoM biosynthesis genes are also plasmid borne (18, 26).
As part of the study that yielded strain JS60, we isolated many other mycobacteria that grew on VC (5). It is not known whether these isolates or similar Mycobacterium strains isolated on ethene (6, 7) possess EaCoMT enzymes. The relationship between strains isolated on VC and ethene is unclear, and the role of factors such as site contamination and geography in the dissemination and evolution of both groups is unknown. On the basis of the recent finding that the EaCoMT gene in Xanthobacter Py2 is carried by a linear plasmid, it might be speculated that similar elements are involved in ethene and VC metabolism. To address these questions, we investigated 10 mycobacteria isolated on VC (6 strains) or ethene (4 strains) from a diverse range of environmental samples. EaCoMT genes and enzyme activities were found in all of the strains. In the VC degraders, the EaCoMT enzymes appear to be encoded on linear megaplasmids.
MATERIALS AND METHODS
Isolation and growth of bacteria.
The growth methods and media used were described previously (5, 5a). Six Mycobacterium strains (JS60, JS61, JS616, JS617, JS619, and JS621) that grow on VC and ethene were previously isolated (5) from groundwater (Plaquemine, La.), activated sludge (Ithaca, N.Y.), pond sediment (Carlyss, La.), activated carbon (Dortmund, Germany), aquifer sediment (Travis Air Force Base, Calif.), and groundwater (Moody Air Force Base, Ga.), respectively. Several mycobacteria that grow on ethene (but not VC) were isolated by enrichment culture as described previously (5), except that ethene was added to the headspace as the sole carbon source (16 ml/160-ml bottle) and incubation was at 30°C. Strains JS622 and JS623 were derived from sandy garden soil (Panama City, Fla.), strain JS624 was from grass rhizosphere soil (Central Park, New York, N.Y.), and strain JS625 was from decomposing tree bark (Washington, D.C.). The isolates were identified by 16S rRNA gene (rDNA) sequencing.
Effects of CoM on growth and enzyme activity.
EaCoMT activity was assayed in cell extracts as described elsewhere (5a). Extracts from strains JS61, JS616, JS617, JS619, and JS621 were prepared from VC-grown cells, whereas extracts from strains JS622, JS623, JS624, and JS625 were prepared from ethene-grown cells. The effect of CoM on growing cultures of strain JS623 was investigated by monitoring ethene consumption and epoxyethane accumulation in 50-ml cultures growing on ethene (10%, vol/vol) as the sole carbon source, either with or without CoM (10 μM) added. Ethene and epoxyethane were quantified by gas chromatography of headspace samples (5a). Control cultures of strain JS623 containing glucose (1%) and Tween 80 (0.05%) in the presence and absence of CoM were monitored by measurement of optical density at 600 nm.
DNA extraction and PCR amplification of EaCoMT genes.
Genomic DNA was extracted either by a previously reported method (5) or by bead beating as follows. Cultures were grown for 3 to 6 weeks on 1/10-strength Trypticase soy agar plates containing 1% glucose (5). Cells scraped from one plate were washed in 1 ml of STE-Tween buffer (100 mM NaCl, 10 mM Tris, 50 mM EDTA, 0.1% Tween 80, pH 8.0), suspended in 1 ml of the same buffer, and added to a screw-cap tube containing 1 ml of STE-Tween-saturated silica-zirconia beads (0.1-mm diameter). The mixture was subjected to bead beating (Mini-Beadbeater; BioSpec Products) at high speed for 2 min, and the lysate was purified by phenol-chloroform extraction and ethanol precipitation (27). The bead beating method was superior to the enzymatic-chemical lysis method (5) in terms of DNA yield and time, but the recovered DNA was of somewhat lower quality, presumably because of shearing forces. PCR was done essentially as described elsewhere (Coleman and Spain, submitted), with annealing at 60°C for 30 s and 5 ng of genomic DNA added as the template. Primers CoM-F1L (5′-AACTACCCSAAYCCSCGCTGGTACGAC-3′) and CoM-R2E (5′-GTCGGCAGTTTCGGTGATCGTGCTCTTGAC-3′) were designed from conserved regions of the EaCoMT genes of Mycobacterium strain JS60, Rhodococcus strain B-276, and Xanthobacter strain Py2 (GenBank accession numbers AY243034, AF426826, and X79863).
