Abstract
Spiralin is the most abundant protein at the surface of the plant pathogenic mollicute Spiroplasma citri and hence might play a role in the interactions of the spiroplasma with its host plant and/or its insect vector. To study spiralin function, mutants were produced by inactivating the spiralin gene through homologous recombination. A spiralin-green fluorescent protein (GFP) translational fusion was engineered and introduced into S. citri by using an oriC-based targeting vector. According to the strategy used, integration of the plasmid by a single-crossover recombination at the spiralin gene resulted in the expression of the spiralin-GFP fusion protein. Two distinct mutants were isolated. Western and colony immunoblot analyses showed that one mutant (GII3-9a5) did produce the spiralin-GFP fusion protein, which was found not to fluoresce, whereas the other (GII3-9a2) produced neither the fusion protein nor the wild-type spiralin. Both mutants displayed helical morphology and motility, similarly to the wild-type strain GII-3. Genomic DNA analyses revealed that GII3-9a5 was unstable and that GII3-9a2 was probably derived from GII3-9a5 by a double-crossover recombination between plasmid sequences integrated into the GII3-9a5 chromosome and free plasmid. When injected into the leafhopper vector Circulifer haematoceps, the spiralinless mutant GII3-9a2 multiplied to high titers in the insects (1.1 × 106 to 2.8 × 106 CFU/insect) but was transmitted to the host plant 100 times less efficiently than the wild-type strain. As a result, not all plants were infected, and symptom production in these plants was delayed for 2 to 4 weeks compared to that in the wild-type strain. In the infected plants however, the mutant multiplied to high titers (1.2 × 106 to 1.4 × 107 CFU/g of midribs) and produced the typical symptoms of the disease. These results indicate that spiralin is not essential for pathogenicity but is required for efficient transmission of S. citri by its insect vector.
Spiroplasma citri is a plant pathogenic bacterium belonging to the class Mollicutes, a group of wall-less organisms phylogenetically related to low-G+C-content gram-positive bacteria (47). Plant pathogenic mollicutes belong to two taxonomically distinct groups, phytoplasmas and spiroplasmas, and are associated with several hundred diseases affecting a wide variety of plants, including fruit trees, ornamentals, vegetables, and grapevine (34, 43, 44). Phytoplasmas and spiroplasmas are restricted to the phloem sieve tubes and are transmitted from plant to plant by sap-feeding leafhoppers or psyllids. However, while phytoplasmas have not been cultured so far, spiroplasmas, and in particular S. citri, have been cultured since 1970 (40, 41). S. citri was first isolated from sweet orange (Citrus sinensis) trees affected by stubborn disease, but it infects many plant species other than citrus, including the Madagascar periwinkle Catharanthus roseus, in which it induces stunting, leaf yellows, and wilting. S. citri is transmitted from plant to plant by the phloem-feeding leafhoppers Circulifer haematoceps and Circulifer tenellus (11). S. citri is the most studied plant mollicute (5, 6, 7, 9). As a result of the development of genetic tools such as plasmid vectors (27, 38, 39, 51), transposon mutagenesis (19), and gene inactivation through homologous recombination (15, 23, 37), genetic studies led to the identification of genes involved in motility (25) and pathogenicity (20). In particular, it was shown that fructose utilization is a key factor of spiroplasmal pathogenicity (22, 9). However, the interactions of S. citri with the host plant and with its insect vector are still poorly understood. In particular, the spiroplasma components involved in these interactions are still to be discovered.
In the cell-wall-free mollicutes, like in walled bacteria, surface proteins are thought to play a key role in the interactions of the bacteria with the environment and/or the infected hosts. They are of primary importance in adherence, invasion, and interaction with the host immune system, and they may also display structural, transport, or enzymatic functions (13, 26, 36, 49). In most plant mollicutes, a single surface protein is immunodominant, and this protein forms the major portion of semipurified cell membrane preparations (1, 2, 4, 53). In spiroplasmas, spiralin is such an immunodominant surface protein and accounts for up to 20 to 30% of the total protein mass of the bacterium (50). Spiralin was the first mollicute protein to be expressed by gene cloning in Escherichia coli (35), and the amino acid sequence has been determined by gene sequencing (14). Spiralin is a lipoprotein of 26 kDa which is conserved among spiroplasmas (8, 17). The preprotein possesses a typical signal peptide sequence upstream of a cysteine residue, a highly conserved central region, an α-helix, and two repeated sequences, which are thought to be specific to the spiroplasma species (17). The mature protein is anchored in the outer leaflet of the membrane lipid bilayer by an N-acylcysteine, while the entire polypeptidic chain is exposed to the outer medium (10, 28). The function of spiralin is unknown. However, because of its abundance and its location at the cell surface, spiralin is a good candidate to be one of the spiroplasma proteins involved in the interactions of the bacterium with its two hosts, the insect vector and the plant. With the aim to study spiralin function, S. citri mutants were produced by spiralin gene disruption and tested for their ability to be transmitted by the leafhopper vector and to produce symptoms in infected plants. The results showed that spiralin is not essential for pathogenicity but is required for efficient transmission of S. citri by its insect vector.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. citri strain GII-3 was originally isolated from its leafhopper vector C. haematoceps, which was captured in Morocco (46). This strain was shown to be phytopathogenic by transmission to periwinkle (C. roseus) through injection to the leafhopper vector C. haematoceps. Spiroplasmas were grown at 32°C in SP4 medium (48) from which fresh yeast extract was omitted. Transformation of S. citri by electroporation was performed as previously described (45). E. coli TG1 [supE hsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB+ lacIq lacZΔM15)], an EcoK− derivative of JM101, was used as the host strain for cloning experiments and for plasmid propagation. Plasmids pC2 and pSD4 have been described elsewhere (27). Plasmid pGUF6 was obtained by inserting successively the spiralin and green fluorescent protein (GFP) genes into pSD4. The spiralin gene was amplified from S. citri genomic DNA with primer pair SR5-SR8, and the 1.1-kbp fragment, restricted with BamHI and BglII, was inserted into the BamHI site of pSD4 to yield pMS4. The 0.8-kbp GFP gene, amplified from pGFPuv (Clontech Laboratories, Inc., Palo Alto, Calif.) with primer pair GFP1-GFP2 and restricted with BglII, was then inserted into the BamHI site of pMS4 to yield pGUF6. The translational fusion of GFP at the 3′end of the spiralin gene was confirmed by sequencing the 1.8-kbp XbaI-EcoRI fragment containing the spiralin-GFP gene fusion. The construction of pCL15 is described in Results.
DNA isolation, Southern blot hybridization, and PCR amplification of DNA.
