Abstract
Bacterial diversity in a deep-sea sediment was investigated by constructing actinobacterium-specific 16S ribosomal DNA (rDNA) clone libraries from sediment sections taken 5 to 12, 15 to 18, and 43 to 46 cm below the sea floor at a depth of 3,814 m. Clones were placed into operational taxonomic unit (OTU) groups with ≥99% 16S rDNA sequence similarity; the cutoff value for an OTU was derived by comparing 16S rRNA homology with DNA-DNA reassociation values for members of the class Actinobacteria. Diversity statistics were used to determine how the level of dominance, species richness, and genetic diversity varied with sediment depth. The reciprocal of Simpson's index (1/D) indicated that the pattern of diversity shifted toward dominance from uniformity with increasing sediment depth. Nonparametric estimation of the species richness in the 5- to 12-, 15- to 18-, and 43- to 46-cm sediment sections revealed a trend of decreasing species number with depth, 1,406, 308, and 212 OTUs, respectively. Application of the LIBSHUFF program indicated that the 5- to 12-cm clone library was composed of OTUs significantly (P = 0.001) different from those of the 15- to 18- and 43- to 46-cm libraries. FST and phylogenetic grouping of taxa (P tests) were both significant (P < 0.00001 and P < 0.001, respectively), indicating that genetic diversity decreased with sediment depth and that each sediment community harbored unique phylogenetic lineages. It was also shown that even nonconservative OTU definitions result in severe underestimation of species richness; unique phylogenetic clades detected in one OTU group suggest that OTUs do not correspond to real ecological groups sensu Palys (T. Palys, L. K. Nakamura, and F. M. Cohan, Int. J. Syst. Bacteriol. 47:1145-1156, 1997). Mechanisms responsible for diversity and their implications are discussed.
Deep-sea sediments cover 63.5% of the Earth's surface (12) and represent the most undersampled marine habitat (5). Deep-sea sediments, once thought of as uniform featureless environments, are extremely heterogeneous; recent reports suggest that species diversity in deep-sea sediments is greater than in coastal sediments (16, 26) and may even rival that of tropical rainforests (38). To date, few studies have focused on microbial diversity in nonhydrothermal vent system deep seas (8, 18, 21, 23, 27, 57), and fewer have investigated diversity through depth profiles (21, 57). Furthermore, all microbial diversity investigations made using marine sediments represent surveys of microbial diversity; none have attempted to estimate and compare bacterial diversity at different sites.
The estimation of bacterial diversity is required for understanding bacterial biogeography, community assembly, and ecological processes (9). To date the inability to accurately measure bacterial diversity has been limited by the size of samples needed to give adequate sample coverage of bacterial communities (11) and by the lack of a clear concept of the bacterial species as an evolving entity (49). Currently, species definition is based on the degree of DNA-DNA relatedness between two bacterial isolates (47). Diversity predictions can be made using statistical approaches that estimate species number from relatively small sample sizes. Hughes et al. (19) recently reviewed both rarefaction and richness estimators that have been applied to microbial data sets and in particular highlighted the utility of nonparametric estimators in predicting and comparing bacterial species number. Rarefaction and richness estimators rely on a species or operational taxonomic unit (OTU) definition. OTU definitions are usually based on 16S ribosomal DNA (rDNA) gene similarity under the assumption that a high degree of similarity will be reflected in the degree of DNA-DNA relatedness, i.e., likely to be the same species (48). The limitations of OTUs have previously been addressed and include inconsistent arbitrary cutoff values (1 to 5% 16S rDNA gene dissimilarities have been used [19]) and the fact that OTUs are counted equivalently despite the fact that some may be highly divergent and phylogenetically unique, whereas others may be closely related and phylogenetically redundant (32). Recently statistical analyses borrowed from population genetics and systematics have been employed and reviewed for use with microbial data sets (32). The benefits of such methods include the fact that OTU definitions are not required and that they do not rely on estimation of the frequency of different sequences i.e., it is phylogenetic diversity and not species richness that is estimated. The estimation of species richness combined with knowledge of phylogenetic diversity will advance our ability to test inferences as to the processes that govern bacterial diversity.
In this study we restricted our investigation to the class Actinobacteria, in order to improve coverage of species in reasonably sized 16S rDNA clone libraries and since diversity estimates are likely to be more applicable when applied to specific groups (17). Members of this class include marine species of value for the production of novel bioactive compounds for biotechnology (3, 33, 45); hence, knowledge of their diversity, distribution, and ecology in marine systems will aid bioprospecting strategies. This is the first study designed to investigate bacterial diversity in a deep-sea sediment by comparing phylogenetic diversity and estimating species number using a number of statistical methods. The principal results show that species richness decreased with sediment depth with a concomitant decrease in genetic diversity and an increase in the level of dominance. Each of the sediment core sections contained unique phylogenetic lineages, and furthermore, correlation of 16S rDNA and DNA-DNA reassociation combined with phylogenetic diversity comparisons showed that clones grouped into operational taxonomic units are likely to be derived from distinct species corresponding to separate ecological units.
MATERIALS AND METHODS
Sediment samples.
Atlantic ocean deep-sea sediment was collected from the edge of the Saharan debris flow near the Canary Islands (27o02.39′N 18o29.02′W) at a depth of 3,814 m using a piston corer during a scientific cruise aboard the RRS Charles Darwin in April 2001. The upper 38 cm of the core was a single turbidite (identified as turbidite A [54]) grading from fine/medium sand at the base to very soft mud at the top; below the turbidite the core was comprised of pelagic clay. Sections were taken at 5 to 12 cm (fine-grained muddy turbidite), 15 to 18 cm (sandy basal layer), and 43 to 46 cm (pelagic clay) below the surface of the sediment. The deposition time of the turbidite was radiocarbon dated according to the method of Thomson and Weaver (54). Approximate dating of the pelagic clay was conducted by visual inspection. Prediction of the age range of the initial sediment from which the turbidite originated was calculated according to the method of Weaver and Thomson (58). Total organic carbon contents were measured using the Walkley-Black method (1).
DNA extraction, amplification, and cloning.
DNA was extracted and concentrated from 10 g of the sediment sections using an UltraClean Mega DNA soil kit (Mo Bio Laboratories, Solana Beach, Calif.) according to the manufacturer's instructions. DNA was subjected to a second round of purification using a Wizard DNA spin-purification column (Promega, Madison, Wis.) and quantified against known concentration standards with Quantity-One software (Bio-Rad, Richmond, Calif.).
PCR amplification of actinobacterial 16S rDNA.
PCR was made in a total volume of 50 μl containing 1× PCR buffer with 1.5 mM MgCl2, deoxynucleoside triphosphates (200 μM [each] dATP, dCTP, dGTP, and dTTP), 0.5 μM [each] S-C-Act-0235-a-S-20 and S-C-Act-0878-a-A-19 primers (46), 2.0 U of DNA polymerase (Expand-Taq; Roche Diagnostics, Mannheim, Germany), and 100 ng of DNA extract. To increase amplification efficiencies from sediment DNA, T4 gene 32 protein (Roche Diagnostics) was added at a concentration of 5 μg per reaction (52). Amplification was made using a “Touchdown” protocol (39), which consisted of an initial denaturation at 95°C for 4 min, followed by denaturation at 95°C for 45 s, annealing at 72°C for 45 s, and extension at 72°C for 1 min; 10 cycles in which the annealing temperature was decreased by 0.5°C/cycle from the preceding cycle; and then 15 cycles of 95°C for 45 s, 68°C for 45 s, and 72°C for 1 min, with the last cycle followed by a 5-min extension at 72°C. PCR products were separated on 2% agarose gels stained with ethidium bromide (41).
