Abstract
Free-living thermotolerant amoebae pose a significant health risk to people who soak and swim in habitats suitable for their growth, such as hot springs. In this survey of 23 different hot springs in Yellowstone and Grand Teton National Parks, we used PCR with primer sets specific for Naegleria to detect three sequence types that represent species not previously described, as well as a fourth sequence type identified as the pathogen Naegleria fowleri.
The vahlkampfiid, or free-living, amoebae are found worldwide in soil and aquatic environments, where they live as phagotrophs feeding on bacteria and detritus (3, 6). Some are thermotolerant and thrive in naturally and artificially heated water in spas, hot springs, stagnant lakes and ponds, rivers, and effluents from power plants. These opportunistic parasites can infect humans and other mammals and cause serious eye or brain infections (8, 17). Because the amoebae are widely distributed in nature, any warm body of water may harbor the organisms and pose a threat. One thermotolerant amoeba, Naegleria fowleri, causes a rare and potentially fatal meningoenchephalitis, contracted from swimming or bathing in contaminated water (9). Therefore, it is important to rapidly detect thermotolerant amoebae in hot springs where people swim and bathe because of their possible impact on human health.
Traditional methods of isolation and culturing are time-consuming and identify only organisms amenable to growth under defined conditions (19). Even if culturable, most species in the genus Naegleria are morphologically indistinguishable (3). Furthermore, microbial isolates that grow in the laboratory may not adequately represent organisms found in natural communities (4). PCR-based approaches for recovery and analysis of rRNA and other gene sequences from the environment provide a relatively rapid means of detection and enhance identification of microorganisms that are not easily distinguished by conventional cultivation and microscopic methods (2).
Our objective in this study was to determine if hot springs in Yellowstone and Grand Teton National Parks harbor pathogenic amoebae. We used PCR amplification, cloning, and sequencing methods to detect taxonomically informative small- and large-subunit-rRNA internal transcribed spacer (ITS) region sequences of Naegleria from samples collected in the parks.
MATERIALS AND METHODS
Sample collection.
The sampling sites were located in Yellowstone and Grand Teton National Parks, in warm water pools and hot springs or sites where heated water flowed into freshwater streams (Table 1).
TABLE 1.
Locations and descriptions of the sites examined
| Sampling site | Date (mo/day/yr) | Temperature (°C) | pHa |
|---|---|---|---|
| Boiling River | 8/23/01 | 35 | 7.3 |
| Nymph Creek | 9/18/01 | 28 | 3.3 |
| Dead Savage Spring | 5/30/02 | 35 | 6.5 |
| Ranger Pool | 5/30/02 | 35 | 6.5 |
| Hillside Springs | 5/30/02 | 35 | 6.8 |
| Seismic Geyser | 5/30/02 | 40 | 7.4 |
| Mallard Lake Trail | 5/30/02 | 38 | 3.3 |
| Madison Campground | 10/2/01 | 37 | 6.7 |
| Firehole Swim Area | 10/2/01 | 14 | ND |
| Terrace Springs | 5/30/02 | 40 | 7.1 |
| Bathtub | 5/30/02 | 36 | 6.1 |
| Spirea Creek | 7/8/02 | 41 | 8.3 |
| Huckleberry Hot Springs | 6/20/02 | 36 | 6.8 |
| Upper Polecat Creek | 6/20/02 | 40 | 6.2 |
| Lower Polecat Creek | 6/20/02 | 37 | 6.5 |
| Kelly Warm Springs | 10/12/01 | 20 | 8.1 |
| Obsidian Creek | 9/6/02 | 37 | 2.2 |
| Water Tower Road | 9/1/02 | 38 | 6.6 |
| Dunanda | 9/23/02 | 36 | ND |
| Three Rivers | 9/20/02 | 39 | ND |
| Scout Pool | 9/27/02 | 21 | ND |
| Morning Falls | 9/28/02 | 25 | ND |
| Sheepeater Cliffs | 10/18/02 | 3 | 7.2 |
ND, no data.
The types of samples collected varied considerably from site to site and included sediments, algal mats, or biofilm scrapings of rock surfaces. Approximately 5 ml of material from at least five sampling locations at each hot pool or stream was collected by using a clean metal spatula. The samples were placed into 50-ml sterile centrifuge tubes, and approximately 30 ml of water from the hot pool or stream was added to the tube. The samples were immediately placed on dry ice and kept frozen until further processing.
