Abstract
The potential for removing uranium from contaminated groundwater by stimulating the in situ activity of dissimilatory metal-reducing microorganisms was evaluated in a uranium-contaminated aquifer located in Rifle, Colo. Acetate (1 to 3 mM) was injected into the subsurface over a 3-month period via an injection gallery composed of 20 injection wells, which was installed upgradient from a series of 15 monitoring wells. U(VI) concentrations decreased in as little as 9 days after acetate injection was initiated, and within 50 days uranium had declined below the prescribed treatment level of 0.18 μM in some of the monitoring wells. Analysis of 16S ribosomal DNA (rDNA) sequences and phospholipid fatty acid profiles demonstrated that the initial loss of uranium from the groundwater was associated with an enrichment of Geobacter species in the treatment zone. Fe(II) in the groundwater also increased during this period, suggesting that U(VI) reduction was coincident with Fe(III) reduction. As the acetate injection continued over 50 days there was a loss of sulfate from the groundwater and an accumulation of sulfide and the composition of the microbial community changed. Organisms with 16S rDNA sequences most closely related to those of sulfate reducers became predominant, and Geobacter species became a minor component of the community. This apparent switch from Fe(III) reduction to sulfate reduction as the terminal electron accepting process for the oxidation of the injected acetate was associated with an increase in uranium concentration in the groundwater. These results demonstrate that in situ bioremediation of uranium-contaminated groundwater is feasible but suggest that the strategy should be optimized to better maintain long-term activity of Geobacter species.
Cold War-era extraction and processing of uranium ore have left many sites around the world contaminated with uranium. Groundwater contamination is of particular concern because oxidized uranium is toxic, generally soluble in groundwater, and therefore mobile within the subsurface at many of these sites. Techniques for removing uranium from groundwater rely on inefficient pump-and-treat technologies or simple groundwater flushing to lower in situ metal concentrations to below acceptable limits (41). Projected treatment times (or natural attenuation) of several decades or longer are not uncommon and have spurred investigation of more-efficient remediation techniques (Natural and Accelerated Bioremediation Research [NABIR] program, Office of Biological and Environmental Research, U.S. Department of Energy [http://www.lbl.gov/NABIR]). In situ immobilization of uranium, which takes advantage of the redox character of uranium, has been suggested as a potential strategy to remove uranium from groundwater (15, 31). U(VI) is the mobile valence state of uranium, particularly in carbonate-containing groundwater, while reduced uranium, U(IV), is insoluble as uraninite (20). Reduction of U(VI) to U(IV) within aquifers could precipitate uranium, preventing further downgradient spread of groundwater contamination (1, 6, 15, 22, 23).
Laboratory studies have suggested that a simple strategy for promoting U(VI) reduction in contaminated aquifers is to add acetate as an electron donor to stimulate the activity of dissimilatory metal-reducing microorganisms (12, 13). Soluble U(VI) persists in subsurface environments because of an insufficient supply of electron donors to consume dissolved oxygen and/or promote active anaerobic respiration. Acetate effectively stimulates U(VI) reduction in subsurface sediments, resulting in removal of uranium from contaminated groundwater. U(VI) is reduced concurrently with Fe(III) and prior to reduction of sulfate (12). Enhanced U(VI) and Fe(III) reduction is associated with an increase in the number of “Geobacteraceae,” by several orders of magnitude. Furthermore, “Geobacteraceae” accounted for ca. 40% of the total microbial community as determined from 16S ribosomal DNA (rDNA)-based clone libraries during the most active period of Fe(III) and U(VI) reduction (18). Detected Geobacter species were the predominant “Geobacteraceae” in groundwaters with freshwater salinities, whereas Desulfuromonas species predominated in groundwaters with marine salinities. Most of the growth of the “Geobacteraceae” could be attributed to electron transfer to Fe(III) because Fe(III) was present in the sediment at millimole-per-kilogram quantities whereas only micromole-per-kilogram quantities of dissolved U(VI) were available (12). No other well-studied dissimilatory metal-reducing microorganisms, such as Shewanella species, could be detected in the anoxic sediments, even with PCR primers specific for the 16S rDNA sequences of these organisms. This result and the fact that Geobacter species available in pure culture are capable of U(VI) reduction (31) suggested that the “Geobacteraceae” in the sediments were responsible for the U(VI) reduction. Although abiotic mechanisms for U(VI) reduction in sediments such as reduction by sulfide, Fe(II), or reduced humic substances have been proposed, each of these abiotic mechanisms was eliminated as a possibility (12).