Cloning and sequencing.
PCR products were purified using QIAquick gel extraction columns according to the manufacturer (Qiagen, Crawley, United Kingdom). Purified DNA was blunt-end ligated into the plasmid vector pST-Blue-1 and used to transform NovaBlue competent cells using a Perfectly Blunt cloning kit (Novagen, Madison, Wis.). Transformed cells were serially diluted and plated onto Luria-Bertani agar containing 50 μg of carbenicillin ml−1, 15 μg of tetracycline ml−1, 70 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside ml−1, and 80 μg of isopropyl-β-d-thiogalactopyranoside ml−1. Small actinobacterial 16S rDNA clone libraries were made for each sediment section by selecting the dilution (for each sediment core section) that yielded between 40 and 70 positive transformants. Transformants were screened by PCR using the primers S-C-Act-0235-a-S-20 and S-C-Act-0878-a-A-19 (46), and plasmids containing actinobacterial 16S rDNA inserts were sequenced commercially (Qiagen) using S-C-Act-0235-a-S-20. Chimera analysis was done as previously described (46).
OTU definition for the class Actinobacteria.
An OTU definition for the class Actinobacteria was made by comparing and plotting DNA-DNA reassociation and 16S rRNA homology from sample data published between 1997 and 2001 (369 separate samples; data available from the corresponding author). Actinobacterial 16S rRNA sequences recovered from each of the sediment sections were placed into OTU groups by comparison using the SIMILARITY_MATRIX function of the Ribosomal Database Project (31). The closest representative for each OTU group was identified using BLAST (2).
Phylogenetic tree construction.
Actinobacterial 16S rDNA sequences recovered from the sediment sections were uploaded to the CLUSTAL X interface, where they were aligned to generate a guide dendrogram from which a final alignment was made (53). All phylogenic calculations were made using programs available in the phylogeny inference package PHYLIP (version 3.5.1; J. Felsenstein, Department of Genetics, University of Washington [http://evolution.genetics.washington.edu/phylip.html]). A distance matrix was constructed from the alignment using the DNADIST program. The phylogenetic tree was generated using the neighbor-joining method from the NEIGHBOR program under the Jukes-Cantor model of nucleotide substitution. Bootstrap analysis was conducted using the SEQBOOT and CONSENSE programs using 100 resamplings of the data.
Sample coverage and species richness estimation.
All species richness and sample coverage calculations were performed with the programs EstimateS (version 5.0.1; R. K. Colwell, University of Connecticut [http://viceroy.eeb.unconn.edu/estimates]) and SPADE (species prediction and diversity estimation; A. Chao and T.-J. Shen [http://chao.stat.nthu.edu.tw]). Each cloned sequence was treated as a separate sample, and 100 randomizations were conducted for all tests. Accumulation curves were calculated by plotting the proportion of individuals sampled against the proportion of OTUs observed. The actinobacterial species richness was calculated for each of the sediment sections using the nonparametric estimators ACE (abundance-based coverage estimator) and Chao1 (6, 7, 19). Since the distribution of estimates is not normal (6), Burnham's log transformation (6) was employed to calculate 95% confidence intervals so that the lower bound of the resulting interval is at least the number of observed species: {S + [(Ñ − S)/C], S + [(Ñ − S)C], where C = exp(1.96{log[1 + σ2/(Ñ − S)2]}0.5), where Ñ is the estimate, S is the number of species observed, and σ is the variance (calculated from the closed-form solutions for the estimators) (6, 7). Extrapolation using best-fit regression analysis was performed (where necessary) to calculate the point at which 95% confidence intervals (CIs) did not overlap (19). Regression analysis was performed using SigmaPlot (version 7.1) (Jandel Scientific, San Rafael, Calif.).
Statistical comparison of coverage.
Actinobacterial 16S rDNA libraries from the three sediment sections were compared using the LIBSHUFF computer program (44) (http://www.arches.uga.edu/∼whitman/libshuff.html), which used the coverage formula of Good (15) to generate homologous and heterologous coverage curves from the 16S rDNA clone libraries. Sequences were randomly shuffled 999 times between samples prior to the distance between the curves being calculated using the Cramér-von Mises test statistic (37). The DNADIST program of PHYLIP (see above URL) using the Jukes-Cantor model for nucleotide substitution was used to generate the distance matrix analyzed by LIBSHUFF.
Reciprocal of Simpson's index.
The reciprocal of Simpson's index (1/D) was used as a measure of diversity, since it has been widely used for ecological studies, has good discriminating ability, and has previously been applied to microbial communities (61).
Lineage-per-time plots.
Plots were made according to the method of Martin (32). Cladograms were made by clustering of genetic distances corrected by the F84 (maximum-likelihood) model of nucleotide substitution. Genetic distances were calculated using DNADIST (see above URL for PHYLIP), and optimization of branch lengths was done using KITSCH (see above URL), enforcing a molecular clock (i.e., under the constraint that all sequences were contemporary).
Comparison of phylogenetic diversity.
The phylogenetic diversity within each community was compared using F statistics (FST) and phylogenetic grouping of taxa (P tests) (32). The FST test was used to compare the genetic diversity within each sediment section community to the total genetic diversity of the communities combined, using the equation FST = (θT − θW)/θT, where θT is the genetic diversity for all samples and θW is the genetic diversity in each community (32). Population differentiation using FST was calculated using the ARLEQUIN program (42); statistical significance was evaluated by randomly assigning sequences to populations and calculating the FST for 1,000 permutations. The P test was used to investigate whether the communities exhibited covariation with phylogeny (32). Neighbor-joining trees constructed under the HKY model of nucleotide substitution in PAUP (51) were used to estimate the minimum number of changes (switch from one community to the other) required to explain observed distribution. The significance of the covariation was determined by establishing the expected number of changes under the null hypothesis (29) that the community did not covary with phylogeny. The P test was conducted using MacCLADE software (28). FST and P tests were also applied to OTU groups to calculate whether such groups are comprised of sequences drawn from the same phylogenetic lineage or if they comprise distinct phylogenetic lineages that are arbitrarily considered as identical.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the National Institute for Biotechnology Information database under the accession numbers AF544254 to AF544369.
RESULTS
Sediment analysis.
Radiocarbon dating of the sediment revealed that the turbidite fraction (0 to 38 cm) of the core was deposited ca. 1,000 years ago (54). However, it is important to note that the turbidite was derived from failure of older, previously deposited sediment from further upslope. Modeling of the coccolith mixture in the turbidite indicted that it consisted of material covering an age range of approximately 200,000 years, suggesting failure of a slab of sediment 15 m thick. Below the turbidite (38 to 60+ cm), the sediment core consisted of pelagic clay. Visual inspection revealed that it contained foraminifera and was comprised of a mixture of fine-grained material: mainly coccoliths, clays, and volcanic minerals. The brown (oxidized) nature of the clay indicated that it was from the Holocene period and was younger than 12,000 years. Total organic carbon contents were 0.92% ± 0.14%, 1.05% ± 0.05%, and 0.98% ± 0.33% percent for the 5- to 12-, 15- to 18-, and 43- to 46-cm sediment sections, respectively.
Construction and analysis of actinobacterial 16S rDNA gene libraries.
DNA extracted from the three sediment sections was amenable to direct PCR amplification. The highest yields of genomic DNA were recovered from the 5- to 12-cm section, and amounts of recovered DNA decreased in proportion to sediment depth (data not shown). The 5- to 12-cm library (61 positive transformants) yielded 57 clones that tested positive for 16S rDNA genes (93%); the 15- to 18-cm library yielded 48 clones (90%); and the 43- to 46-cm library yielded 62 clones (85%). None of the actinobacterial 16S rDNA genes were considered to be chimeras under the methods employed. Sequencing revealed that all 167 16S rDNA clones belonged to the class Actinobacteria, giving a specificity of 100% for the S-C-Act-0235-a-S-20 and S-C-Act-0878-a-A-19 primers.