In the laboratory, the samples were thawed at room temperature, vortexed vigorously, and centrifuged at 2,000 × g to pellet the material. Supernatant fluids were saved for pH measurements. The pellets were suspended by vortexing in 2 ml of saved supernatant fluids. One milliliter of each resuspended sample solution was placed in 2-ml screw-cap microcentrifuge tubes and mixed vigorously with 1 ml of a 1:1 mixture of absolute ethanol and 10 mM Tris-HCl (pH 7.5)-10 mM NaCl-1 mM EDTA (pH 8). The samples were stored at −20°C.
For remote sampling sites, where transport of dry ice was not feasible, the following method was used. Sediment or algal mat samples were collected to fill about one-half the volume of 2-ml sterile screw-cap microcentrifuge tubes, each containing 1 ml of a 1:1 mixture of absolute ethanol and 10 mM Tris-HCl (pH 7.5)-10 mM NaCl-1 mM EDTA (pH 8). The samples were shaken vigorously, kept at ambient temperature, returned to the laboratory, and stored at −20°C.
Nucleic acid extraction and amplification.
DNA was extracted as previously described (14). The quantity and yield of DNA were visualized with UV light on 1% agarose gels in 1× Tris-borate-EDTA buffer (50 mM Tris-HCl, 50 mM boric acid, 1 mM EDTA) containing ethidium bromide and a 100-bp DNA ladder (Promega Corporation, Fitchburg, Wis.) as a standard.
PCR amplification was performed according to the manufacturer's instructions by using a Taq Gold DNA polymerase kit (Applied Biosystems, Foster City, Calif.) and a deoxynucleoside triphosphate mix (Promega).
Two primer sets were used in this study. One set, referred to as the ITS primer set, 5′-GAACCTGCGTAGGGATCATTT-3′ and 5′-TTTCTTTTCCTCCCCTTATTA-3′, complements sites on the 18S rRNA gene adjacent to the ITS-1 region and on the 23S rRNA gene approximately 70 nucleotides from the ITS region (10). This region includes a contiguous segment containing an ITS-1 region that is highly variable in length and nucleotide sequence, followed by a 5.8S rRNA gene that is relatively conserved in length and nucleotide sequence, followed by an ITS-2 region that is highly variable in length and nucleotide sequence. This set yields a PCR product that includes the ITS-1, 5.8S rRNA gene, and ITS-2 regions.
A second primer set specific for N. fowleri, FW, 5′-GTGAAAACCTTTTTTCCATTTACA-3′ and 5′-AAATAAAAGATTGACCATTTGAAA-3′, complements sites in the ITS-1 and ITS-2 regions (10).
Reaction conditions for both primer sets were 95°C for 5 min, followed by 30 cycles of 95°C for 15 s, 55°C for 90 s, 72°C for 90 s, and an extension at 72°C for 10 min.
DNA from an N. fowleri isolate (American Type Culture Collection accession number 30863) was extracted as described above and used as a positive control for all PCRs.
Clone library construction.
PCR amplicons were cloned with a TA cloning kit by using pCR2.1-TOPO and Escherichia coli TOP10 competent cells (Invitrogen, Carlsbad, Calif.) to produce libraries (18). Individual colonies were selected, and their plasmids were extracted by using the Promega Plus SV miniprep DNA purification system. The plasmids were assayed for inserts by restriction digestion according to the directions in the TA cloning kit.
Sequencing and phylogenetic analysis.
Plasmid inserts were sequenced by using M13 forward and M13 reverse primers with an ABI Prism BigDye Terminator reaction kit and an ABI 310 DNA sequencer (Applied Biosystems).
The cloned sequences were compared to a current database of genetic sequences (GenBank) by using BLAST (1). The sequences were aligned by using Sequencher 3.1.1 (Gene Codes Corporation, Ann Arbor, Mich.) and Clustal X (5). A phylogenetic tree illustrating the relationships of the Yellowstone and Grand Teton National Park sequence types to each other and to the sequences of cultivated Naegleria spp. was constructed by using the neighbor-joining algorithm (13) within PAUP* 4.0b8 (16) with distance correction set to the Kimura two-parameter model (7). The tree included nucleotides corresponding to the complete ITS-1 (42 nucleotides) region and 5.8S rRNA gene (175 nucleotides) and the first 32 nucleotides of the ITS-2 region of N. fowleri G1-3-e3a (GenBank accession number AJ132019). In some cases, the ITS-1 sequences of the reference Naegleria spp. were much longer than our sequences of interest. These regions were not used in constructing the tree since they were nonhomologous and, in our analyses, could not be unambiguously aligned. Similarly, a large portion of the ITS-2 region of each sequence was not used to generate the tree. Small gaps of 1 to 4 nucleotides were present in the alignments and occurred between regions of high homology. These gaps were interpreted as real deletion or insertion events that represent divergence between sequences. They were included in the tree analysis.