To determine if results from laboratory sediment incubations could be extrapolated to in situ uranium bioremediation in a contaminated aquifer, an in situ acetate injection experiment was conducted at a field site in Rifle, Colo. A field experiment was important because static incubations of sediments in the laboratory do not adequately replicate hydrogeochemical conditions in aquifers. An additional reason for concern is that it was recently suggested that microbial U(VI) reduction may not be an effective strategy for in situ treatment of uranium contamination in aquifers because the initial U(IV) products of U(VI) reduction are small (nanoparticulate) and may be mobile in the subsurface (39). Here we report that, as in the laboratory incubations, acetate addition stimulates the growth of Geobacter species and the effective removal of U(VI) from the groundwater in situ. However, further optimization is required in order to promote the long-term growth and activity of Geobacter species because the sulfate-reducing microorganisms that became predominant with continued acetate injection appeared to be less effective at U(VI) reduction.
MATERIALS AND METHODS
Site description and test plot design.
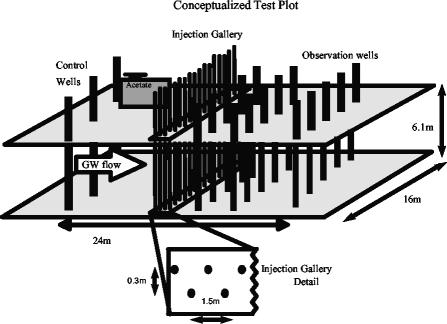
An in situ test plot was constructed on the grounds of a former uranium ore processing facility in Rifle, Colo. This site, designated the Old Rifle site, is part of the Uranium Mill Tailings Remedial Action (UMTRA) program of the U.S. Department of Energy (41). The former processing facility contained large piles of mill tailings on site from which residual uranium leached into the subsurface. All surface structures and contaminated soil have since been removed from the site, leaving only residual groundwater contamination within the local aquifer. This unconfined aquifer lies within an alluvial deposit in the floodplain of the Colorado River and is underlain by an impermeable layer of the Wasatch formation at a depth of ca. 6.1 m (41). Groundwater enters the aquifer from upgradient sources above the floodplain and exits into the Colorado River. Uranium concentrations in the groundwater within the test area range from 0.4 to 1.4 μM and are above the UMTRA maximum contaminant limit of 0.18 μM. Nitrate was not detected in Old Rifle groundwater, and dissolved oxygen concentrations averaged less than 0.2 mg/liter throughout the site.
Groundwater flow is generally toward the Colorado River at an approximate linear rate of 0.82 m/day (hydraulic conductivity, 54 m/day; porosity, 0.27; hydraulic gradient, 0.004 m/m). An injection gallery composed of 20 3.2-cm-diameter wells (schedule 40 polyvinyl chloride [PVC]) was installed in two rows (0.3 m apart) of 10 wells each positioned perpendicular to groundwater flow. All injection wells in each row were spaced 1.5 m apart, and the two rows were offset from each other by 0.8 m for a total gallery width of 16 m (Fig. 1). All wells were installed to a depth of 6.1 m and were screened from a depth of 1.5 to 6.1 m, encompassing the entire saturated interval of the aquifer (2.4 m). Each injection well contained three injection points composed of 0.3-cm-diameter stainless steel tubing (Supelco, Bellefonte, Pa.) positioned at three different depths within the saturated subsurface. Each injection point was connected to a flowmeter (59 total; Cole-Parmer Instrument Company, Vernon Hills, Ill.) at the injection well head, which controlled the flow of acetate solution from a manifold (10.2-cm-diameter schedule 40 PVC) spanning the entire width of the injection gallery. The injection manifold was connected to a stainless steel tank placed within a storage shed at one end of the injection gallery via 0.6-cm-diameter stainless steel tubing (Supelco).