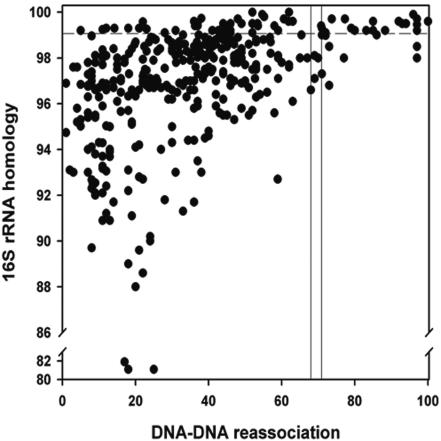
In order to conduct richness estimation, clones were placed into OTUs. OTUs were defined by comparing 16S rRNA homology with DNA-DNA reassociation values for members of the class Actinobacteria, using the ∼70% DNA-DNA reassociation cutoff point as the phylogenetic definition of a species (48). A 16S rRNA homology value of >99% minimized DNA-DNA reassociation values of <70% and incorporated 70% of all values of >70% (Fig. 1). Thus, an actinobacterial OTU was defined as a 16S rDNA sequence group in which sequences differed by ≤1%. Previous work has shown that 16S rRNA homology values calculated using the region amplified by the present primers are conservative by approximately 0.7% (46). Seventy-two percent of species with >99% 16S rRNA homology had <70% DNA-DNA reassociation values. Of the actinobacterial 16S rDNA sequences recovered, 116 OTUs were identified (Table 1). The closest relatives of representative actinobacterial OTUs, identified by searching in the GenBank database, are given in Table 1. The prefix T indicates that the 16S rDNA gene sequences was recovered from the 5- to 12-cm sediment section, M indicates the 15- to 18-cm section, and D indicates the 43- to 46-cm section. Phylogenetic analysis indicated that actinobacterial OTUs were most closely related to members of the suborders Corynebacterineae, Frankineae, and Streptomycineae (Fig. 2). However, only 9% of the OTUs showed 100 to 99% homology with cultured actinobacteria (i.e., likely to be known species), 85% had 98 to 95% homology with known species, and 17% had 94 to 89% homology. Using the DNA-DNA reassociation comparison with 16S rDNA homology, it is probable that 91% of the OTUs recovered from the deep-sea sediment represent novel species or genera.
FIG. 1.
Comparison of 16S rRNA homology and DNA-DNA reassociation values in the class Actinobacteria. The bar indicates the threshold value for species delineation (48). The horizontal line indicates the 16S rDNA homology value for which an actinomycete OTU was defined.
TABLE 1.
Actinomycete 16S rDNA sequences identified in a deep-sea sediment core in three clone libraries
| Representative clone (accession no.) | No. of clones in library
|
Closest relative
|
||||
|---|---|---|---|---|---|---|
| 5-12 cm | 15-18 cm | 43-46 cm | Actinomycete | Accession no. | % Identity | |
| T01 (AF544299) | 1 | Verrucosispora sp. strain IM-6907 | AF131631 | 97 | ||
| T02 (AF544256) | 1 | Streptomyces thermocarboxydovorans | U94489 | 98 | ||
| T03 (AF544259) | 1 | Streptomyces thermocarboxydovorans | U94489 | 95 | ||
| T04 (AF544298) | 1 | Streptomyces sp. strain KACC 20159 | AF345862 | 89 | ||
| T05 (AF544297) | 1 | Uncultured actinobacterium clone ASb07 | AY124393 | 96 | ||
| T06 (AF544294) | 1 | Streptomyces sp. strain 19504 | AJ315072 | 92 | ||
| T07 (AF544296) | 1 | Streptomyces capensis | AF452714 | 97 | ||
| T08 (AF544295) | 1 | Dietzia maris | Y18883 | 94 | ||
| T09 (AF544279) | 1 | Uncultured actinobacterium clone SDe09 | AY124457 | 92 | ||
| T10 (AF544293) | 1 | Corynebacterium ulcerans | X84256 | 96 | ||
| T11 (AF544254) | 1 | Streptomyces sp. strain IM-6899 | AF131529 | 95 | ||
| T12 (AF544255) | 1 | Streptomyces sp. strain IM-6960 | AF131527 | 95 | ||
| T13 (AF544292) | 1 | Kineococcus-like bacterium AS2978 | AF060676 | 95 | ||
| T14 (AF544291) | 1 | Rhodococcus sp. strain ARG-BN062 | AF420423 | 95 | ||
| T15 (AF544290) | 1 | Frankia sp. strain | AF063642 | 93 | ||
| T16 (AF544257) | 1 | Dietzia maris | Y18883 | 95 | ||
| T17 (AF544298) | 1 | Mycobacterium sp. strain | U46146 | 97 | ||
| T19 (AF544258) | 1 | Streptomyces sp. strain IM-1436 | AF131505 | 96 | ||
| T21 (AF544288) | 1 | Geodermatophilus sp. strain BC509 | AJ296063 | 96 | ||
| T22 (AF544260) | 1 | Streptomyces sp. strain IM-1436 | AF131505 | 97 | ||
| T23 (AF544287) | 1 | Streptomyces thermocarboxydovorans | U94489 | 99 | ||
| T24 (AF544286) | 1 | Corynebacterium tuberculostearicum | AF510731 | 98 | ||
| T26 (AF544285) | 1 | Streptomyces sp. strain IM-1436 | AF131505 | 96 | ||
| T27 (AF544261) | 1 | Streptomyces sp. strain IM-7585 | AF131604 | 96 | ||
| T28 (AF544284) | 1 | Amycolatopsis sp. strain GY109 | AF466094 | 96 | ||
| T29 (AF544262) | 1 | Uncultured bacterium ARFS-3 | AJ277836 | 98 | ||
| T30 (AF544263) | 1 | Tsukamurella inchonensis | AF283281 | 93 | ||
| T31 (AF544283) | 1 | Micromonospora sp. strain IM-7020 | AF131416 | 95 | ||
| T32 (AF544264) | 1 | Streptomyces sp. strain NK1057 | AF492844 | 98 | ||
| T33 (AF544282) | 3 | 1 | Uncultured actinobacterium clone ASb07 | AY124393 | 99 | |
| T34 (AF544265) | 1 | Dietzia maris | Y18883 | 94 | ||
| T35 (AF544281) | 1 | Blastococcus sp. strain BC448 | AJ316571 | 98 | ||
| T36 (AF544266) | 1 | Streptomyces sp. strain IM-8073 | AF131609 | 97 | ||
| T37 (AF544280) | 1 | Streptomyces sp. strain 11AG8 | AF233375 | 95 | ||
| T38 (AF544267) | 1 | Streptomyces capensis | AF452714 | 98 | ||
| T40 (AF544268) | 1 | Blastococcus sp. strain BC448 | AJ316571 | 97 | ||
| T41 (AF544269) | 1 | 2 | Dietzia maris | Y18883 | 94 | |
| T42 (AF544300) | 1 | Streptomyces pallidus | AJ399492 | 94 | ||
| T43 (AF544270) | 1 | Rhodococcus tukisamuensis | AB067734 | 95 | ||
| T44 (AF544301) | 1 | Amycolatopsis sp. strain GY109 | AF466094 | 96 | ||