This project was conducted under the direction of the Yellowstone Center for Resources following the guidelines for research in Yellowstone National Park.
Nucleotide sequence accession numbers.
All rDNA sequences from the clone libraries were deposited in GenBank (accession numbers AY274812, AY274813, and AY274814).
RESULTS AND DISCUSSION
We used molecular techniques to survey for Naegleria from 23 different aquatic geothermal sites in Yellowstone and Grand Teton National Parks. Temperatures and pH ranged from 3 to 41°C and 2.2 to 8.3, respectively (Table 1). PCR amplifications were performed on 81 samples with the ITS and FW primer sets. Eleven of the sites yielded PCR products with the ITS primer set. The Boiling River site was the only site that yielded a PCR product with both primer sets. We prepared rDNA PCR clone libraries for each of these products. Seventy-five ITS clones and nine FW clones were sequenced (Table 2).
TABLE 2.
Number of clones found at various locations in Yellowstone and Grand Teton National Parks for four sequence types of Naegleriaa
| Sampling site | No. of clones with indicated sequence type
|
|||
|---|---|---|---|---|
| AHHS1-4 | ANC11-1 | ASG3-1 | AUPC1-8 | |
| Boiling River | 2 | 1 | 9 | 0 |
| Nymph Creek | 3 | 10 | 0 | 0 |
| Hillside Springs | 7 | 1 | 0 | 0 |
| Seismic Geyser | 3 | 0 | 2 | 0 |
| Mallard Lake Trail | 4 | 0 | 1 | 0 |
| Madison Campground | 1 | 4 | 0 | 0 |
| Terrace Spring | 8 | 0 | 0 | 0 |
| Bathtub | 5 | 1 | 0 | 0 |
| Huckleberry Hot Spring | 15 | 0 | 0 | 0 |
| Upper Polecat Creek | 6 | 0 | 0 | 1 |
| Total | 54 | 17 | 12 | 1 |
The number of clones for ASG3-1 was determined by using the FW primer set. For all other types, the ITS primer set was used.
Four different Naegleria-like sequence types were detected. These were designated types AGS3-1, ANC11-1, AHHS4-1, and AUPC1-8. Type ASG3-1 was identical to the cultivated N. fowleri G1-3-e3a strain (GenBank accession number AJ132019) over the entire ITS-1 region, 5.8S rRNA gene, and ITS-2 region.
When BLAST searches were performed with the entire ITS-1 region, 5.8s rRNA gene, and ITS-2 region of the remaining three sequence types, only the 5.8S rRNA gene portion of the sequences matched sequences in the database. BLAST searches using only the ITS-1 or ITS-2 regions did not return results indicating high homology to other ITS-1 or ITS-2 sequences in the database. Based on 5.8S rRNA gene sequences, the ANC11-1 type was most similar (∼98%) to N. sturti (Y10195) and N. niuginiensis (Y10193). The AHHS1-4 sequence type was most similar (∼91%) to N. fowleri (AJ132028) and N. carteri (Y10191). The AUPC1-8 sequence type was represented by a single clone and was most similar (∼98%) to AHHS1-4.
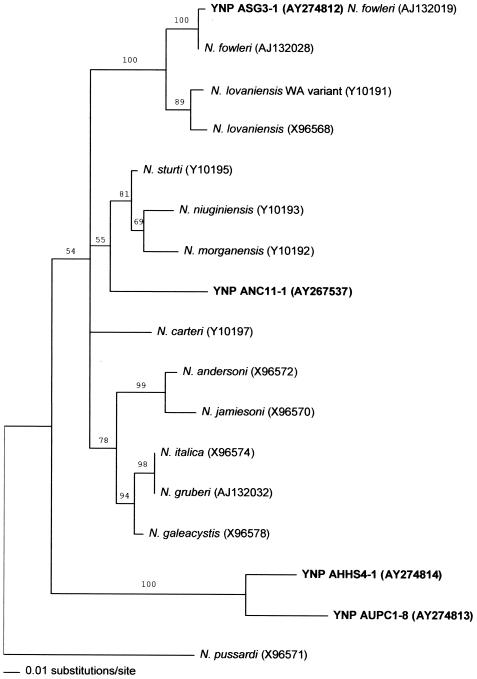
A phylogenetic tree based on the ITS-1 and 5.8S rRNA gene regions and approximately the first 30 nucleotides of the ITS-2 region was constructed to illustrate the relationships of the Yellowstone and Grand Teton National Park cloned sequence types to each other and to the sequences of cultivated Naegleria spp. (Fig. 1). As discussed above, the ASG3-1 sequence type matched that of N. fowleri (AJ132019). The ANC11-1 sequence formed a clade with N. sturti, N. niuginiensis, and N. morganensis, but bootstrap values for its inclusion were low. The AHHS1-4 and AUPC1-8 sequences formed a clade with high bootstrap support. These results suggest that the organisms harboring the sequence types ANC11-1, AHHS1-4, and AUPC1-8 are not represented in the database by known Naegleria species. Thus, the exact phylogenetic relationships of these types with other Naegleria species are unclear.