Purification of plasmid DNA, cloning experiments, and DNA manipulations in E. coli were performed by standard procedures (42). S. citri genomic DNA was prepared from 10-ml cultures (approximately 1010 cells) with the Wizard genomic DNA purification kit from Promega Biosciences Inc. (San Luis Obispo, Calif.). Total DNA from insects was isolated essentially as described by Maixner et al. (31), using a cetyltrimethylammonium bromide extraction method. Probes for Southern blot hybridizations were generated by PCR in the presence of digoxigenin-11-dUTP (Roche Diagnostics, Meylan, France). Hybridization was performed at 42°C in 5× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50% formamide, 0.02% sodium dodecyl sulfate, 0.1% N-lauroylsarcosine, 1.5% blocking reagent, and 200 μg of denatured salmon sperm DNA per ml, with 50 ng of denatured probe. After washing under standard stringent conditions, hybridization signals were detected with antidigoxigenin-alkaline phosphatase conjugate and CDP-Star as the substrate, according to the manufacturer's instructions. Amplification was carried out in a 50-μl reaction mixture containing 5 to 20 ng of target DNA and 2.5 U of Taq DNA polymerase (Invitrogen SARL, Cergy Pontoise, France) or Vent DNA polymerase (New England Biolabs, Inc., Beverly, Mass.) with the buffer recommended by the supplier. Amplification was achieved in over 35 cycles, each consisting of 45 s at 95°C, 45 s at the annealing temperature, and 1 min at 72°C. The annealing temperature was set according to the melting temperatures of the primers used for amplification. Primer sequences are listed in Table 1.
TABLE 1.
Nucleotide sequences and positions of primers used in this study
| Primera | Sequenceb | Position | GenBank accession no. |
|---|---|---|---|
| GFP1 | 5′-TAGCAGATCTAAAATGAGTAAAGGAGAAGAAC-3′ | 286-307 | U62636 |
| GFP2 | 5′-TCGGAGATCTGTTGGAATTCATTATTTGTAGAG-3′ | 994-1026 | U62636 |
| SB2 | 5′-GTGAAGCACCAGTTTCTGAACC-3′ | 4184-4205 | AF012877 |
| SR5 | 5′-GGTCAGATCTAGCCAGAATTAAAAGTTAGTG-3′ | 4141-4171 | AF012877 |
| SR8 | 5′-AAGTTCGGATCCTGCTTTTGGTGGTGC-3′ | 5184-5210 | AF012877 |
| SR12 | 5′-GTGTATCAGCTGTCGGAAC-3′ | 4513-4531 | AF012877 |
| SR14 | 5′-TGTCGGATCCACATCAGTGGTTGCATGTAAT-3′ | 4523-4553 | AF012877 |
| SR16 | 5′-CTTTTTGATCTGAATAGTTTCCCGCATCAC-3′ | 5068-5097 | AF012877 |
| SR17 | 5′-TGTCGGAACAAGATCTGTGGTTGCATGTAAT-3′ | 4523-4553 | AF012877 |
| SR23 | 5′-CTTGTGAATCACCACCAGATG-3′ | 5380-5400 | AF012877 |
| SR24 | 5′-GCAAGTCAAGCGGGAC-3′ | 4069-4084 | AF012877 |
| TET8 | 5′-GGAGAAATCCCTGCTCGGTG-3′ | 1957-1976 | X56353 |
| CJ6 | 5′-CAATTACCAACCATGTTAGC-3′ | 3576-3595 | U89875 |
| CJ8 | 5′-GGTTAGTAATGCTGATCGC-3′ | 2328-2346 | U89875 |
| U | 5′-GTAAAACGACGGCCAGT-3′ | 379-395 | L09137 |
| R | 5′-GGAAACAGCTATGACCATG-3′ | 461-479 | L09137 |
Primers GFPn, SB2 and SRn, Tet8, CJn, and U and R refer to GFP, the spiralin gene of S. citri, the tetM gene of Tn916, the scm1 gene of S. citri, and the universal and reverse primers of pUC19, respectively.
Restriction sites used for cloning are underlined.
Western immunoblotting, colony blot immunoassay, and immunofluorescence.
Spiroplasma cells from a 50-ml culture were collected by centrifugation and washed three times in HS buffer (8 mM HEPES [pH 7.4], 280 mM sucrose). Proteins were solubilized in 0.25 ml of solubilization buffer and separated by electrophoresis through a sodium dodecyl sulfate-15% polyacrylamide gel according to standard procedures (42). Proteins were electroblotted to nitrocellulose in transfer buffer containing 25 mM Tris base, 190 mM glycine, and 20% methanol. The nitrocellulose sheet was saturated in TBS (50 mM Tris-HCl [pH 7.4], 200 mM NaCl) containing 1% nonfat dry milk for 2 h and incubated overnight with the primary antibody. After three washes in TBS containing 0.2% Triton X-100 and two additional washes in TBS, polypeptides reacting with the primary antibody were visualized by using a goat anti-rabbit or a rabbit anti-mouse immunoglobulin G-alkaline phosphatase conjugate with 5-bromo-4-chloro-3-indolylphosphate and 1-nitroblue tetrazolium chloride as chromogenic substrates. For direct colony blotting, nitrocellulose filters (0.45-μm pore size) were gently placed on spiroplasma colonies on the surface of agar plates. After 10 min, the filters were carefully removed and treated as described above for Western blots. Immunofluorescence assays were carried out as described previously (33). Both thin sections of spiroplasma-infected plants and spiroplasma cultures were probed with the anti-GFP (polyclonal) and antispiralin (monoclonal) primary antibodies.
Experimental transmission assay.
Microinjection of S. citri cultures into C. haematoceps leafhoppers and transmission to periwinkle plants were carried out as previously described (18, 20). Briefly, the injected insects were caged on young periwinkle plants (12 insects per plant, five plants per spiroplasma strain) for a 2-week transmission period. Culture of S. citri from plants and insects and transmission through Parafilm membranes have also been described previously (18, 20). For transmission through Parafilm membranes, leafhoppers were injected with S. citri cultures and caged on healthy periwinkle plants for 2 weeks to allow the spiroplasmas to multiply. The insects were then transferred to small cages (five insects per cage) in which a Parafilm membrane separated the insects from the culture medium (18, 20). When feeding through the membrane, infected leafhoppers injected S. citri cells into the medium. After 24 h at 20°C, the culture medium was removed from the cages and plated on solid SP4 medium for CFU counts.
RESULTS
Transformation of S. citri GII-3 with pCL15.