Preparation of cell plugs and PFGE.
Cultures (700 ml) of Mycobacterium strains JS60, JS61, JS616, JS617, JS619, and JS621 were grown on ethene until mid-exponential phase, glycine (0.5%) and ampicillin (200 μg/ml) were added, and then incubation was continued overnight. Cells were pelleted and frozen in aliquots at −80°C. After thawing, the cell pellets (50 to 100 μl) were washed in STE-Tween, suspended in molten agarose (0.5 to 1.0 ml, 1% in 0.5× TBE buffer) (27), and pipetted into plug molds. The optical density at 600 nm of cells in the plugs ranged from 20 to 80, requiring individual optimization for each strain. Plugs (typically 5 to 10) were treated with lysozyme (50 mg in 5 ml of STE) at 37°C overnight, rinsed in wash buffer (20 mM Tris, 50 mM EDTA, pH 8), and then treated with proteinase K (5 mg in 5 ml of sodium lauryl sarcosinate [1%], sodium deoxycholate [0.2%], EDTA [0.1 M, pH 8]) for 24 h at 60°C. Plugs were rinsed three times in wash buffer and stored in the same buffer at 4°C. Contour-clamped homogeneous electric field (CHEF) pulsed-field gel electrophoresis (PFGE) was done in 1% agarose gels (0.5× TBE) at 14°C, 6 V/cm, and a 120° angle, with the switching time ramped from 10 to 40 s over 18 h (CHEF-DRIII system; Bio-Rad). Lambda phage concatemers (Bio-Rad) used as molecular weight markers were regenerated by heating at 45°C for 25 min before use.
Southern blotting.
Southern blot assays were performed via standard methods (28), and the ECL kit (Amersham-Pharmacia) was used for detection. In Southern blots from standard agarose gels, acid depurination was omitted, while for PFGE gels, this step was replaced with a nicking procedure (CHEF-DRIII instructions; Bio-Rad), in which the gel was stained for 30 min in 1 μg of ethidium bromide per ml and then subjected to UV radiation at 60 mJ/cm2 (Stratagene UV cross-linker). The probe used for Southern blotting in all cases was an 891-bp region of the strain JS60 EaCoMT gene, amplified by PCR with the CoM-F1L and CoM-R2E primers.
RESULTS AND DISCUSSION
Isolation of ethene-assimilating bacteria.
To complement a previously isolated set of VC-assimilating bacteria (5), four bacteria that grow on ethene were isolated from soil samples with no known exposure to chlorinated ethenes. Partial sequencing of 16S rDNA indicated that all four isolates were distinct and were strains of Mycobacterium. On the basis of a 421-bp alignment of the 16S rDNAs, the closest species matches were JS622-M. rhodesiae (98.3% identity), JS623-M. smegmatis (96.7% identity), JS624-M. peregrinum (97.9% identity), and JS625-M. mageritense (99.0% identity). The GenBank accession numbers of the 16S rDNA sequences of the four strains are AY162027 to AY162030. None of the four isolates grew on VC (data not shown).
EaCoMT activity is widespread in alkene-assimilating mycobacteria.
CoM-dependent epoxyethane metabolism was found in cell extracts of all of the VC- and ethene-assimilating Mycobacterium strains examined (Table 1). Very little epoxyethane transformation occurred in reaction mixtures without added CoM. The low activities in some reaction mixtures that were not supplemented with CoM may have been due to endogenous levels of CoM in the cell extracts or to the activity of other epoxide-transforming enzymes. Extracts from VC-grown cells of the strains originally isolated on VC had generally higher levels of EaCoMT activity (1,550 ± 821 nmol/min/mg of protein) than extracts from ethene-grown cells of the strains isolated on ethene (592 ± 241 nmol/min/mg of protein).