FIG. 1.
Neighbor-joining analysis using the 5.8S rRNA gene sequences and portions of the adjacent ITS-1 and ITS-2 regions of Naegleria species from GenBank and those PCR-amplified from bulk DNA extracted from hot springs in Yellowstone and Grand Teton National Parks. The tree was generated by using PAUP*, distances were calculated by using the Kimura two-parameter model, and bootstrap values of >50% are indicated. YNP, Yellowstone National Park.
The ANC11-1 sequence (GenBank accession number AY267537) was previously identified as a possible new species in our study of cloned ITS region sequences at a low-pH site (Nymph Creek) (14). The sequence was detected again at Nymph Creek and at additional sites in this study (Table 2). A total of 17 clones contained the ANC11-1 sequence type. This sequence was the most prevalent type (10 of 13 clones) in the low-pH Nymph Creek site and in the Madison Campground site (4 of 5 clones), where the pH was 6.7. Its abundance in this nearly neutral pH site suggests that the organism harboring this sequence is not limited to low-pH springs.
The AHHS1-4 sequence type was detected in every sample that yielded a PCR product (Table 2). This type was the most abundant sequence in 8 of 10 clone libraries and was the most frequent clone detected overall (54 of 75 clones). It occurred over a wide pH range. This finding suggests that this sequence type may represent a particularly abundant species in the parks' geothermal features. Because of its prevalence and the potential for some Naegleria spp. to cause infections, obtaining isolates and assessing the virulence of the organism harboring this sequence type are of particular interest.
The ASG3-1 sequence (an N. fowleri-like sequence) was rare among the clones detected with the ITS primer set. It was retrieved from two clones from the Seismic Geyser site and from one clone from the Mallard Lake Trail site. In contrast, the ASG3-1 sequence was the only sequence detected by using the N. fowleri-specific FW primer set. All the ASG3-1 sequences detected by using the FW primer set (a total of nine) were retrieved from the Boiling River site, which was the only site that yielded a PCR product with the FW primer set. The detection of this N. fowleri sequence in the low-pH Mallard Lake site suggests that the organism harboring this sequence can tolerate low-pH environments.
We found seven instances with unique sequences represented by single clones. In all but one case (see below), these unique sequences were <1.0% different from one another or from the defined sequence types (ASG3-1, ANC11-1, or AHHS4-1) over their entire ITS regions. Studies have shown that such sequence microvariation is inherent in a PCR-amplification-cloning-sequencing approach to microbial community analysis (11, 15). The best evidence that a cloned sequence from an environmental sample is not an artifact is its repeated detection in clone libraries from different sites or its identical match to a cultivated strain. In this study, unique clones with <1% sequence differences from definite sequence types were included as part of the defined group. One sequence, AUPC1-8, represented by a single clone, was >1.0% different (98% similar over the 5.8S rRNA gene sequence) from the AHHS4-1 sequence type. Since this degree of variation falls outside that exhibited by other unique clones in this study, we do not consider this to be a methodological artifact.
The ASG3-1 sequence type matches a known pathogenic species, N. fowleri. This implies that N. fowleri is present in the Boiling River, Seismic Geyser, and Mallard Lake Trail sites. However, only the Boiling River site yielded a PCR product with the N. fowleri-specific (FW) primer set. This finding is inconsistent with the detection of N. fowleri (ASG3-1) sequences from the Seismic Geyser and Mallard Lake Trail sites with the ITS primer set, since the ITS-generated clones contained regions complementary to the FW primers. Similarly, the absence of the ASG3-1 type in the Boiling River ITS clone library was inconsistent with the detection of the ASG3-1 type at this site with the FW primer set. These inconsistencies may reflect variability in sample composition, the presence of inhibitory substances in the DNA preparations, or suboptimal PCR conditions for some samples.