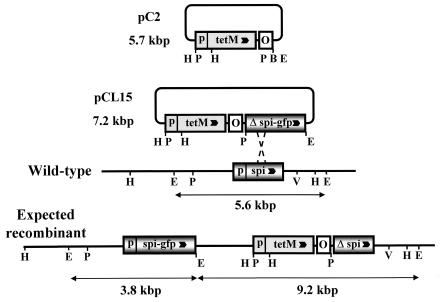
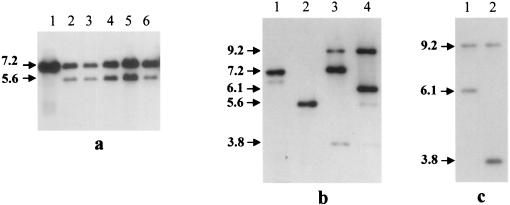
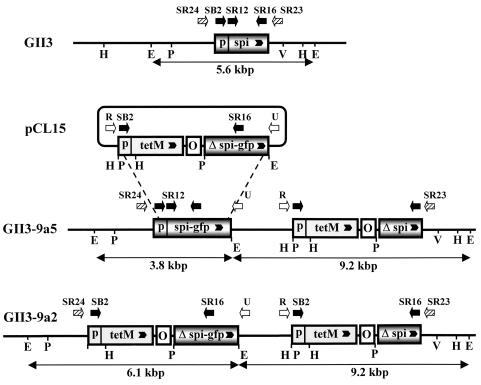
To construct the targeting vector pCL15, the spiralin-GFP translational fusion was amplified from pGUF6 with primers SR17 and GFP2. After restriction with BglII, the 1,403-bp fragment was inserted into the BamHI site of pC2 to yield pCL15 (Fig. 1). In this plasmid construct, the 5′-truncated spiralin gene lacks the transcription promoter, the ribosome binding site, and the sequences corresponding to the first 18 amino acids of the signal peptide, including the ATG initiation codon. According to this strategy, the spiralin-GFP fusion protein can be expressed only after the plasmid has integrated into the chromosome through a single-crossover recombination at the spiralin gene. As a result of plasmid integration, the recombinant chromosome possesses a deletion copy of the spiralin gene (which cannot be expressed) and a spiralin-GFP gene fusion, the transcription of which is driven by the chromosomal spiralin gene promoter (Fig. 1). No wild-type spiralin can be produced by such a transformant. Transformation of S. citri GII-3 with pCL15 yielded tetracycline-resistant colonies at an efficiency of 1.2 × 102 transformants/μg of DNA. Twelve individual transformants were grown in the presence of tetracycline and subcultured for 15 passages. During passaging, integration of the plasmid into the host chromosome was checked by Southern blot hybridization of genomic DNAs with the spiralin probe (Fig. 2). At the fifth passage, all five clones tested were found to contain free pCL15 as revealed by the 7.2-kbp fragment hybridizing with the spiralin probe. Accordingly, the 5.6-kbp chromosomal spiralin fragment was also detected (Fig. 2a). At the ninth passage, plasmid integration at the spiralin gene was revealed, in clone 9, by the presence of the expected two EcoRI fragments of 9.2 and 3.8 kbp hybridizing with the spiralin probe (Fig. 2b, lane 3) and the absence of the 5.6-kbp signal corresponding to the wild-type fragment. At this stage however, free plasmid (7.2-kbp signal) was still detected (Fig. 2b, lane 3). In contrast, after five additional passages, no free plasmid was detected (Fig. 2b, lane 4). Surprisingly, while the 9.2-kbp fragment was still present, the 3.8-kbp fragment was replaced by a 6.1-kbp signal. Therefore, the ninth passage of clone 9 was propagated for five additional passages, without tetracycline to cure the free plasmid, and then plated on tetracycline agar medium to select recombinants. The resulting subclones displayed two types of hybridization profiles, one type (GII3-9a5) with the expected EcoRI fragments of 9.2 and 3.8 kbp (Fig. 2c, lane 2) and the other (GII3-9a2) with two fragments of 9.2 and 6.1 kbp (Fig. 2c, lane 1).
FIG. 1.
Partial restriction maps of plasmids pC2 and pCL15 and schematic representation of pCL15 integration by recombination at the spiralin gene. O, S. citri oriC; p, spiralin gene promoter; spi, spiralin gene; Δ spi, 5′-truncated spiralin gene; gfp, GFP gene; tetM, tetM gene of Tn916; B, BamHI; E, EcoRI; H, HindIII, P, PstI; V, EcoRV. Arrowheads indicate direction of transcription.
FIG. 2.
Southern blot hybridization between EcoRI-restricted DNA extracted from pCL15 transformants and the spiralin probe. (a) Lane 1, purified pCL15; lanes 2 to 6, DNAs from five individual clones extracted at the fifth passage. (b) Lane 1, purified pCL15; lane 2 DNA extracted from untransformed cells; lanes 3 and 4, DNAs extracted from clone 9 after 9 and 14 passages, respectively. (c) DNAs extracted from two individual subclones isolated from the ninth passage of clone 9. Lane 1, subclone 9a2; lane 2, subclone 9a5. Sizes are indicated in kilobase pairs.
Immunodetection of the spiralin-GFP fusion protein in mutant GII3-9a5.
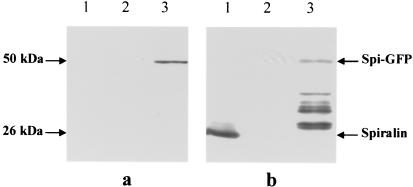
To determine whether the spiralin-GFP fusion protein was expressed in the S. citri transformants, proteins were separated by polyacrylamide gel electrophoresis and probed with antibodies directed against GFP or spiralin (Fig. 3). As shown by the immunoblots, GII3-9a5 did produce the fusion protein, which reacted with both anti-GFP and antispiralin antibodies (Fig. 3, lanes 3). Interestingly, GFP epitopes were also detected by direct immunoblotting of GII3-9a5 colonies and by immunofluorescence (data not shown), indicating that the fusion protein and, in particular, GFP epitopes were exposed at the spiroplasma cell surface. However, because of a Gly→Cys mutation at position 33 of the GFP sequence, no spontaneous fluorescence of spiroplasma cells was observed. As expected, the 50-kDa fusion protein was also detected with the antispiralin monoclonal antibody (Fig. 3b, lane 3). However, most of the translation products were detected as degraded or nonterminated polypeptides, which did not react with the anti-GFP antibodies (Fig. 3, lanes 3). GII3-9a5 produced no wild-type spiralin (Fig. 3b, lane 3). Unexpectedly, proteins from GII3-9a2 did not react with anti-GFP and antispiralin antibodies, indicating that neither the fusion protein nor the wild-type spiralin was produced (Fig. 3, lanes 2).
FIG. 3.
Western immunoblotting of total proteins from S. citri GII-3, GII3-9a2, and GII3-9a5. Proteins were probed with anti-GFP (a) or antispiralin (b) antibodies. The arrows indicate positions of spiralin and spiralin-GFP fusion protein.
Spiralin gene region of mutant GII3-9a2.