FIG. 1.
Concept and layout of the in situ test plot installed at the Old Rifle UMTRA site in Rifle, Colo.
A 2120L (560-gal) stainless steel tank (Rain-for-Rent, Rifle, Colo.) served as a storage tank for the acetate solution. The tank was filled periodically over the course of the experiment with native groundwater collected from an upgradient well (∼50 m upgradient from the gallery) and amended with sodium acetate (Sigma Chemical Company, St. Louis, Mo.) and potassium bromide (Sigma Chemical Company) at concentrations of ca. 100 and 10 mM, respectively. During filling and chemical addition, groundwater within the tank was continuously sparged with nitrogen gas to remove residual traces of dissolved oxygen. The solution of acetate and bromide was stored under a 0.1-atm (1.5-lb/in2) N2 headspace to prevent entry of air into the tank over time and to provide additional head for injecting the acetate solution into the subsurface. During operation of the injection gallery all flowmeters were set to provide 1 to 3 ml of acetate solution/min to the subsurface, which corresponds to approximately a 1 to 3% volume addition (1 to 3 mM acetate, 100 to 300 μM bromide in situ) to the aquifer per day.
To evaluate stimulated changes within the subsurface, a total of 15 monitoring wells were installed downgradient in three rows of five wells, each spaced 2.8 m apart and centered on the injection gallery (Fig. 1). Each row of monitoring wells was positioned at fixed distances from the injection gallery roughly corresponding to groundwater travel times of approximately 4, 9, and 18 days (ca. 3.7, 7.3, and 14.6 m from the gallery). An additional three wells were installed 3.7 m upgradient from the injection gallery to serve as control wells. All monitoring and control wells were installed to the same depth and were screened over the same interval as the injection wells. The entire test plot sits within a uranium-contaminated portion of the aquifer. Groundwater containing U(VI) flows past the control wells and injection gallery from upgradient sources and out into the monitoring-well field.
Analysis of groundwater samples.
Acetate was continuously injected into the Old Rifle aquifer for 3 months between June and October 2002. Groundwater samples were systematically collected at regular intervals from all monitoring wells and the storage tank during acetate injection. Groundwater parameters from each well, including pH, dissolved oxygen, conductivity, and redox potential, were monitored by using a peristaltic pump (Cole-Parmer Instrument Company) connected to a flow cell attached to multiprobe data sonde (Hydrolab-Hach Company, Chicago, Ill.). All wells were purged until these groundwater parameters stabilized (ca. 12-liter purge at 0.5 liters/min). After purging, the data sonde was disconnected and groundwater samples were taken directly from the pump outlet. All groundwater samples were filtered with 0.2-μm-pore-size PTFE (Teflon) syringe filters (Alltech Associates Inc., Deerfield, Ill.). Samples (15 ml) for U(VI) and anion (bromide, nitrate, and sulfate) analysis were placed into plastic 15-ml sterile conical tubes (VWR International, Inc., Bridgeport, N.J.). Samples (19 ml) for acetate were collected into no-headspace glass sample containers (Eagle Picher 20-ml amber vials; VWR International, Inc.) and preserved with 1 ml of 0.1 M H2SO4. Samples (19 ml) for Fe(II) and sulfide analyses were also collected into no-headspace glass sample containers (Eagle Picher 20-ml amber vials; VWR International, Inc.) and preserved with 1 ml of 10 M HCl and 0.1 M KOH, respectively. All samples collected in the field were shipped back to the laboratory via overnight courier and stored at 4°C prior to analysis. Uranium was measured by kinetic phosphorescence analysis as previously described (12). Acetate was measured with high-pressure liquid chromatography on a Hewlett-Packard series 1100 high-pressure liquid chromatograph (Agilent Technologies, Inc., Albany, N.Y.) using a fast-acid analysis column (Bio-Rad, Hercules, Calif.) with a 0.5 mM H2SO4 eluent and absorbance detection (210 nm). Sulfate, nitrate, and bromide concentrations were measured from filtered samples with a Dionex DX-100 ion chromatograph as previously described (28). Fe(II) and sulfide concentrations were determined by previously described spectrophotometric techniques (5, 9, 29).