TABLE 1.
EaCoMT activity in cell extracts of alkene-assimilating Mycobacterium strains
| Strain | Isolation and growth substrate | EaCoMT activitya (nmol/min/mg of protein)
|
|
|---|---|---|---|
| CoM added | No cofactor | ||
| JS60 | VC | 980 (930-1,060)b | 0 (0)b |
| JS61 | VC | 2,910 (2,330-3,490) | 110 (70-160) |
| JS616 | VC | 1,800 (1,710-1,890) | 10 (0-20) |
| JS617 | VC | 900 (710-1,090) | 30 (20-40) |
| JS619 | VC | 1,920 (1,880-1,960) | 130 (130) |
| JS621 | VC | 790 (780-800) | 110 (110) |
| JS622 | Ethene | 930 (850-1,000) | 50 (0-100) |
| JS623 | Ethene | 590 (400-780) | 60 (0-120) |
| JS624 | Ethene | 470 (440-500) | 10 (0-20) |
| JS625 | Ethene | 380 (290-460) | 0 (0) |
The results are averages of two assays, and the ranges are in parentheses. Reaction mixtures (1 ml) contained 0.1 mg of protein, 5 μmol of epoxyethane, and 10 μmol of CoM. Specific activities were calculated from the epoxyethane depletion rate after subtraction of the appropriate abiotic rate of epoxyethane loss (either 2.5 nmol/min in Tris buffer or 13 nmol/min in Tris buffer plus CoM).
From a previous study (5a).
Previous work (5a) indicated almost identical levels of EaCoMT activity in ethene- and VC-grown cells of strain JS60. Therefore, the higher activities in extracts from VC-grown cells (Table 1) are more likely due to the fact that these strains were originally isolated on VC, rather than because VC was used as the growth substrate in this particular experiment. Higher EaCoMT activities might be expected in the VC-assimilating strains if EaCoMT plays a role in chlorooxirane metabolism (5a) because chlorooxirane is much more unstable and reactive than epoxyethane (aqueous half lives of 90 s [15] and 13 days [35], respectively). Higher EaCoMT activity is unlikely to be the sole reason for the ability of some strains to grow on VC in addition to ethene. For example, strain JS622 displayed higher EaCoMT activity than did strains JS617 and JS621, yet the latter two strains can grow on VC, whereas strain JS622 cannot.
CoM stimulates ethene metabolism in growing cultures of strain JS623.
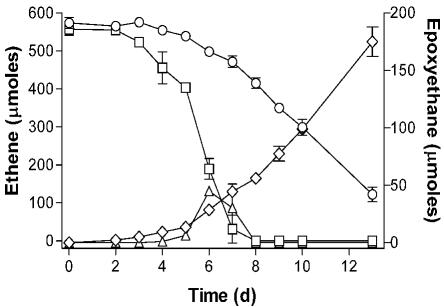
Cultures of Mycobacterium strain JS623 accumulated large amounts of epoxyethane during growth on ethene, in contrast to almost all of the other VC and ethene degraders. On the basis of the finding of EaCoMT in all of the strains (above), we hypothesized that an inadequate supply of CoM in the culture might be responsible for epoxyethane accumulation. Addition of 10 μM CoM to the MSM minimal medium accelerated ethene consumption in cultures of strain JS623 and eliminated the accumulation of epoxyethane (Fig. 1). CoM did not stimulate the growth of strain JS623 on glucose (data not shown), suggesting that the effect was specific to ethene metabolism. While an EaCoMT enzyme is present in all of the VC and ethene degraders examined, the ability of the strains to biosynthesize CoM may vary. It should therefore be noted that the EaCoMT activities measured in cell extract experiments with excess CoM (Table 1) may be higher than the in vivo activities in growing cultures, where limitations in the biosynthesis or recycling of CoM could affect the reaction rate.
FIG. 1.