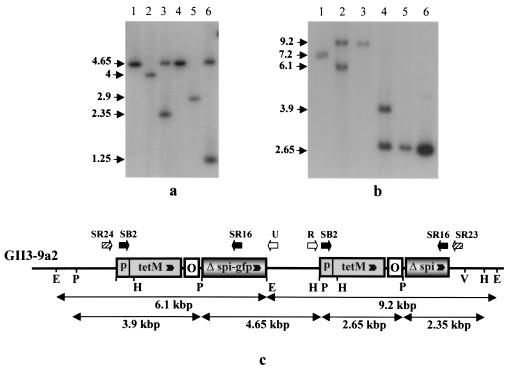
To understand why GII3-9a2 did not produce spiralin, the spiralin gene region was characterized by Southern blot hybridizations with spiralin-, tetM-, and oriC-specific probes; PCR with various combinations of primers; and sequencing of PCR fragments. As shown in Fig. 2c, mutant 9a2 contained two EcoRI fragments of 9.2 and 6.1 kbp hybridizing with the spiralin probe. Interestingly, these two fragments were also found to hybridize with the tetM probe (Fig. 4b, lane 2), whereas a single copy of the tetM gene was present in GII3-9a5 (Fig. 4b, lane 3). Similar results were obtained when DNAs were digested with PstI (Fig. 4b, lanes 4 and 5). The whole set of results (those in Fig. 4a and b and others not shown) led us to establish the map presented in Fig. 4c. Because such a rearrangement was unexpected, this map was further confirmed by PCR with primer pairs SB2-SR16, SR24-U, R-SR23, and SR12-SR16 and by sequencing the R-SR23 and SR24-U amplification products. Figure 5 shows that the 9.2-kbp EcoRI fragment of GII3-9a2 is identical to that of GII3-9a5 (see also Fig. 2c). In contrast, the 6.1-kbp EcoRI fragment does not carry the spiralin-GFP gene fusion but instead carries a fragment containing the tetM gene, the oriC, and the truncated copy of the spiralin gene fused to GFP, identical to that of plasmid pCL15. As indicated by this map, the occurrence in GII3-9a2 of two truncated copies (lacking the 5′ end sequences) of the spiralin gene clearly explains why this mutant did not produce spiralin.
FIG. 4.
(a and b) Southern blot hybridization between restricted DNA extracted from S. citri GII-3, GII3-9a2, or GII3-9a5 and the spiralin (a) or tetM (b) probe. (a) Purified pCL15 (lanes 1 and 4) and genomic DNAs from GII-3 (lanes 2 and 5) and GII3-9a2 (lanes 3 and 6) were restricted by HindIII plus PstI (lanes 1 to 3) or EcoRV plus PstI (lanes 4 to 6). (b) Purified pCL15 (lanes 1 and 6) and genomic DNAs from GII3-9a2 (lanes 2 and 4), and GII3-9a5 (lanes 3 and 5) were restricted by EcoRI (lanes 1 to 3) or PstI (lanes 4 to 6). Sizes are indicated in kilobase pairs. (c) Partial restriction map and gene organization of the spiralin gene region of GII3-9a2. The map is not to scale. Thick arrows indicate positions of primers. For abbreviations, see the legend to Fig. 1.
FIG. 5.
Partial restriction maps and gene organization of the spiralin gene regions of S. citri GII-3, GII3-9a5, and GII3-9a2 and schematic representation of double-crossover recombination between pCL15 and the chromosome of GII3-9a5. The maps are not to scale. Thick arrows indicate the positions of primers. For abbreviations, see the legend to Fig. 1.
Helicity, motility, and growth of mutants GII3-9a2 and GII3-9a5.
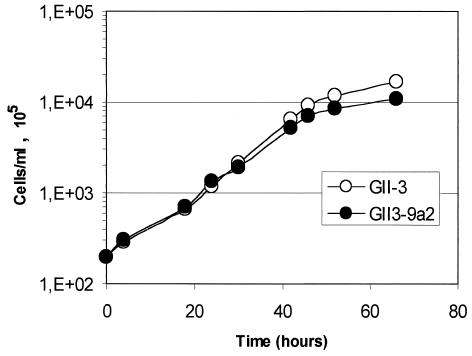
The isolation and growth of GII3-9a2 and GII3-9a5 in SP4 medium proved that spiralin was not essential for the spiroplasma to grow in vitro. In addition, dark-field microscopy observation of spiroplasma cultures revealed that cells of GII3-9a2 and GII3-9a5 still displayed helical morphology and motility characteristic of the wild-type strain GII-3. Mutant GII3-9a5, however, showed a higher proportion of elongated cells than GII-3. As indicated by the growth curve presented in Fig. 6, the spiralinless mutant GII3-9a2 grew in vitro at approximately the same rate as the wild-type strain GII-3, at least during the first 48 h. Within 72 h, the spiroplasma titer reached 109 CFU/ml, a titer that was not significantly different from that of GII-3 (1.7 × 109 CFU/ml). In agar medium, the fact that GII3-9a2 formed fuzzy colonies similar to those of the wild-type strain strongly suggested that motility was not affected. In contrast, GII3-9a5 grew at a lower rate and formed smaller colonies (data not shown). In vitro propagation of GII3-9a5 proved this mutant to be unstable, leading to subclones which did not produce the spiralin-GFP fusion protein.
FIG. 6.
Multiplication of S. citri GII-3 and GII3-9a2 in SP4 medium.
Transmission and pathogenicity of mutants GII3-9a2 and GII3-9a5.
To determine whether the absence of spiralin had an effect on the transmissibility and/or pathogenicity of S. citri, experimental transmission assays were carried out. Spiroplasmas were transmitted to periwinkle plants through injection to the leafhopper vector C. haematoceps as described in Materials and Methods. After the 2-week transmission period, the insects were removed and symptom production was monitored for at least 12 weeks. In the experiments of Table 2, we compared four spiroplasma strains, including the two spiralin mutants, GII3-9a2 and GII3-9a5, as well as two control strains, the wild-type strain GII-3 and strain GII3-11c1, in which the pCL15 plasmid had integrated at the oriC rather than at the spiralin gene (data not shown). Strain GII3-11c1, like S. citri GII-3, did produce spiralin. In the case of the control strains (both GII-3 and GII3-11c1), each plant developed severe symptoms within 2 weeks after transmission, showing that the presence of pCL15 sequences in the GII3-11c1 chromosome did not affect its transmission and pathogenicity. In contrast, in the case of the spiralin mutants, only one or two plants out of five showed symptoms, while the others did not. In addition, symptoms were produced with a delay of 2 to 7 weeks compared to those induced in the plants infected by the control strains. As shown in Table 2, two independent experiments yielded similar results. These data suggested that in the case of the spiralin mutants, very few spiroplasma cells were introduced into the plants by the leafhoppers. Therefore, depending on whether the number of spiroplasmas injected into the plants was above or below the threshold required to induce infection, plants developed or did not develop symptoms. Accordingly, spiroplasmas could be cultured only from symptomatic plants and not from plants without symptoms. In these experiments, the low transmission efficiencies of the mutants could not be explained by the inability of the mutants to multiply in the insects. As indicated in Table 2, the spiroplasma titer of the spiralin mutant GII3-9a2 (2.8 × 106 CFU/insect), was not significantly different from those determined for the control strains (1 × 106 and 1.9 × 106 for GII-3 and 7.5 × 105 and 1.2 × 106 for GII3-11c1). In addition, these titers were significantly higher than the minimum value (104 to 105 CFU/insect) required for efficient transmission under our experimental conditions (18). Interestingly, despite the delay of symptom appearance, the plants infected by the spiralin mutants developed severe symptoms (leaf yellowing and deformation, stunting) that were undistinguishable from those produced by the wild-type strain. In addition, the spiroplasma titers in symptomatic plants (ranging from 1.2 × 106 to 1.4 × 107 CFU/g of midribs) were not significantly different from those of the control strains (ranging from 4.5 × 106 to 1.2 × 107 CFU/g of midribs). However, because of the delay of infection, stunting of GII3-9a2-infected plants was less pronounced than that observed for the control strains (Fig. 7). To further confirm that symptom expression was due to the multiplication of the spiralin mutant, spiroplasmas were isolated from symptomatic plants and characterized by Southern blot hybridization. In the case of GII3-9a2, the hybridization profiles were identical to those of the spiralin mutant injected into the insects, indicating that no contaminants and no revertants were present in these plants (data not shown). In addition, the absence of spiralin in the spiroplasmas isolated from GII3-9a2-infected plants was confirmed by Western and colony immunoblotting. In the case of GII3-9a5, the detection of additional hybridizing fragments indicated the presence in the plant of a mixed population, which probably occurred because of the GII3-9a5 instability. These variants were also shown not to possess spiralin.