16S rDNA-based microbial community analysis.
Microorganisms in the groundwater were collected in the field either on sterile flat filters (0.2-μm pore size; Supor-200; Pall-Gelman Laboratory, Ann Arbor, Mich.) or cartridge filters (0.2-μm pore size; Sterivex-GP; Millipore Corporation, Bedford, Mass.). The filters were immediately frozen in the field and shipped on dry ice back to the laboratory for analysis. Microbial community analyses were performed as previously described (18). Briefly, DNA from filters was extracted with either the FastDNA SPIN kit (Bio101, Inc., Carlsbad, Calif.) or by a modified phenol-chloroform DNA extraction method (34, 37). Comparable community analyses were obtained regardless of filter type or DNA extraction method utilized (data not shown). 16S rDNA was amplified with two primer sets: (i) 8F (AGAGTTTGATCMTGGCTCAG) and 519R (GWATTACCGCGGCKGCTG) and (ii) 338F (ACTCCTACGGGAGGCAGC) and 907R (CCGTCAATTCMTTTRAGTTT) (4, 11, 19). PCR mixtures (100 μl) contained ∼5 ng of template DNA, 10 μl of 10× polymerase buffer (Qiagen, Valencia, Calif.), 20 μl of 5× Q buffer (Qiagen), 3 mM MgCl2 (Qiagen), 1 μl of bovine serum albumin (0.5 mg/ml), 200 μM deoxyribonucleoside triphosphates (Sigma-Aldrich Co., St. Louis, Mo.), 25 pmol of forward and reverse primers (Sigma-Genosys, The Woodlands, Tex.), and 1.25 U of Taq polymerase (Qiagen). PCR mixtures containing all components except the template and Taq polymerase were UV irradiated for 5 min to ensure sterility. Reactions were carried out in a Peltier thermal cycler (PTC 200; MJ Research Inc., Waltham, Mass.), beginning with a 5-min denaturation at 95°C and then 30 cycles of 94°C (30 s), 45°C (1 min), 72°C (1 min) and a final 7-min elongation at 72°C. The PCR products from the two primer sets were isolated by gel extraction (Qiagen) and pooled prior to cloning. Clone libraries were constructed by insertion of the amplified 16S rDNA sequences into the TOPO TA vector pCR 2.1 and cloning into chemically competent Escherichia coli TOP10 cells according to the manufacturer's instruction (Invitrogen, Carlsbad, Calif.).
Inserts of 16S rDNA from at least 30 clones from each clone library were amplified with M13 forward and reverse primers (33, 42). PCR products were cleaned with the QIAquick PCR purification kit (Qiagen) and sequenced with the M13F primer. Sequences were compared to those compiled in GenBank with the BLAST suit of programs (2, 3). Alignments and identity matrix comparisons of clone and deposited sequences were performed in BioEdit (17).
Phospholipid fatty acid (PLFA) analysis.
For lipid-based community analysis, microorganisms in the groundwater were collected on Bio-Sep polymer beads retained in perforated Teflon perfluoroalkoxy tubes (length, 4 cm; outside diameter, 1.25 cm) that were suspended in selected wells (B-02, M-03, M-08, and M-13) within the test plot during acetate injection. At selected time intervals the tubes were recovered, frozen on site with dry ice, and shipped via overnight courier to the laboratory. Lipid biomarkers were analyzed as previously described (36, 44). Briefly, the biocarrier beads were extracted with a single-phase chloroform-methanol buffer system (43). Total extracted lipids were fractionated into neutral lipids, glycolipids, and polar lipids by silicic acid column chromatography (16). The polar lipid fraction was transesterified to the fatty acid methyl esters with a mild alkaline methanolysis (16) and was analyzed by gas chromatography-mass spectroscopy (6890 series gas chromatograph interfaced to a 5973 series mass-selective detector; Agilent Technologies).
RESULTS
Operation of injection gallery.