| T45 (AF544271) | 1 | Streptomyces sp. strain IM-8073 | AF131609 | 95 | ||
| T47 (AF544272) | 2 | Streptomyces thermocarboxydovorans | U94489 | 99 | ||
| T48 (AF544302) | 1 | Mycobacterium sp. strain STR-12 | AF236839 | 96 | ||
| T49 (AF544273) | 1 | Streptomyces somaliensis | AJ007403 | 95 | ||
| T50 (AF544274) | 1 | Uncultured actinobacterium clone SDb12 | AY124440 | 91 | ||
| T51 (AF544303) | 1 | Streptomyces sp. strain IM-7585 | AF131604 | 95 | ||
| T52 (AF544275) | 1 | Streptomyces somaliensis | AJ007401 | 90 | ||
| T54 (AF544304) | 1 | Geodermatophilus sp. strain | X92358 | 96 | ||
| T56 (AF544276) | 1 | Geodermatophilus sp. strain IM-1092 | AF131367 | 93 | ||
| T57 (AF544305) | 1 | Streptomyces thermocarboxydovorans | U94489 | 99 | ||
| T58 (AF544277) | 1 | Mycobacterium sp. strain STR-18 | AF236841 | 95 | ||
| T59 (AF544306) | 1 | Rhodococcus opacus | Y11892 | 92 | ||
| T60 (AF544278) | 1 | Streptomyces thermocarboxydovorans | U94489 | 98 | ||
| M03 (AF544336) | 1 | Corynebacterium appendicis | AJ314919 | 99 | ||
| M06 (AF544335) | 2 | Rhodococcus sp. strain SRB1948-Z40 | AB010913 | 96 | ||
| M07 (AF544334) | 1 | Corynebacterium striatum | X81910 | 94 | ||
| M08 (AF544308) | 1 | Rhodococcus ruber | X80625 | 95 | ||
| M09 (AF544333) | 1 | Rhodococcus fascians | X79186 | 95 | ||
| M11 (AF544307) | 1 | Pseudonocardia aurantiaca | AF325727 | 96 | ||
| M12 (AF544332) | 1 | Dietzia maris | Y18883 | 95 | ||
| M14 (AF544322) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| M15 (AF544331) | 1 | Rhodococcus koreensis | AF124342 | 96 | ||
| M16 (AF544321) | 1 | Streptomyces sp. strain CN732 | AF101415 | 92 | ||
| M17 (AF544330) | 1 | Pseudonocardia alaniniphila | AF325726 | 98 | ||
| M20 (AF544320) | 1 | Nocardioides sp. strain V4.BO.15 | AJ244655 | 93 | ||
| M22 (AF544319) | 1 | Rhodococcus fascians | X79186 | 95/PICK> | ||
| M23 (AF544329) | 1 | Turicella otitidis | X73976 | 99 | ||
| M24 (AF544318) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| M25 (AF544328) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| M26 (AF544317) | 1 | Mycobacterium manitobense | AY082001 | 94 | ||
| M27 (AF544327) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| M28 (AF544316) | 1 | Turicella otitidis | X73976 | 92 | ||
| M29 (AF544326) | 1 | Actinosynnema sp. strain IM-1402 | AF131351 | 95 | ||
| M30 (AF544315) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| M33 (AF544325) | 1 | Rhodococcus fascians | X79186 | 96 | ||
| M35 (AF544324) | 1 | Rhodococcus zopfii | AF191343 | 93 | ||
| M36 (AF544314) | 1 | Uncultured actinobacterium clone ASb09 | AY124394 | 94 | ||
| M37 (AF544323) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| M38 (AF544337) | 1 | Mycobacterium manitobense | AY082001 | 96 | ||
| M39 (AF544313) | 1 | Frigoribacterium sp. strain 301 | AF157479 | 96 | ||
| M40 (AF544312) | 1 | Rhodococcus fascians | X79186 | 95 | ||
| M41 (AF544338) | 2 | Rhodococcus sp. strain UFZ-B520 | AF235011 | 98 | ||
| M43 (AF544311) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| M44 (AF544339) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| M45 (AF544310) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| M46 (AF544340) | 1 | Mycobacterium manitobense | AY082001 | 96 | ||
| M47 (AF544309) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| D02 (AF544341) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| D05 (AF544342) | 1 | 1 | Mycobacterium manitobense | AY082001 | 98 | |
| D06 (AF544343) | 1 | Kitasatospora sp. strain IM-6832 | AF131378 | 96 | ||
| D11 (AF544344) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| D13 (AF544345) | 1 | Rhodococcus fascians | X79186 | 99 | ||
| D16 (AF544346) | 2 | Rhodococcus sp. strain S9 | AF260713 | 98 | ||
| D17 (AF544347) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| D22 (AF544348) | 1 | Rhodococcus fascians | X79186 | 96 | ||
| D25 (AF544349) | 1 | Arthrobacter sp. strain “SMCC G960” | AF197025 | 96 | ||
| D26 (AF544350) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| D27 (AF544351) | 1 | Mycobacterium sp. strain STR-21 | AF236837 | 96 | ||
| D32 (AF544352) | 1 | Rhodococcus fascians | X79186 | 99 | ||
| D35 (AF344353) | 1 | Uncultured actinobacterium clone ASc02 | AY124397 | 95 | ||
| D36 (AF544354) | 1 | Rhodococcus sp. strain YK2 | AB070458 | 96 | ||
| D41 (AF544355) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| D42 (AF544356) | 1 | Mycobacterium sp. strain STR-11 | AF236838 | 97 | ||
| D43 (AF544357) | 1 | Dietzia maris | Y18883 | 95 | ||
| D44 (AF544358) | 2 | Rhodococcus fascians | AF205371 | 98 | ||
| D45 (AF544359) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| D47 (AF544360) | 1 | Dietzia maris | Y18883 | 94 | ||
| D48 (AF544361) | 1 | Rhodococcus fascians | X79186 | 99 | ||
| D50 (AF544362) | 1 | Rhodococcus fascians | X79186 | 97 | ||
| D52 (AF544363) | 1 | Blastococcus sp. strain BC412 | AJ316574 | 98 | ||
| D54 (AF544364) | 1 | Rhodococcus fascians | X79186 | 98 | ||
| D55 (AF544365) | 1 | Rhodococcus fascians | X79186 | 96 | ||
| D57 (AF544366) | 11 | 31 | Rhodococcus fascians | X79186 | 100 | |
| D58 (AF544367) | 1 | Mycobacterium manitobense | AY082001 | 96 | ||
| D61 (AF544368) | 1 | Dietzia maris | Y18883 | 95 | ||
| D62 (AF544369) | 1 | Rhodococcus fascians | X79186 | 97 | ||
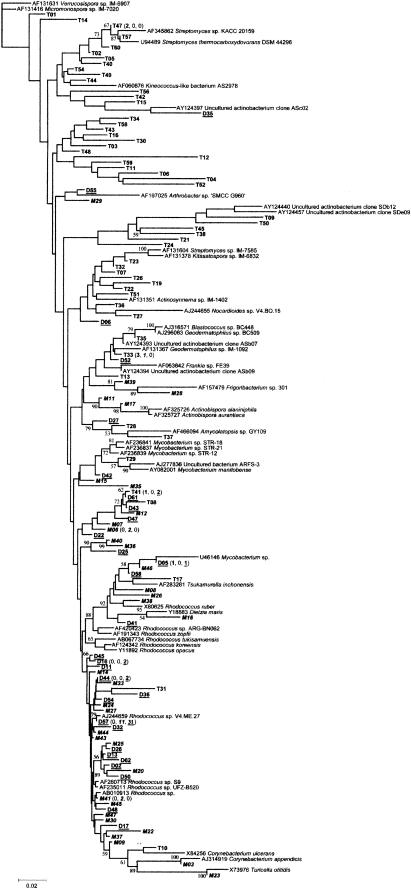
FIG.2.