TABLE 2.
Experimental transmission of S. citri GII-3, GII3-9a2, GII3-9a5, and GII3-11cl to periwinkle (C. roseus) through injection to the leafhopper vector C. haematoceps
| Expt | Inoculant | Plant | Living insectsa | CFU/ insectb | Symptomsc | CFU/g of midribsd |
|---|---|---|---|---|---|---|
| 1 | SP4 medium | P1 | 5 | − | ||
| P2 | 4 | − | NDe | |||
| P3 | 8 | − | ND | |||
| GII3 (wild type) | P1 | 6 | 1 × 106 | + | 5.7 × 106 | |
| P2 | 4 | + | ND | |||
| P3 | 2 | + | ND | |||
| GII3-9a2 (Δspi) | P1 | 10 | 2.8 × 106 | + (2) | 1.2 × 106 | |
| P2 | 12 | − | ||||
| P3 | 1 | − | ||||
| P4 | 9 | − | ||||
| P5 | 7 | + (4) | 2.3 × 106 | |||
| GII3-9a5 (spi-gfp) | P1 | 9 | 4.9 × 105 | + (7) | 7.3 × 105f | |
| P2 | 10 | − | ||||
| P3 | 4 | − | ||||
| P4 | 7 | − | ||||
| P5 | 12 | − | ||||
| GII3-11c1 (ori) | P1 | 5 | 7.5 × 105 | + | 4.5 × 106 | |
| P2 | 7 | + | 5.3 × 106 | |||
| P3 | 10 | + | ND | |||
| P4 | 11 | + | ND | |||
| P5 | 2 | + | ND | |||
| 2 | SP4 medium | P1 | 8 | − | ||
| P2 | 1 | ND | ||||
| P3 | 5 | ND | ||||
| GII3 (wild type) | P1 | 7 | 1.9 × 106 | + | 1.2 × 107 | |
| P2 | 7 | + | ||||
| P3 | 6 | + | ||||
| GII3-9a2 (Δspi) | P1 | 7 | 2.9 × 106 | − | ||
| P2 | 4 | − | ||||
| P3 | 11 | + (3) | 1.4 × 107 | |||
| P4 | 5 | − | ||||
| P5 | 9 | − | ||||
| GII3-11c1 (ori) | P1 | 6 | 1.2 × 106 | + | 1 × 107 | |
| P2 | 9 | + | ND | |||
| P3 | 9 | + | 9.5 × 106 | |||
| P4 | 9 | + | ND | |||
| P5 | 9 | + | 8.5 × 106 |
Number of living insects at the end of the 2-week transmission period. All plants were initially subjected to infection by 12 injected insects.
Average of four independent determinations, each from a group of five insects taken at the end of the transmission period.
Numbers in parentheses indicate the delay (in weeks) in symptom appearance compared to that for the control strain GII-3.
Spiroplasma titers were determined at 6 (experiment 1) or 7 (experiment 2) weeks posttransmission.
ND, not done.
In this plant, spiroplasma titers were 2.2×102 and 7.3×105 CFU/g of midribs at 8 and 16 weeks posttransmission, respectively.
FIG. 7.
Symptoms induced in periwinkle plants infected by S. citri GII-3, GII3-9a2, and GII3-11c1. Plants were photographed 7 weeks after transmission.
To further study the ability of the spiralin mutants to be transmitted by the insect vector, infected leafhoppers were subjected to tests involving transmission through a Parafilm membrane. Leafhoppers were infected through injection of S. citri GII-3 and spiralin mutants GII3-9a2 and GII3-9a5. After 2 weeks on healthy periwinkles to allow spiroplasma multiplication, the insects were transferred to small cages and fed through a Parafilm membrane on spiroplasma-free culture medium for 24 h. At the end of the transmission period, the number of spiroplasmas cells injected by the insects into the medium was determined as CFU by plating on SP4 agar medium. The results of two independent experiments are presented in Table 3. In the case of S. citri GII-3, all cages yielded positive cultures, whereas only 3 out of 10 (experiment 1) and 1 out of 4 (experiment 2) were positive for GII3-9a2. In the case of the spiralin mutant, the average number of CFU transmitted per insect was roughly 100 times less than that of S. citri GII-3 (0.2 versus 22 in experiment 1 and 0.1 versus 19 in experiment 2). These results clearly indicated that despite multiplication to a high titer in the insect, the spiralinless mutant GII3-9a2 was poorly transmitted compared to the wild-type strain. For mutant GII3-9a5 the results are more difficult to analyze, as this mutant proved to be unstable, leading to a mixed population of cells harboring different genotypes (see above).
TABLE 3.
Transmission of S. citri GII-3, GII3-9a2, and GII3-9a5 through Parafilm membranes
| Expt | Strain | Spiroplasma titer in insects (CFU/insect) | No. of caged insects | No. of cages with positive culture/total | Spiroplasmas inoculated into medium
|
|
|---|---|---|---|---|---|---|
| CFU | CFU/insecta | |||||
| 1 | GII-3 | 1.6 × 106 | 40 | 8/8 | 891 | 22 |
| GII3-9a2 | 1.1 × 106 | 50 | 3/10 | 11 | 0.2 | |
| GII3-9a5 | 1.5 × 106 | 50 | 3/10 | 19 | 0.4 | |
| 2 | GII-3 | 9 × 105 | 19 | 4/4 | 358 | 19 |
| GII3-9a2 | 1.9 × 106 | 20 | 1/4 | 2 | 0.1 | |
Ratio of total number of CFU to total number of insects fed in the cages.