There are few reported studies of the occurrence of Naegleria species, other than N. fowleri, from continents besides Europe and Australia (3). A previous survey of Naegleria-like amoebae in hot springs in Yellowstone and Grand Teton National Parks utilized cultivation and microscopy approaches for detection, identification, and determination of virulence (12). Ramaley et al. (12) found thermotolerant amoebae in numerous hot springs and runoff channels in the parks and surrounding areas. Naegleria species isolates accounted for almost half of the amoebae observed. Although no pathogenic isolates of N. fowleri were found in the study, isolates of N. australiensis that were virulent in mice were identified and verified by using isoenzyme analysis. No amoebae were isolated from acidic springs in their study.
Our study is the first to use a PCR-based approach to document the presence of N. fowleri, as well as Naegleria sequence types not previously described, in a variety of aquatic geothermal sites found in Yellowstone and Grand Teton National Parks. Three of the four sequence types detected were not represented by cultivated species in GenBank. When considered with the Ramaley et al. study, our results provide evidence that thermotolerant amoebae, including N. fowleri, are not transient organisms but thrive in some hot springs. These amoebae may pose health risks to people who use the hot springs for recreation.
Acknowledgments
We are grateful to the National Science Foundation (Microbial Observatory grant 9977922); the National Park Service, Department of the Interior; and the Thermal Biology Institute at Montana State University for financial support.
Thanks to all who assisted in the field and laboratory: Tai Takenaka, Emily Kuhn, Dean Snow, Bob Seibert, Wes Miles, Kathleen O'Leary, Lane Baker, David Daniels, Mike Keller, Mark Sheehan, Mike Sheehan, and the Bechler Ranger Station staff. Thanks to Mary Bateson for sequencing. We thank John Varley, Tom Oliff, Anne Deutch, Christie Hendrix, and Liz Cleveland for their assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral Zettler, L. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 3.De Jonckheere, J. F. 2002. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 41:309-342. [Google Scholar]
- 4.Ferris, M. J., A. L. Ruff-Roberts, E. D. Kopczynski, M. M. Bateson, and D. M. Ward. 1996. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl. Environ. Microbiol. 62:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeanmougin, F., J. D. Thompson, M. Guoy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 6.John, D. T. 1993. Opportunistically pathogenic free-living amebae, p. 143-246. In J. P. Kreier and J. R. Baker (ed.), Parasitic protozoa. Academic Press, Inc., San Diego, Calif.
- 7.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 8.Kollars, T. M., Jr., and W. E. Wilhelm. 1996. The occurrence of antibodies to Naegleria species in wild mammals. J. Parasitol. 82:73-77. [PubMed] [Google Scholar]
- 9.Martinez, A. J., and G. S. Visvesvara. 1997. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 7:583-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pélandakis, M., S. Serre, and P. Pernin. 2000. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 47:116-121. [DOI] [PubMed] [Google Scholar]
- 11.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaley, R. F., P. L. Scanlan, and W. D. O'Dell. 2001. Presence of thermophilic Naegleria isolates in the Yellowstone and Grand Teton National Parks, p. 41-50. In A.-L. Reysenbach (ed.), Thermophiles: biodiversity, ecology, and evolution. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 13.Saitou, N., and M. Nei. 1997. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 14.Sheehan, K. B., M. J. Ferris, and J. M. Henson. 2003. Detection of Naegleria sp. in a thermal, acidic stream in Yellowstone National Park. J. Eukaryot. Microbiol. 50:263-265. [DOI] [PubMed] [Google Scholar]
- 15.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. D. Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swofford, D. L. 2001. PAUP* 4.0b8. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Inc., Sutherland, Mass.
- 17.Szénási, Z., T. Endo, K. Yagita, and E. Nagy. 1998. Isolation, identification and increasing importance of ‘free-living’ amoebae causing human disease. J. Med. Microbiol. 47:5-16. [DOI] [PubMed] [Google Scholar]
- 18.Vergin, K. L., M. S. Rappe, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-940. [DOI] [PubMed] [Google Scholar]
- 19.Ward, D. M., M. M. Bateson, R. Weller, and A. L. Ruff-Roberts. 1992. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 12:219-286. [Google Scholar]