Effect of CoM on ethene metabolism in growing cultures of strain JS623. Symbols: ○, ethene in cultures without CoM; □, ethene in cultures with 10 μM CoM; ⋄, epoxyethane in cultures without CoM; ▵, epoxyethane in cultures with 10 μM CoM. The data shown are averages of three replicate cultures, and the error bars show the standard deviations. d, days.
PCR amplification of EaCoMT genes from Mycobacterium strains.
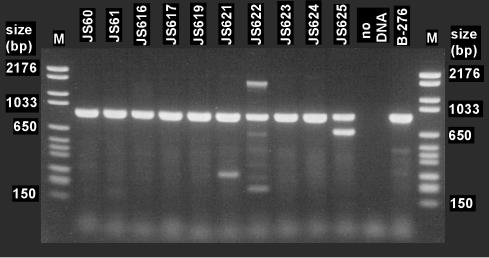
PCR with primers CoM-F1L and CoM-R2E yielded strong products of the expected size (891 bp) from all of the VC- and ethene-assimilating Mycobacterium strains tested (Fig. 2). The 891-bp PCR products were cloned and sequenced, and all were revealed to be partial EaCoMT genes (GenBank accession numbers AY243035 to AY243043). The 680-bp amplicon from strain JS625 was primed at both ends with CoM-F1L and bore no resemblance to EaCoMT genes. This sequence contained one large partial open reading frame (183 amino acids) that was somewhat similar to hypothetical protein Rv2624c from Mycobacterium tuberculosis (26.4% identity). The faint secondary products seen in amplifications with the strain JS621 and JS622 DNAs were not investigated further.
FIG. 2.
PCR amplification of EaCoMT gene fragments from ethene- and VC-assimilating mycobacteria with the CoM-F1L and CoM-R2E primers. The propene degrader Rhodococcus strain B-276 was included as a positive control. Molecular size markers are in lanes M.
Only one EaCoMT sequence was obtained from each of the 891-bp PCR products (two or three cloned amplicons were sequenced in each case), but it is possible that multiple EaCoMT genes exist in the strains. Thus, the enzyme activities in Table 1 cannot be attributed rigorously to the EaCoMT genes that were sequenced. To address this issue, we prepared Southern blots from EcoRV or SphI digests of genomic DNAs from the strains and probed them with the 891-bp EaCoMT PCR product from strain JS60. In the EcoRV digests of strains JS61, JS617, JS619, and JS621 and the SphI digests of strains JS61 and JS621, a single strongly hybridizing band was seen (data not shown). In blots from restriction digests of the other strains, a smear of hybridizing DNA was seen but there were no discrete bands. Such results could be due to bead beater-induced DNA shearing or to nonspecific nuclease activity. Further experiments with higher-quality DNA preparations and a wider range of restriction enzymes are required to clarify our initial data, but the results obtained thus far suggest that a single EaCoMT allele is present in strains JS61, JS617, JS619, and JS621. Similar methods used in a previous study (5a) indicated that only a single EaCoMT gene was present in strain JS60.
Sequence analysis of Mycobacterium EaCoMT genes.