DISCUSSION
In this study, two S. citri mutants were generated by disrupting the spiralin gene through homologous recombination, and characterized. Mutant GII3-9a5 was obtained through a single-crossover recombination between a spiralin-GFP translational fusion carried by the targeting plasmid pCL15 and the spiralin gene present in the chromosome. Even though most of the translation products were detected as truncated or nonterminated polypeptides, this mutant produced detectable amounts of the full-length spiralin-GFP fusion protein. Interestingly enough, the detection of GFP epitopes by direct immunoblotting of colonies and immunofluorescence of intact spiroplasma cells labeled with anti-GFP antibodies strongly suggested that the fusion protein, like the wild-type spiralin, was exposed at the cell surface, probably anchored in the membrane by the N-acylated cysteine of the spiralin moiety. Thus, this work illustrates the feasibility of expressing foreign epitopes at the cell surface of a mollicute through fusion with the major surface lipoprotein.
Mutant GII3-9a2, which has no spiralin, was obtained fortuitously during isolation of GII3-9a5. Comparison of the spiralin gene regions of these mutants led to the conclusion that GII3-9a2 probably arose from GII3-9a5 through a double-crossover recombination between free pCL15 and pCL15 sequences carried by the GII3-9a5 chromosome. The occurrence of such a recombination event implies that free plasmid is present in cells having plasmid sequences integrated into their chromosomes. In previous studies, we have shown that S. citri oriC plasmids with minimal oriC sequences exert less incompatibility than those with large oriC fragments and are therefore maintained as free plasmids for many generations. Consequently, copies of free plasmid are still detected after the plasmid has integrated into the host chromosome (27; S. Duret and J. Renaudin, unpublished data). In agreement with these observations, the presence of free pCL15 was indeed detected in cells of clone 9 at the ninth passage (Fig. 2b, lane 3), from which mutants GII3-9a2 and GII3-9a5 were derived. Nevertheless, because the frequency of recombination in S. citri is very low (15, 32), the selection of a double-crossover recombination event can be explained only if it confers a strong selective advantage to the resulting mutant. In this particular case, the selective advantage of GII3-9a2 over GII3-9a5 might be the loss of GFP, the expression of which seems to be toxic for the spiroplasma. Mutant GII3-9a5, producing the spiralin-GFP fusion protein, displayed elongated cells and formed smaller colonies in agar plates, suggesting altered cell division. In addition, this mutant proved to be unstable during propagation, leading to subclones, none of which expressed the GFP epitopes. Also in agreement with a possible toxicity of GFP, several attempts to express GFP in S. citri were unsuccessful because of GFP gene deletions (S. Duret and J. Renaudin, unpublished results).
Spiralin is the most abundant membrane protein of S. citri. However, the isolation of an S. citri mutant that does not possess spiralin proved that this surface lipoprotein is not essential for cell survival. Because the whole polypeptide is exposed at the cell surface (10), it is thought that spiralin is more likely involved in the interactions of the spiroplasma with its environment than in structural features (such as morphology) of the spiroplasma cell. Indeed, studying GII3-9a2 showed that the lack of spiralin had no significant effect on the growth, motility, or helical morphology of S. citri. Furthermore, the appearance of severe symptoms in GII3-9a2-infected periwinkles indicated that spiralin is not essential for pathogenicity. In previous studies, the reversion of S. citri mutants (obtained through either transposition or targeted gene disruption) to the wild-type phenotype has made pathogenicity tests more difficult to analyze (20, 22). Interestingly, the fact that GII3-9a2 lacked sequences of the 5′ end of the spiralin gene precluded the possible occurrence of recombination events leading to a wild-type phenotype. Accordingly, no spiralin-producing revertants were detected in GII3-9a2-infected plants.
In nature, S. citri is transmitted in a propagative manner by the phloem-feeding leafhoppers C. haematoceps and C. tenellus (21, 30). For transmission to occur, the ingested spiroplasmas must infect gut epithelial cells, where they multiply before crossing into the hemocoel. Spiroplasmas continue to multiply in the hemolymph and then invade other organs, including the salivary glands, from which they are injected into the phloem via salivary secretions during insect feeding (16, 30). Therefore, the ability of S. citri to be transmitted relies on its ability to multiply in the insect tissues and to cross physical barriers such as the intestinal epithelium and the salivary gland membranes. Experimental transmission of GII3-9a2 to periwinkle through injection to the leafhopper vector showed that, despite its ability to multiply to a high titer in the insect, the spiralinless mutant was poorly transmitted. As further documented by insect feeding through Parafilm membranes, transmission of the spiralinless mutant was 100 times less efficient than that of the wild-type strain GII-3. Due to the small number of spiroplasmas introduced into the plants by the leafhoppers, symptoms were delayed for several weeks compared to infection with S. citri GII-3 or GII3-11c1, and some plants even failed to develop symptoms. As expected, no spiroplasma could be cultured from these asymptomatic plants. These results are reminiscent of previous observations showing that the progressive loss of transmissibility, due to extensive passaging in vitro, was associated with a significant reduction of the spiralin content (S. Beistero and J. Renaudin, unpublished results). These data suggest that the absence of spiralin dramatically reduces the ability of the spiroplasma to invade the salivary glands or its ability to survive in the insect saliva. However, the molecular mechanisms by which spiralin is involved are unknown.
In pathogenic mycoplasmas, adhesion to host cell membranes is the first step in the initiation of infection and is mediated by surface proteins called adhesins (26, 36). Transmission of S. citri by its leafhopper vector also involves adherence and invasion of insect host cells (30). Based on virology studies on transmission of luteovirus by aphids (24, 29), it was hypothesized that leafhopper transmission is mediated by recognition of specific spiroplasma membrane proteins, which leads to a process of receptor-mediated endocytosis (16). Recently, an adhesion-related protein (SARP1) has been characterized (3, 52). However, the involvement of SARP1 in transmission of S. citri by its insect vector has not been documented. In this respect, spiralin, which is the most abundant surface protein, might function as a ligand to interact with insect protein receptors, allowing the spiroplasma to cross cellular barriers. Therefore, it would be of interest to determine whether spiralin specifically interacts with proteins from the leafhopper vector C. haematoceps. Alternatively, spiralin might have a protecting role. Recently, a model in which spiralin could form a protein “carpet” covering most if not all lipids of the spiroplasma outer membrane bilayer was proposed (12). Such a protection of the spiroplasma membrane by spiralin might be crucial for spiroplasma survival in the insect tissues. If so, the spiralinless mutant should be more sensitive to degrading enzymes than the wild-type strain. These two hypotheses are currently being investigated. However, the finding that the spiralinless mutant GII3-9a2 displayed residual transmissibility suggests that spiroplasmal components other than spiralin are also involved in transmission of S. citri by its leafhopper vector. Indeed, very recent studies in our laboratory (A. Boutareaud, J. L. Danet, M. Garnier, and C. Saillard, submitted for publication) suggest that a lipoprotein similar to solute binding proteins of ABC transporters might also be involved.