Phylogenetic relationship of partial 16S rDNA sequences generated in this study. See Materials and Methods for description of tree construction. Scale bar represents the number of changes per base position. Numbers at tree nodes represent the number of times the topology to the right of the node was recovered in 100 bootstrap resamplings. Clone prefixes: T = 5- to 12-cm (bold) actinomycete community; M = 15 to 18 cm (bold italics), and D = 43 to 46 cm (bold underlined). The numbers in parentheses indicate the number of times the OTU was found in each of the libraries e.g., D57 (0, 11, 31) was not found in the 5- to 12-cm community and was present 11 and 31 times in the 15-to-18-cm and 43- to 46-cm communities, respectively.
The OTU representation was distinct for each of the sediment sections. The 5- to 12-cm sediment actinobacterium community was dominated by bacteria most closely related to Streptomyces species (45%), though OTUs from this sediment section were distributed throughout the phylogenetic tree (Fig. 2). OTUs from the 15- to 18-cm sediment section were dominated by bacteria most closely related to Rhodococcus species (56%), with only one OTU (M16) being most closely related to a Streptomyces species. The 43- to 46-cm community was also dominated by Rhodococcus species (62%), with no Streptomyces species present. No OTU was found in all sediment sections; however, two OTUs (represented by T41 and D05) were present in both the 5- to 12-cm and 43- to 46-cm sediment sections; one OTU (represented by T33) was present in both the 5- to 12-cm and 15- to 18-cm sections; and one OTU (represented by D57) was present in the 15- to 18-cm and 43- to 46-cm sections. The most dominant OTU in the 5- to 12-cm community was T33 (most closely related to the uncultured actinobacterium clone ASb07 [99%]), which comprised 5% of all actinobacterial 16S rDNA sequences recovered from this section. The 15- to 18-cm and 43- to 46-cm sediment communities were dominated by an OTU represented by D57 (most closely related to Rhodococcus fascians [100%]), comprising 23 and 50% of the communities, respectively, and 34% of all OTUs described.
Actinobacteria species richness.
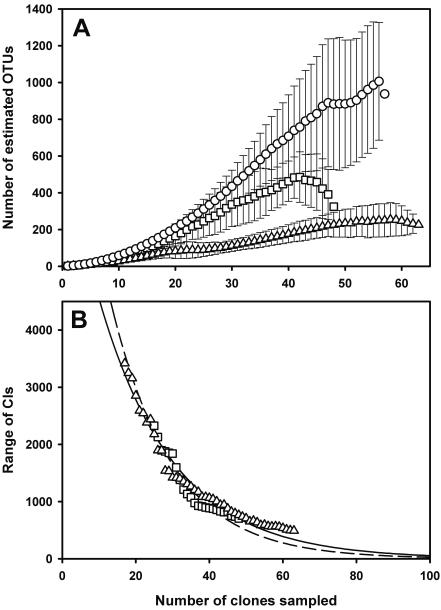
Typical accumulation curves for microbial communities (19) were found by plotting the cumulative number of OTUs observed against the sampling effort. Curves indicated a slight increase in sample coverage, in proportion to total richness, with depth. However, the curves closely matched those expected when every sequence identified is an individual; hence, each sediment section was sampled with roughly equal intensity relative to its overall richness (19). Nonparametric estimation revealed a trend of decreasing species richness concomitant with increasing sediment depth (Table 2; Fig. 3A). The 95% CIs for the last estimate in each sediment section overlap and therefore it is not possible to reject the null hypothesis, at the significance level of 0.05, that there is no difference between species richness in the three sediment communities. The range of CIs in the 5- to 12-cm community continued increasing with sample size, and therefore it was not possible to determine how much more sampling was required to detect a significant difference in species number between this community and the two others. However, the range of CIs stabilized in the 15- to 18-cm and 43- to 46-cm communities allowing estimation of the effort required to detect a significant difference between these two communities by estimating the size of the CIs in larger samples (19). Thus, approximately 100 samples would be needed to detect a significant difference between these two communities (Fig. 3B).
TABLE 2.
Comparison of diversity indices in three deep-sea sediment core communities
| Community | Diversity estimate
|
|||||
|---|---|---|---|---|---|---|
| No. of distinct sequences | No. of OTUsa
|
1/Db | Nucleotide diversity | θ (π)c | ||
| ACE | Chao1 | |||||
| 5-12 cm | 53 | 616 (289, 3,379) | 1,406 (307, 7,280) | 51 | 0.11 ± 0.05 | 63.9 ± 31.0 |
| 15-18 cm | 36 | 325 (114, 1,105) | 308 (106, 1,103) | 15 | 0.07 ± 0.04 | 42.1 ± 20.7 |
| 43-46 cm | 30 | 226 (82, 765) | 212 (75, 762) | 4 | 0.05 ± 0.03 | 32.4 ± 15.9 |
The values in parentheses are the 95% confidence intervals.
Reciprocal of Simpson's index.
Total genetic variation.
FIG. 3.
(A) Nonparametric (ACE) estimates of actinobacteria species richness as a function of sample size; 5 to 12 cm (○) (n = 56), 15 to 18 cm (n = 48), and 43 to 46 cm (▵) (n = 63). Error bars are standard deviations. Curves are averaged over 100 simulations using EstimateS. (B) Average size of the 95% CIs of the Chao1 estimate for the 15- to 18-cm and 43- to 46-cm communities. The curves are fitted negative exponential curves [15 to 18 cm, f(x) = 9,861 × 10−0.0596x and r2 = 0.97; 43 to 46 cm, f(x) = 7,433 × 10−0.0491x and r2 = 0.99].
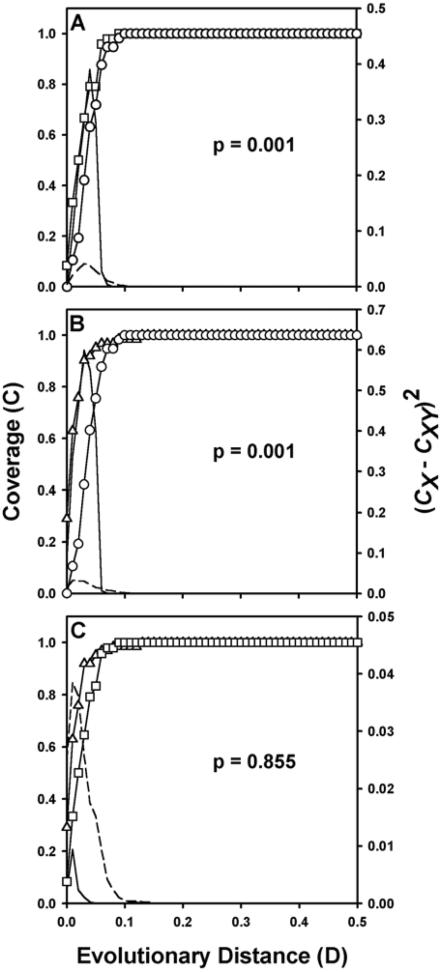
Significance of difference between coverage curves.