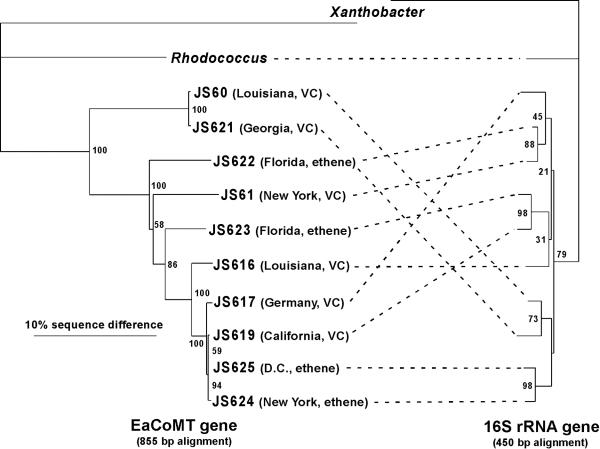
In silico translation of the EaCoMT gene fragments revealed a 278-amino-acid open reading frame in all cases. The histidine and the first cysteine residue of the His-X-Cys-Xn-Cys motif involved in zinc binding in the Xanthobacter strain Py2 EaCoMT enzyme (17) were identified in each of the deduced amino acid sequences (the second cysteine residue lay outside the region amplified). Phylogenetic analysis of the Mycobacterium, Xanthobacter strain Py2, and Rhodococcus strain B-276 EaCoMT genes (Fig. 3) showed that the Mycobacterium sequences clustered together, having 80.5 to 99.9% sequence identity with each other, 68.7 to 70.5% identity with the B-276 gene, and 52.6 to 55.9% identity with the strain Py2 gene (percentages from a PHYLIP DNA distance similarity table (10). In several cases, almost identical EaCoMT sequences were obtained from strains derived from geographically distant samples. For example, three base changes or fewer (0.12 to 0.36% difference) separated the EaCoMT sequences of strains JS619 (California), JS624 (New York), and JS625 (Washington, D.C.), whereas the 16S rDNA sequences of these three strains were separated by 1.79 to 5.13%. Similar observations of highly conserved catabolic genes in phylogenetically diverse bacterial strains are not uncommon in the biodegradation literature (13, 23).
FIG. 3.
Comparative phylogeny of EaCoMT and 16S rDNAs from alkene-oxidizing bacteria. The geographical origins and isolation substrates of the JS strains (all Mycobacterium spp.) are shown. Alignments and neighbor-joining trees were generated with ClustalX and TreeView, with Xanthobacter as the outgroup. The 16S rDNA alignment was manually adjusted by removing nucleotides at ambiguous positions. Percent bootstrap values from 100 trees are shown at the nodes. Because 16S rDNA sequences for Rhodococcus strain B-276 and Xanthobacter strain Py2 were not available, the sequences from Rhodococcus rhodochrous ATCC 271T and Xanthobacter autotrophicus JW33 were substituted.
The tree topologies estimated from the EaCoMT and 16S rDNA sequences have numerous differences (Fig. 3). Assuming vertical inheritance of 16S rDNA, this suggests that lateral gene transfer (LGT) of EaCoMT genes has occurred (30). The LGT hypothesis is supported by the fact that the EaCoMT genes are in many cases more similar to each other than are the (slow-evolving) 16S rDNAs. We used the maximum-likelihood-based Shimodaira-Hasegawa test (28) in PAUP*b10 (29) to test the null hypothesis that the two genes in each strain have the same evolutionary history. On the basis of the 16S rDNA data set, the likelihood scores of the two topologies shown in Fig. 3 were estimated (GTR+I+G model in Modeltest) (24) to be −1,508.37 (16S rDNA) and −1,550.33 (EaCoMT). The latter is significantly lower than the former (P < 0.0001), and thus it is unlikely that the two genes have the same evolutionary history. On the basis of bootstrap values (Fig. 3), the strongest case for LGT concerns the EaCoMT genes of strains JS623 and JS619. Indeed, when these strains were removed from the data sets and the Shimodaira-Hasegawa test was repeated, no significant differences between the likelihood scores were found (likelihood scores of −1,419.10 and −1,418.65; P = 0.381). We conclude that rigorous statistical support for LGT only exists in the cases of strains JS619 and JS623.
Analysis of plasmids by PFGE and Southern blotting.