Acknowledgments
This work was funded by INRA and the Université Victor Segalen Bordeaux 2.
We are grateful to A. André for critical reading of the manuscript and helpful discussions. We also thank P. Bonnet and J. B. Reynaud for growing plants and insects.
REFERENCES
- 1.Barbara, D. J., A. Morton, M. F. Clark, and D. L. Davies. 2002. Immunodominant membrane proteins from two phytoplasmas in the aster yellows clade (chlorante aster yellows and clover phyllody) are highly divergent in the major hydrophilic region. Microbiology 148:157-167. [DOI] [PubMed] [Google Scholar]
- 2.Berg, M., D. L. Davies, M. F. Clark, H. J. Vetten, G. Maier, C. Marcone, and E. Seemüller. 1999. Isolation of the gene encoding an immunodominant membrane protein of the apple proliferation phytoplasma, and expression and characterization of the gene product. Microbiology 145:1937-1943. [DOI] [PubMed] [Google Scholar]
- 3.Berg, M., U. Melcher, and J. Fletcher. 2001. Characterization of Spiroplasma citri adhesion related protein SARP1, which contains a domain of a novel family designated sarpin. Gene 275:57-64. [DOI] [PubMed] [Google Scholar]
- 4.Blomquist, C. L., D. J. Barbara, D. J. Davies, M. F. Clark, and B. C. Kirkpatrick. 2001. Cloning and characterization of a major membrane protein of the X-disease phytoplasma. Microbiology 147:571-580. [DOI] [PubMed] [Google Scholar]
- 5.Bové, J. M. 1993. Molecular features of mollicutes. Clin. Infect. Dis. 17(Suppl. 1):S10-S31. [DOI] [PubMed] [Google Scholar]
- 6.Bové, J. M. 1997. Spiroplasmas: infectious agents of plants, arthropods, and vertebrates. Wien Klin. Wochenschr. 109:604-612. [PubMed] [Google Scholar]
- 7.Bové, J. M., P. Carle, M. Garnier, F. Laigret, J. Renaudin, and C. Saillard. 1989. Molecular and cellular biology of spiroplasmas, p. 243-364. In R. F. Whitcomb and J. G. Tully (ed.), The mycoplasmas, vol. V. Academic Press, Inc., New York, N.Y.
- 8.Bové, J. M., X. Foissac, and C. Saillard. 1993. Spiralins, p. 203-223. In S. Rottem and I. Kahane (ed.), Subcellular biochemistry, vol 20. Mycoplasma cell membrane. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 9.Bové, J. M., J. Renaudin, C. Saillard, X. Foissac, and M. Garnier. 2003. Spiroplasma citri, a plant pathogenic mollicute: relationships with its two hosts, the plant and the leafhopper vector. Annu. Rev. Phytopathol. 41:483-500. [DOI] [PubMed]
- 10.Brenner, C., H. Duclohier, V. Krchnak, and H. Wroblewski. 1995. Conformation, pore-forming activity, and antigenicity of synthetic peptide analogs of a spiralin amphipathic α-helix. Biochim. Biophys. Acta 1235:161-168. [DOI] [PubMed] [Google Scholar]
- 11.Calavan, E. C., and J. M. Bové. 1989. Ecology of Spiroplasma citri, p. 425-487. In R. F. Whitcomb and J. G. Tully (ed.), The mycoplasmas, vol. V. Academic Press, Inc., New York, N.Y.
- 12.Castano, S., D. Blaudez, B. Desbat, J. Dufourcq, and H. Wroblewski. 2002. Secondary structure of spiralin in solution, at the air/water interface, and in interaction with lipid monolayers. Biochim. Biophys. Acta 1562:45-56. [DOI] [PubMed] [Google Scholar]
- 13.Chambaud, I., H. Wroblewski, and A. Blanchard. 1999. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 7:493-499. [DOI] [PubMed] [Google Scholar]
- 14.Chevalier, C., C. Saillard, and J. M. Bové. 1990. Organization and nucleotide sequences of the Spiroplasma citri genes for ribosomal protein S2, elongation factor Ts, spiralin, phosphofructokinase, pyruvate kinase, and an unidentified protein. J. Bacteriol. 172:2693-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duret, S., J. L. Danet, M. Garnier, and J. Renaudin. 1999. Gene disruption through homologous recombination in Spiroplasma citri: an scm1-disrupted motility mutant is pathogenic. J. Bacteriol. 181:7449-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher, J., A. Wayadande, U. Melcher, and F. Ye. 1998. The phytopathogenic mollicute-insect vector interface: a closer look. Phytopathology 88:1351-1358. [DOI] [PubMed] [Google Scholar]
- 17.Foissac, X., J. M. Bové, and C. Saillard. 1997. Sequence analysis of Spiroplasma phoeniceum and Spiroplasma kunkelii spiralin genes and comparison with other spiralin genes. Curr. Microbiol. 35:240-243. [DOI] [PubMed] [Google Scholar]
- 18.Foissac, X., J. L. Danet, C. Saillard, R. F. Whitcomb, and J. M. Bové. 1996. Experimental infection of plants by spiroplasmas, p. 385-389. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. 2. Academic Press, Inc., New York, N.Y.