LIBSHUFF (44) analysis of homologous and heterologous coverage curves indicated that the 5- to 12-cm community was significantly different from both the 15- to 18-cm and 43- to 46-cm communities (P = 0.001 [Fig. 4A and B ]). Comparison of the 15- to 18-cm and 43- to 36-cm communities suggested that they were not significantly different (P = 0.855 [Fig. 4C]). Further information on the differences between the 5- to 12-cm community and the two other communities was gained by examination of the distribution of (CX − CXY)2 with D (44). At a D value of <0.10, the actual values of (CX − CXY)2 are greater than the comparable values at a P value of 0.05 obtained during the calculation of ΔC (Fig. 4A and B). This result suggests that the 5- to 12-cm library differs greatly from the other two libraries at low levels of genetic distance but shares all deep taxa (D > 0.10). Comparison of the 15- to 18-cm and 43- to 46-cm libraries show that the actual values of (CX − CXY)2 do not exceed the comparable values at a P value of 0.05 and differ only at D < 0.01, suggesting that nearly all taxa are present in both libraries (Fig. 4C).
FIG. 4.
LIBSHUFF comparisons of the three actinobacterial 16S rDNA libraries. Solid lines indicate the value of (CX − CXY)2 for samples at each value of D. D is equal to the Jukes-Cantor evolutionary distance determined by the DNADIST program of PHYLIP. Broken lines indicate the P = 0.05 value of (CX − CXY)2 for the randomized samples. (A) Comparison of 5- to 12-cm library (○ [homologous]) with 15- to 18-cm library (□ [heterologous]). (B) Comparison of 5- to 12-cm library (○ [heterologous]) with 43- to 46-cm library (▵ [homologous]). (C) Comparison of 15- to 18-cm library (□ [heterologous]) with 43- to 46-cm library (▵ [homologous]).
Reciprocal of Simpson's index.
The reciprocal of Simpson's index is sensitive to the level of dominance in a community and indicated that diversity decreased in proportion to depth. The 5- to 12-, 15- to 18-, and 43- to 46-cm communities had 1/D index values of 51, 15, and 4, respectively. Zhou et al. (61) suggest that values for 1/D below about 50 indicate typical dominance profiles; therefore, both the 15- to 18- and 43- to 46-cm communities show dominance, and the 5- to 12-cm community lies on the border between a dominant profile and a uniform profile.
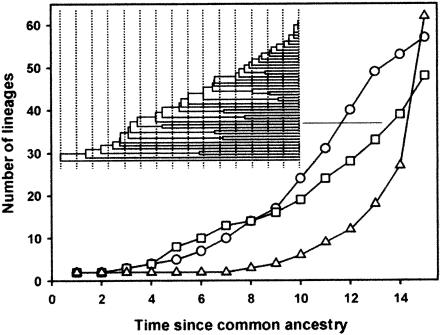
Phylogenetic diversity.
Phylogenetic diversity between the three communities (i.e., methods not based on OTUs) was made initially using lineage-per-time plots (32). The plots were similar for the 5- to 12-cm and 15- to 18-cm communities (Fig. 5) and showed correspondence with trees resulting from a constant birth and death rate model (32); the only difference between the curves was a slight excess of divergent lineages in the 5- to 12-cm community. The 43- to 46-cm community plot displayed a convex shape indicating an excess of closely related lineages (Fig. 5). Phylogenetic diversity in the three samples was further compared using genetic differentiation tests. Table 2 shows the diversity estimates calculated using OTU definitions for each community compared with the diversity statistics generated from population genetics. In general, the statistics show the same pattern; distinct OTUs, estimated number of OTUs, nucleotide diversity, and average sequence divergence all decreased in proportion to depth. Genetic differentiation and covariation with phylogeny among the three sediment section communities were assessed by using FST and P tests (32). Use of the FST test showed differentiation between all sediment sections: 5 to 12 cm and 15 to 18 cm (FST = 0.14858; P < 0.00001), 5 to 12 cm and 43 to 46 cm (FST = 0.24842; P < 0.00001), and 15 to 18 cm and 43 to 46 cm (FST = 0.06293; P < 0.00001). In addition, the P test was highly significant (P < 0.001) for the three communities, indicating that the actinobacterial communities present at different depths covaried with phylogeny. Significance for both FST and P tests signified that there was less genetic diversity present in each individual community than there was for the three communities combined and that each community harbored distinct phylogenetic lineages (32). The OTU group represented by sequence D57 contained 11 and 31 sequences from the 15- to 18-cm and 43- to 46-cm communities, respectively. FST and P tests applied to the whole OTU group showed that the group was in fact comprised of clades of closely related sequences that were unique to either the 15-to-18- or 43- to 46-cm communities and that these clades were interspersed throughout the OTU group (FST, P < 0.00001; P test, P = 0.21). Separate analysis of the sequences in the OTU group from either the 15- to 18- or 43- to 46-cm communities indicated that they were drawn from the same pool of sequences (FST, P > 0.9; P test, P ≥ 0.5 for both).
FIG. 5.
Lineage-per-time plot for the 5- to 12-cm (○), 15- to 18-cm (□), and 43- to 46-cm (▵) actinobacteria 16S rDNA libraries. The ordinate is the number of lineages; the abscissa is time (arbitrary units) measured from the common ancestor. See Materials and Methods for a description of plot construction.
DISCUSSION
Diversity estimates and statistical comparisons.
The methods employed in this study enabled us to (i) compare quantitatively the coverage of actinobacterial 16S rDNA genes in the separate libraries; (ii) estimate species richness; and (iii) compare the phylogenetic diversity in each community (i.e., the genetic relatedness of the actinobacteria in the different sediment sections). The nonparametric estimators, the LIBSHUFF calculations, and the reciprocal of Simpson's index are all reliant on an OTU definition. In this study we used experimental data to define an actinobacterial OTU to provide the highest likelihood that sequences placed into OTU groups were generated from identical species. Some limitations in the use of OTUs for diversity statistics have been highlighted previously (19, 32), and our study prompts a further caveat: the use of 16S rRNA homology and DNA-DNA reassociation correlations for actinobacteria showed that even with a nonconservative OTU definition (i.e., ≤1% sequence divergence) the majority of species diversity will be overlooked. It is evident from the present study that when an OTU definition of ≤1% sequence divergence is applied, >70% of sequences placed into an OTU group originated from different species. Moreover, this observation is supported by empirical predictions of DNA-DNA relatedness when the similarity of 16S rDNA genes is known; for 99.8% similarity there is only a 50% probability that DNA-DNA relatedness is >70% (24). Palys et al. (36) state that species cutoff values for DNA-DNA reassociation (such as 70%) are arbitrary and are not guaranteed to yield groups of bacteria that correspond to real ecological units. The results of this study support this contention; FST and P tests applied to an OTU group revealed that it was comprised of clades unique to either sampling location. Thus, species with >99% 16S rRNA homology from two differing environments can clearly be distinguished. The application of phylogenetic diversity statistics (32) may enable researchers to identify species that form monophyletic clades that are unique to their environment, thereby enabling practical implementation of microbial species concepts where the environment is implicit in the definition, i.e., the phylospecies (49). Clearly, it is not possible to state definitively what level of DNA-DNA reassociation should be used for defining bacterial OTUs; hence, diversity estimates based on OTUs should be interpreted cautiously. However, the use of OTUs is appropriate for comparing relative richness when applied to data sets representing the same length and region of the 16S rDNA gene.
Phylogenetic diversity comparisons recently proposed for microbial studies require no definition of an OTU; the phylogenetic tree provides the information necessary for comparison (32). These methods are complementary to species richness estimates and provide a way of assessing whether the communities under investigation contain different phylogenetic groups regardless of the species number. However, phylogenetic comparisons are dependent on sequencing; diversity comparisons involving thousands of clones (61) are currently too costly to achieve using sequence analysis alone. Martin (32) compared phylogenetic diversity in previously published 16S rDNA clone libraries and showed that conclusions regarding diversity based solely on the frequency of different species (30) do not account for actual differences in the phylogenetic composition of the communities.