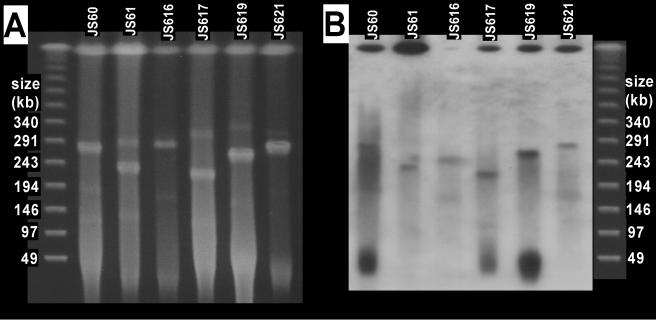
On the basis of the phylogenetic evidence of lateral transfer of Mycobacterium EaCoMT genes (above) and the finding of plasmid-borne propene metabolism genes in Rhodococcus strain B-276 and Xanthobacter strain Py2 (18, 26), we hypothesized that catabolic plasmids were present in the VC- and ethene-assimilating strains and searched for such plasmids by PFGE. Because of difficulties in preparing DNA plugs from some of the strains isolated on ethene, we focused our analysis on the six VC-assimilating strains. CHEF-PFGE revealed one strong plasmid band and several fainter plasmid bands in each of the six strains investigated (Fig. 4A). All of the bands migrated at the same positions relative to the linear markers when the pulse conditions were altered (20 to 80 s over 18 h; data not shown), indicating that the plasmids were linear rather than circular (4).
FIG. 4.
(A) Ethidium bromide-stained CHEF-PFGE gel showing plasmids in VC-assimilating Mycobacterium strains. (B) Southern blot prepared from a PFGE gel by using the JS60 EaCoMT gene as the probe. Lambda concatemer markers are shown on the left and right and apply to both the blot and the gel.
After Southern blotting of the pulsed-field gels, probing with the strain JS60 EaCoMT gene resulted in hybridization signals with the strong plasmid band in most of the strains (Fig. 4B). The simplest hypothesis (i.e., that the genes are plasmid borne) is complicated by the additional strong probe binding at the “compression zone” (3, 20) and because of the weak probe hybridization with some of the smaller plasmids. The multiple signals could be due to multiple copies of the EaCoMT gene in the Mycobacterium strains, the resolution of different forms of the same plasmid in the gel, or nonspecific binding of the probe. Southern blots from restriction digests of total DNA (above) seem to eliminate the possibility of multiple EaCoMT gene locations, but duplication of a large region of DNA would be consistent with both results. The presence of multiple plasmid forms is also a possibility. Concatemers of circular plasmids occur in Streptomyces strains (14), and a combination of linear and circular forms of the same plasmid in a single bacterium is common (3). Signals from both the compression zone and plasmid band were seen in Southern blotting experiments with Rhodococcus strain B-276 (26), in which the alkene monooxygenase genes were clearly plasmid borne (on the basis of the lack of plasmid in propene-negative mutants). The authors of that study attributed the probing result to nonspecific binding at the compression zone because of the larger amount of DNA there. It is possible that the signal from the compression zone in our study is due to the EaCoMT probe hybridizing to chromosomal methionine synthase (metE), which encodes a zinc-dependent enzyme with some sequence similarity to EaCoMT (17).
Implications of this study.
We found EaCoMT activity and genes in all of the alkene-oxidizing mycobacteria that we examined, including strains isolated on both VC and ethene. EaCoMT genes were not found in BLAST database searches of Mycobacterium genomes (or any other genomes) that have been completed to date, indicating that EaCoMT is specific to the alkene-assimilation pathway. It remains to be determined whether EaCoMT is involved in the VC and ethene assimilation pathways of Pseudomonas (33, 34) and Nocardioides (5) strains. The EaCoMT gene primers we have developed could be used to address this question. In addition, such primers will be very useful for culture-independent monitoring of microbial populations during the natural attenuation or bioremediation of chlorinated ethenes. Such methods are particularly relevant for mycobacteria, which are slow growing and sometimes difficult to isolate in pure culture (5).