- 19.Foissac, X., C. Saillard, and J. M. Bové. 1997. Random insertion of Tn4001 in the genome of Spiroplasma citri strain GII-3. Plasmid 37:80-86. [DOI] [PubMed] [Google Scholar]
- 20.Foissac, X., C. Saillard, J. L. Danet, P. Gaurivaud, C. Paré, F. Laigret, and J. M. Bové. 1997. Mutagenesis by insertion of transposon Tn4001 into the genome of Spiroplasma citri: characterization of mutants affected in plant pathogenicity and transmission to the plant by the leafhopper vector Circulifer haematoceps. Mol. Plant-Microbe Interact. 10:454-461. [Google Scholar]
- 21.Fos, A., J. M. Bové, J. Lallemand, C. Saillard, J. C. Vignault, Y. Ali, P. Brun, and R. Vogel. 1986. La cicadelle Neoaliturus haematoceps (Mulsant and Rey) est vecteur de Spiroplasma citri en méditerranée. Ann. Inst. Pasteur/Microbiol. (Paris) 137A:97-107. [DOI] [PubMed] [Google Scholar]
- 22.Gaurivaud, P., J. L. Danet, F. Laigret, M. Garnier, and J. M. Bové. 2000. Fructose utilization and pathogenicity of Spiroplasma citri. Mol. Plant-Microbe Interact. 13:1145-1155. [DOI] [PubMed] [Google Scholar]
- 23.Gaurivaud, P., F. Laigret, E. Verdin, M. Garnier, and J. M. Bové. 2000. Fructose operon mutants of Spiroplasma citri. Microbiology 146:2229-2236. [DOI] [PubMed] [Google Scholar]
- 24.Gildow, F. E. 1993. Evidence for receptor-mediated endocytosis regulating luteovirus acquisition by aphids. Phytopathology 83:270-277. [Google Scholar]
- 25.Jacob, C., F. Nouzières, S. Duret, J. M. Bové, and J. Renaudin. 1997. Isolation, characterization, and complementation of a motility mutant of Spiroplasma citri. J. Bacteriol. 179:4802-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unraveling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 27.Lartigue, C., S. Duret, M. Garnier, and J. Renaudin. 2002. New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48:149-159. [DOI] [PubMed] [Google Scholar]
- 28.Le Hénaff, M., and C. Fontenelle. 2000. Chemical analysis of processing of spiralin, the major lipoprotein of Spiroplasma melliferum. Arch. Microbiol. 173:339-345. [DOI] [PubMed] [Google Scholar]
- 29.Li, C., D. Cox-Foster, S. M. Gray, and F. Gildow. 2001. Vector specificity of barley yellow dwarf virus BYDV-MAV in the aphid, Sitobion avenae. Virology 286:125-133. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H. Y., D. J. Gumpf, G. N. Oldfield, and E. C. Calavan. 1983. The relationship of Spiroplasma citri and Circulifer tenellus. Phytopathology 73:585-590. [Google Scholar]
- 31.Maixner, M., U. Arhens, and E. Seemüller. 1995. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alterwild-type hosts, and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 101:241-250. [Google Scholar]
- 32.Marais, A., J. M. Bové, and J. Renaudin. 1996. Spiroplasma citri virus SpV1-derived cloning vector: deletion formation by illegitimate and homologous recombination in a spiroplasmal host strain which probably lacks a functional recA gene. J. Bacteriol. 178:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Gros, G., M. L. Iskra, M. Garnier, J. Gandar, and J. M. Bové. 1987. Production of monoclonal antibodies against phloem-limited prokaryotes of plants: a general procedure using extracts from infected periwinkles as immunogen. Ann. Inst. Pasteur/Microbiol. (Paris) 138:625-637. [DOI] [PubMed] [Google Scholar]
- 34.McCoy, R. E., A. Caudwell, C. J. Chang, T. A. Chen, L. N. Chiykowski, M. T. Cousin, J. L. Dale, G. T. DeLeeuw, D. A. Golino, K. J. Hackett, B. C. Kirkpatrick, R. Marwitz, H. Petzold, R. C. Sinha, M. Sugiura, R. F. Whitcomb, I. L. Yang, B. M. Zhu, and E. Seemüller. 1989. Plant diseases associated with mycoplasma-like organisms, p. 545-640. In R. F. Whitcomb and J. G. Tully (ed.), The mycoplasmas, vol. V. Academic Press, Inc., New York, N.Y.
- 35.Mouchès, C., T. Candresse, G. Barroso, C. Saillard, H Wroblewski, and J. M. Bové. 1985. Gene for spiralin, the major membrane protein of the helical mollicute Spiroplasma citri: cloning and expression in Escherichia coli. J. Bacteriol. 164:1094-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renaudin, J. 2002. Extrachromosomal elements and gene transfer, p. 347-370. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 38.Renaudin, J., and J. M. Bové. 1995. Plasmid and viral vectors for gene cloning and expression in Spiroplasma citri, p. 167-178. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology. Academic Press, San Diego, Calif.
- 39.Renaudin, J., A. Marais, E. Verdin, S. Duret, X. Foissac, F. Laigret, and J. M. Bové. 1995. Integrative and free Spiroplasma citri oriC plasmids: expression of the Spiroplasma phoeniceum spiralin in Spiroplasma citri. J. Bacteriol. 177:2800-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saglio, P., D. Laflèche, C. Bonissol, and J. M. Bové. 1971. Culture in vitro des mycoplasmes associés au stubborn des agrumes et leur observation au microscope électronique. C. R. Acad. Sci. 272:1387-1390. [Google Scholar]
- 41.Saglio, P., M. Lhospital, D. Laflèche, G. Dupont, J. M. Bové, J. G. Tully, and E. A. Freundt. 1973. Spiroplasma citri gen. and sp. n.: a mycoplasma-like organism associated with “stubborn” disease of citrus. Int. J. Syst. Bacteriol. 23:191-204. [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Seemüller, E., M. Garnier, and B. Schneider. 2002. Mycoplasmas of plants and insects, p. 91-115. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 44.Seemüller, E., C. Marcone, U. Lauer, A. Ragozzino, and M. Göschl. 1998. Current status of molecular classification of the phytoplasmas. J. Plant Pathol. 80:3-26. [Google Scholar]
- 45.Stamburski, C., J. Renaudin, and J. M. Bové. 1991. First step toward a virus-derived vector for gene cloning and expression in spiroplasmas, organisms which read UGA as a tryptophan codon: synthesis of chloramphenicol acetyltransferase in Spiroplasma citri. J. Bacteriol. 173:2225-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignault, J. C., J. M. Bové, C. Saillard, R. Vogel, A. Faro, L. Venegas, W. Stemmer, S. Aoki, R. E. McCoy, A. S. Al-Beldawi, M. Larue, O. Tuzcu, M. Ozsan, A. Nhami, M. Abassi, J. Bonfils, G. Moutous, A. Fos, F. Poutiers, and G. Viennot-Bourgin. 1980. Mise en culture de spiroplasmes à partir de matériel végétal et d'insectes provenant de pays circum méditerranéens et du Proche Orient. C. R. Acad. Sci. Ser. III 290:775-780. [Google Scholar]
- 47.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. Van Etten, J. Maniloff, and C. R. Woese. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitcomb, R. F. 1983. Culture media for spiroplasmas. Methods Mycoplasmol. 1:147-159. [Google Scholar]
- 49.Wieslander, A., and M. Rosén. 2002. The cell membrane and transport, p. 131-161. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 50.Wroblewski, H., K. E. Johansson, and S. Hjerten. 1977. Purification and characterization of spiralin, the main protein of the Spiroplasma citri membrane. Biochim. Biophys. Acta 465:275-289. [DOI] [PubMed] [Google Scholar]
- 51.Ye, F., J. Renaudin, J. M. Bové, and F. Laigret. 1994. Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr. Microbiol. 29:23-29. [DOI] [PubMed] [Google Scholar]
- 52.Yu, J., A. C. Wayadande, and J. Fletcher. 2000. Spiroplasma citri surface protein P89 implicated in adhesion to cells of the vector Circulifer tenellus. Phytopathology 90:716-722. [DOI] [PubMed] [Google Scholar]
- 53.Yu, Y.-L., K.-W Yeh, and C.-P Lin. 1998. An antigenic protein gene of a phytoplasma associated with sweet potato witches' broom. Microbiology 144:1257-1262. [DOI] [PubMed] [Google Scholar]