This study is the first to apply both microbial species richness and phylogenetic diversity estimates to a marine environment. Comparison of all diversity measures (Table 2) combined with the FST and P tests clearly shows that diversity in the sediment section decreases with depth and that each sediment section was comprised of distinct actinobacterial lineages. The only exception was the LIBSHUFF result for the 15- to 18-cm and 43- to 46-cm communities, predicting that both communities were composed of the same taxa. The LIBSHUFF program is a good test of overlap, since it considers the distribution of pairwise differences instead of the mean and variance (i.e., the FST test [A. P. Martin, personal communication]). However, it is not sensitive to the phylogenetic grouping of taxa (P test). Results of the LIBSHUFF program are dependent on sample size; the minimum number of sequences necessary to distinguish between two dissimilar libraries increases with library complexity and decreases with the magnitude of dissimilarity (44). Therefore, libraries composed of closely related taxa will be treated as equal within small sample sizes; this was tested by constructing two small artificial actinobacterial communities in which equal numbers of identical genera were present but in which the species represented in each library differed (data not shown). Results of the LIBSHUFF comparison for these communities indicated that the communities were identical (P = 1.00). Therefore, the results of LIBSHUFF for the 15- to 18-cm and 43- to 46-cm communities are probably due to the fact that the two libraries are composed largely of members of the same genera. A greater number of sequences are needed for LIBSHUFF to distinguish between these communities.
Mechanisms responsible for diversity.
Four mechanisms were recently proposed for the production of noncompetitive diversity profiles (61): (i) superabundant resources, (ii) resource heterogeneity, (iii) spatial isolation, and (iv) nonequilibrium conditions. An absence of these mechanisms will result in competitive diversity profiles. It is reasonable to assume that in marine sediments spatial isolation is not a factor, due to constant aqueous contact. Rank abundance profiles represented by uniform low population densities (e.g., 5- to 12-cm section) are consistent with resource limitation (61); hence, resource superabundance is unlikely to be maintaining species diversity. Furthermore, in order for resource superabundance to maintain diversity it must be such that species saturation never occurs, i.e., continuously abundant, or species spatially isolated (both unlikely in deep-sea sediments). However, it is possible that all species are maintained, but at suboptimal growth rates, thereby maintaining a species-unsaturated environment, eliminating competition and thus supporting diversity.
The 5- to 12-cm community displayed a borderline noncompetitive diversity profile (1/D = 51) with high species diversity estimates, indicating that the diversity of the 5- to 12-cm community is maintained by nonequilibrium and heterogeneous resource conditions. Thus, the two deeper communities, with proportionally lower estimates, are exposed to a decreasing gradient of the two above mechanisms. These factors seem tenable, since the majority of the carbon in the sediment is deposited by “marine snow” (settling of organisms living in the photic zone), which varies in its composition and deposition frequency (55). The surface sediment will also be subjected to hydrodynamics (deep-sea storms) and patch dynamics (carcass falls). These factors contribute to the nonequilibrium and resource heterogeneity present in the surface layers that are ameliorated in the deeper sections. Carbon inputs that do not originate from marine snow (i.e., upwellings) are likely to be homogeneous (e.g., methane) and serve to increase competition in the deeper sediments. The fact that the total organic carbon concentrations in the sediment sections were equal indicates that if the carbon resource directly influences diversity, it is due to heterogeneity rather than abundance. Other resources, such as the type and abundance of electron acceptors, may also influence diversity. Oxygen penetration into marine sediments is dependent on a number of factors including water flow rate and topography at the sediment-water boundary layer (14, 62), with values of <3 mm to >200 mm being reported (4, 59). Therefore, decreasing oxygen abundance in proportion to depth may be responsible for the concomitant decrease in diversity, with the fluxes at the boundary contributing to nonequilibrium conditions and hence increased diversity.
Lack of competition is not the only proposed model for diversity maintenance; other models for supersaturated coexistence invoke competition as the promoting force of diversity. Huisman et al. (20) analyzed multispecies models incorporating different physiological scenarios and plausible tradeoffs and showed that >100 species could be maintained on just three resources. Similarly, models computed using nonlinear formulae showed that competition could either promote or reduce diversity depending on resource utilization rates (56). Furthermore, and of particular interest to bioprospectors, the production of antimicrobial compounds may promote stable coexistence of huge numbers of species (10).
Selection of different populations.
The highest similarity for two clones present in both the 5- to 12- and 15- to 18-cm sediment sections was 99.8% (OTU group represented by T33). Therefore, the two species from which the clones originate diverged approximately 5 million to 11 million years ago (depending on the model for 16S rDNA nucleotide base substitution employed) (34, 35). Thus, it is likely that the differing populations observed in these two sediment sections did not diverge after their secondary sedimentation. Some of the phylogenetically distinct populations present in these two turbidite sections would have diverged during the deposition of the initial sediment from which the turbidite originated (15 m thick covering 250 million years), and others (with >4.5% 16S rDNA difference) would have diverged prior to the deposition of that sediment. Distinct populations would have been mixed during the formation and deposition of the turbidite, which occurs in a matter of days. Therefore, distinct ecological groups that evolved in a specific niche would survive and proliferate upon reformation of that niche (or an analogous one) after sedimentation. As mentioned above, the number of exploitable niches will decrease with depth, and therefore, burial of populations upon sedimentation will cause a selective sweep of the total population being deposited. The lineage-per-time plots give preliminary evidence for this selective sweep, showing a trend toward an excess of closely related species. The fact that clones D57 and M04 were identical suggests possible movement of species between the turbidite and pelagic clay, although it is not possible to state in which direction. It is important to note that divergence of species post-turbidite formation (i.e., in the last 1,000 years) has probably occurred through periodic selection and that it is the use of the 16S rDNA gene that prevents us from observing divergence on this time scale. Palys et al. (36) used protein-coding genes as a means to delineate ecologically distinct bacteria. These authors concluded that the inability of 16S rRNA sequence data to distinguish between some taxa was a consequence of the low evolutionary rate of the 16S rRNA genes. Thus, the use of markers with rapid evolutionary rates would be required if we wished to examine divergence after turbidite formation, such as insertion sequence fingerprinting on bacteria cultured from different sediment sections that have identical 16S rRNA genes (60).
Implications of diversity estimates.
The combined use of species richness and diversity estimates provides information that enables a deeper understanding of microbial biodiversity in areas such as diversity management, biogeography, and bioprospecting. To date it is still not clear how microbial diversity influences environmental functions, such as nutrient cycling, degradation of xenobiotics, and ecosystem stability. Therefore, it is difficult to predict how to successfully manage microbial diversity or even if management is necessary (i.e., species redundancy). Statistical diversity estimates that account both for phylogeny and richness of species applied in concert with hypotheses that test ecological theory may provide new insights into how diversity affects ecosystem function.
Staley and Gosink (50) give three reasons for the importance of bacterial biogeography: determination of how many bacterial species exist, species preservation, and identification of ecological roles through knowledge of bacterial distribution. Phylogenetic diversity estimates aid in the identification of bacterial species that are cosmopolitan or endemic. Endemism is suggested when clades of bacteria unique to a particular environment are identified, i.e., the P test. Furthermore, the FST test enables species to be identified that contribute to differences in the bacterial genetic diversity between two environments. Once cosmopolitan or endemic species are identified, their distribution and number can be estimated using richness estimates.
The implementation of techniques like those described in this study will enhance bioprospecting strategies in several respects. Thus, the molecular census of species gives information on species presence or absence in the environment being sampled; this information can be employed in the design of cultivation strategies. This methodology is not novel, but the combination of such information with the statistical diversity estimation methods provides the bioprospector with additional resources. First, coverage curves and richness estimators provide a means to assess how much further sampling is required. Second, comparison of the cultured subset to the molecular data provides a means to assess how many of the target species are present in the screening stage, and correlation of these data to the success of the screening (i.e., how many novel compounds have been identified) will determine whether or not continued cultivation and screening efforts should be undertaken and allow analogous environments to be identified for future screens.