The 10 VC and ethene degraders we studied were scattered throughout the genus Mycobacterium, with no apparent correspondence seen between phylogeny and alkene growth substrate. Although EaCoMT activities tended to be higher in the VC degraders than in the ethene degraders, it is difficult to compare these results because of the use of different growth substrates in experiments with each group of strains. Sequencing of EaCoMT genes did not reveal any signature regions that discriminated between the VC- and ethene-assimilating strains, and indeed, in the case of strains JS619 (VC) and JS625 (ethene), only a single nucleotide difference separated the EaCoMT PCR products. An important question arising from our work therefore concerns the distinction between bacteria that can grow on both VC and ethene and those that grow on ethene alone.
There is good evidence in at least one case that VC degraders evolved directly from ethene-degraders—this was observed in vitro with Pseudomonas strain DL1 (34). The transition from ethene to VC assimilation could be due to recruitment of an additional catabolic enzyme (23), changes in enzyme specificity (25), or alteration of enzyme expression levels (31). With respect to enzyme specificity and expression level, it is interesting that although the EaCoMT gene sequences of strains JS619 and JS625 were almost identical, the EaCoMT activity in cell extracts was fivefold higher in strain JS619. Sequencing of complete EaCoMT genes and flanking DNA and analysis of the activity of other catabolic enzymes may help to shed light on the factors that distinguish ethene- and VC-assimilating bacteria and yield insights into the possible evolution of the latter group from the former.
The EaCoMT gene appears to be carried on large linear plasmids in VC-degrading strains. On the basis of the gene organization in Mycobacterium strain JS60 (5a), it is likely that the alkene monooxygenase genes are also present on the same plasmids. Cryptic linear plasmids have previously been identified in various pathogenic Mycobacterium strains (19, 21, 22) and in ethene-oxidizing Mycobacterium strain E-1-57 (26). Only one previous study (36) has associated a specific catabolic function (morpholine biodegradation) with plasmids in mycobacteria. Our results indicate that catabolic plasmids are more widespread in this genus than was previously believed. Plasmid-borne genes for ethene and VC biodegradation could potentially be transferred among bacteria in the environment (8, 13), and thus, further investigation of such elements is warranted in light of the potential importance of the alkene-degrading phenotype to bioremediation and natural attenuation processes (5).
Acknowledgments
We thank Nathan Lo for help with phylogenetic analyses and Crystal Henley for assistance with the isolation of ethene degraders. Scott Ensign kindly provided strain B-276.
This work was funded by the U.S. Strategic Environmental Research and Development Program. N.V.C. was supported by a postdoctoral fellowship from the Oak Ridge Institute for Science and Education (U.S. Department of Energy).
REFERENCES
- 1.Allen, J. R., and S. A. Ensign. 1997. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272:32121-32128. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. R., D. D. Clark, J. G. Krum, and S. A. Ensign. 1999. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. USA 96:8432-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 4.Birren, B., and E. Lai. 1993. Pulsed-field gel electrophoresis: a practical guide. Academic Press., San Diego, Calif.
- 5.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Coleman, N. V., and and J. C. Spain. 2003. Epoxyalkane:coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185:5536-5545. [DOI] [PMC free article] [PubMed]
- 6.de Bont, J. A. M. 1976. Oxidation of ethylene by soil bacteria. Antonie van Leeuwenhoek 42:59-71. [DOI] [PubMed] [Google Scholar]
- 7.de Bont, J. A. M., and W. Harder. 1978. Metabolism of ethylene by Mycobacterium E 20. FEMS Microbiol. Lett. 3:89-93. [Google Scholar]
- 8.Dejonghe, W., J. Goris, S. El Fantroussi, M. Hofte, P. De Vos, W. Verstraete, and E. M. Top. 2000. Effect of dissemination of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation plasmids on 2,4-D degradation and on bacterial community structure in two different soil horizons. Appl. Environ. Microbiol. 66:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensign, S. A. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845-5853. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 11.Habets-Crützen, A. Q. H., L. E. S. Brink, C. G. van Ginkel, J. A. M. de Bont, and J. Tramper. 1984. Production of epoxides from gaseous alkenes by resting-cell suspensions and immobilized cells of alkene-utilizing bacteria. Appl. Microbiol. Biotechnol. 20:245-250. [Google Scholar]
- 12.Hartmans, S., and J. A. M. de Bont. 1992. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl. Environ. Microbiol. 58:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 15.Kline, S. A., J. J. Solomon, and B. L. van Duuren. 1978. Synthesis and reactions of chloroalkene epoxides. J. Org. Chem. 43:3596-3600. [Google Scholar]
- 16.Krum, J. G., and S. A. Ensign. 2000. Heterologous expression of bacterial epoxyalkane:coenzyme M transferase and inducible coenzyme M biosynthesis in Xanthobacter strain Py2 and Rhodococcus rhodochrous B276. J. Bacteriol. 182:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krum, J. G., H. Ellsworth, R. R. Sargeant, G. Rich, and S. A. Ensign. 2002. Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase. Biochemistry 41:5005-5014. [DOI] [PubMed] [Google Scholar]
- 18.Krum, J. G., and S. A. Ensign. 2001. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J. Bacteriol. 183:2172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183:2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew, M. K., C. L. Smith, and C. R. Cantor. 1988. High-resolution separation and accurate size determination in pulsed-field gel electrophoresis of DNA. 2. Effect of pulse time and electric field strength and implications for models of the separation process. Biochemistry 27:9210-9216. [DOI] [PubMed] [Google Scholar]
- 21.Picardeau, M., and V. Vincent. 1997. Characterization of large linear plasmids in mycobacteria. J. Bacteriol. 179:2753-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picardeau, M., and V. Vincent. 1998. Mycobacterial linear plasmids have an invertron-like structure related to other linear replicons in actinomycetes. Microbiology 144:1981-1988. [DOI] [PubMed] [Google Scholar]
- 23.Poelarends, G. J., M. Zandstra, T. Bosma, L. A. Kulakov, M. J. Larkin, J. R. Marchesi, A. J. Weightman, and D. B. Janssen. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 25.Pries, F., A. J. van den Wijngaard, R. Bos, M. Pentenga, and D. B. Janssen. 1994. The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution. J. Biol. Chem. 269:17490-17494. [PubMed] [Google Scholar]
- 26.Saeki, H., M. Akira, K. Furuhashi, B. Averhoff, and G. Gottschalk. 1999. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology 145:1721-1730. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Shimodaira, H., and H. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inferences. Mol. Biol. Evol. 16:964-969. [Google Scholar]
- 29.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 30.Syvanen, M. 1994. Horizontal gene flow: evidence and possible consequences. Annu. Rev. Genet. 28:237-261. [DOI] [PubMed] [Google Scholar]
- 31.van der Ploeg, J. R., J. Kingma, E. J. De Vries, J. G. Van der Ven, and D. B. Janssen. 1996. Adaptation of Pseudomonas sp. GJ1 to 2-bromoethanol caused by overexpression of an NAD-dependent aldehyde dehydrogenase with low affinity for halogenated aldehydes. Arch. Microbiol. 165:258-264. [DOI] [PubMed] [Google Scholar]
- 32.Verce, M. F., C. K. Gunsch, A. S. Danko, and D. L. Freedman. 2002. Cometabolism of cis-1,2-dichloroethene by aerobic cultures grown on vinyl chloride as the primary substrate. Environ. Sci. Technol. 36:2171-2177. [DOI] [PubMed] [Google Scholar]
- 33.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2001. Transition from cometabolic to growth-linked biodegradation of vinyl chloride by a Pseudomonas sp. isolated on ethene. Environ. Sci. Technol. 35:4242-4251. [DOI] [PubMed] [Google Scholar]
- 35.Washington, J. W. 1995. Hydrolysis rates of dissolved volatile organic compounds: principles, temperature effects and literature review. Ground Water 33:415-424. [Google Scholar]
- 36.Waterhouse, K. V., A. Swain, and W. A. Venables. 1991. Physical characterization of plasmids in a morpholine-degrading Mycobacterium. FEMS Microbiol. Lett. 80:305-310. [DOI] [PubMed] [Google Scholar]