Niche adaptation in microbes through periodic selection implies that the use of composite samples (one sample containing distinct niches) for the cultivation of microbes will bias recovery in favor of those best adapted to the culture medium; those with large cell volumes, high ribosome concentrations, and multiple rRNA operons (13, 25). Application of diversity estimators coupled with diversity indices sensitive to the level of dominance in a community enables the discrimination of environments, either geographically isolated or as specific niches within one sample, and therefore, appropriate methods can be employed to reduce culture bias, such as physical separation of the delineated environments, use of low-nutrient or polymeric growth substrates (22, 40), or dilution to extinction culture (43).
Acknowledgments
We thank the crew of the RRS Charles Darwin for collection of the deep-sea sediments, Jennifer Hughes for helpful advice on nonparametric estimators, Andrew Martin for introducing us to methods for genetic diversity comparison and for his guidance in their application, and Anne Chao for advice on closed-form variance solutions and access to the SPADE program.
This work was supported by UK Natural Environment Research Council (NERC) grants NER/T/S/2000/00614 and NER/T/S/2000/00616.
REFERENCES
- 1.Allison, L. E. 1965. Organic carbon, p. 1367-1378. In C. A. Black (ed.), Methods of soil analysis, part 2: chemical and microbiological properties. American Society of Agronomy, Madison, Wis.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, C. M. 1999. Molecular diffusive fluxes of oxygen in sediments of Port Phillip Bay in south-eastern Australia. Mar. Freshw. Res. 50:557-566. [Google Scholar]
- 5.Butman, C. A., and J. T. Carlton. 1995. Marine biological diversity—some important issues, opportunities and critical research needs. Rev. Geophys. 33:1201-1209. [Google Scholar]
- 6.Chao, A. 1987. Estimating the population-size for capture recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 7.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 8.Colquhoun, J. A., J. Mexson, M. Goodfellow, A. C. Ward, K. Horikoshi, and A. T. Bull. 1998. Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie Leeuwenhoek 74:27-40. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czaran, T. L., R. F. Hoekstra, and L. Pagie. 2002. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. USA 99:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emery, K. O. 1969. The continental shelves. Sci. Am. 221:106-122. [Google Scholar]
- 13.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster, S., M. Huettel, and W. Ziebis. 1996. Impact of boundary layer flow velocity on oxygen utilisation in coastal sediments. Mar. Ecol. Prog. Ser. 143:173-185. [Google Scholar]
- 15.Good, I. J. 1953. The population frequency of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 16.Gray, J. S. 2002. Species richness of marine soft sediments. Mar. Ecol. Prog. Ser. 244:285-297. [Google Scholar]
- 17.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisman, J., A. M. Johansson, E. O. Folmer, and F. J. Weissing. 2001. Towards a solution of the plankton paradox: the importance of physiology and life history. Ecol. Lett. 4:408-411. [Google Scholar]
- 21.Inagaki, F., Y. Sakihama, A. Inoue, C. Kato, and K. Horikoshi. 2002. Molecular phylogenetic analyses of reverse-transcribed bacterial rRNA obtained from deep-sea cold seep sediments. Environ. Microbiol. 4:277-286. [DOI] [PubMed] [Google Scholar]
- 22.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, C., L. N. Li, J. Tamaoka, and K. Horikoshi. 1997. Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles 1:117-123. [DOI] [PubMed] [Google Scholar]
- 24.Keswani, J., and W. B. Whitman. 2001. Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int. J. Syst. E vol. Microbiol. 51:667-678. [DOI] [PubMed] [Google Scholar]
- 25.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin, L. A., R. J. Etter, M. A. Rex, A. J. Gooday, C. R. Smith, J. Pineda, C. T. Stuart, R. R. Hessler, and D. Pawson. 2001. Environmental influences on regional deep-sea species diversity. Annu. Rev. Ecol. Syst. 32:51-93. [Google Scholar]
- 27.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 28.Maddison, W. P., and D. R. Maddison. 2002. MacClade, v. 4.05. Sinauer Press, Sunderland, Mass.
- 29.Maddison, W. P., and M. Slatkin. 1991. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution 45:1184-1197. [DOI] [PubMed] [Google Scholar]
- 30.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mincer, T. J., P. R. Jensen, C. A. Kauffman, and W. Fenical. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. Ser. B Biol. Sci. 253:167-171. [Google Scholar]
- 35.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria—evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 36.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 37.Pettitt, A. N. 1982. Cramer-von Mises statistic, p. 220-221. In S. Kotz and N. L. Johnson (ed.), Encyclopedia of statistical sciences. Wiley-Interscience, New York, N.Y.
- 38.Poore, G. C. B., and G. D. F. Wilson. 1993. Marine species richness. Nature 361:597-598. [Google Scholar]
- 39.Roux, K. H. 1995. Optimization and troubleshooting in PCR, p. 53-62. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 40.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schneider, S., J. Kueffer, D. Roessli, and L. Excoffier. 1997. Arlequin ver. 1.1: a software for population genetic data analysis. Genetics and Biometry Lab, University of Geneva, Geneva, Switzerland.
- 43.Schut, F., E. J. Devries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sponga, F., L. Cavaletti, A. Lazzarini, A. Borghi, I. Ciciliato, D. Losi, and F. Marinelli. 1999. Biodiversity and potentials of marine-derived microorganisms. J. Biotechnol. 70:65-69. [Google Scholar]
- 46.Stach, J. E. M., L. A. Maldonado, A. C. Ward, M. Goodfellow, and A. T. Bull. 2003. New primers specific for Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828-841. [DOI] [PubMed]
- 47.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kämpfer, M. C. J. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Trüper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. E vol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 48.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 49.Staley, J. Speciation and bacterial phylospecies, p. 40-48. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington D.C., in press.
- 50.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 51.Swofford, D. L. 1993. PAUP—a computer-program for phylogenetic inference using maximum parsimony. J. Gen. Physiol. 102:A9. [Google Scholar]
- 52.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant-DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson, J., and P. P. E. Weaver. 1994. An AMS radiocarbon method to determine the emplacement time of recent deep-sea turbidites. Sediment. Geol. 89:1-7. [Google Scholar]
- 55.Turley, C. 2002. The importance of ′marine snow.' Microbiol. Today 29:177-179. [Google Scholar]
- 56.Vandermeer, J., M. A. Evans, P. Foster, T. Hook, M. Reiskind, and M. Wund. 2002. Increased competition may promote species coexistence. Proc. Natl. Acad. Sci. USA 99:8731-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver, P. P. E., and J. Thomson. 1993. Calculating erosion by deep-sea turbidity currents during initiation and flow. Nature 364:136-138. [Google Scholar]
- 59.Wenzhofer, F., O. Holby, and O. Kohls. 2001. Deep penetrating benthic oxygen profiles measured in situ by oxygen optodes. Deep-Sea Res. Part I Oceanogr. Res. Pap. 48:1741-1755. [Google Scholar]
- 60.Yates, M. D., F. A. Drobniewski, and S. M. Wilson. 2002. Evaluation of a rapid PCR-based epidemiological typing method for routine studies of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, J. Z., B. C. Xia, D. S. Treves, L. Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziebis, W., M. Huettel, and S. Forster. 1996. Impact of biogenic sediment topography on oxygen fluxes in permeable seabeds. Mar. Ecol. Prog. Ser. 140:227-237. [Google Scholar]