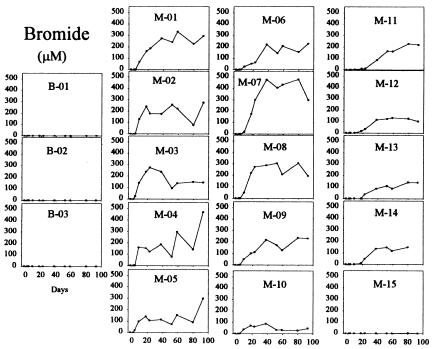
Potassium bromide added to the injected acetate solution served as a groundwater tracer within the test plot and as a control on the volume of solution added to the subsurface. During the experiment, bromide was not found in any of the upgradient control wells and was detected in all the downgradient monitoring wells, except one well (M-15) positioned at the furthest downgradient corner of the test plot (Fig. 2). This confirmed initial estimates of the direction of groundwater flow and indicated injection of solution across a broad front within the subsurface. Breakthrough of bromide was detected within 4, 9, and 18 days after initiation of injection in the first, second, and third monitoring-well rows, respectively, corresponding to a groundwater travel time of 0.82 m/day (linear distances from gallery, 3.7, 7.3, and 14.6 m respectively). Averaged bromide concentrations from within the first row of monitoring wells from day 17 through day 59 (186 μM) compared with the concentration of bromide within the storage tank (9.7 mM) indicate a 2% (average) volume addition of solution to the aquifer per day.
FIG. 2.
Bromide within the monitoring-well field. B-01 to B-03, upgradient control wells; M-01 to M-15, downgradient monitoring wells. Groundwater flow is from left to right, and the injection gallery is positioned between the control wells and the first row of monitoring wells.
Impact of acetate injection on aquifer geochemistry.
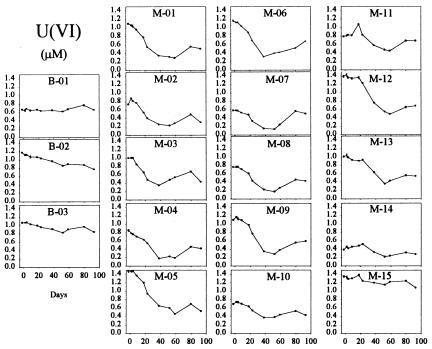
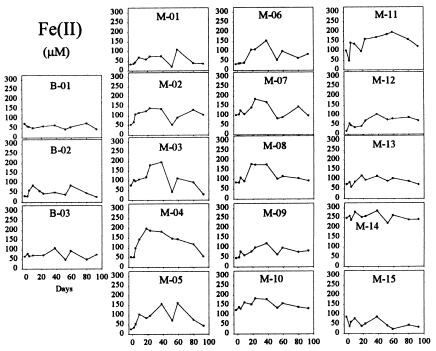
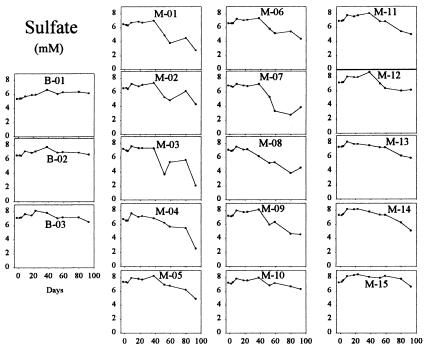
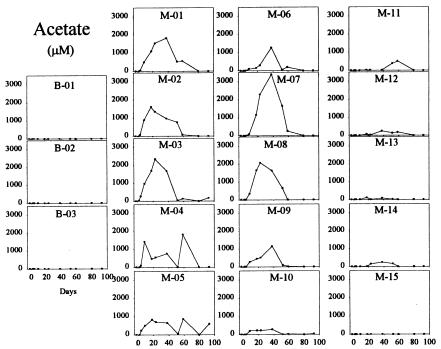
Soluble U(VI) concentrations began to decrease within the monitoring-well field relative to upgradient control wells within 9 days after the start of the acetate injection (Fig. 3). U(VI) concentrations decreased to levels at or below the UMTRA-prescribed limit of 0.18 μM within 50 days in some of the wells. This initial loss of U(VI) was not the result of a pH change within the test plot as all pH values remained essentially constant throughout acetate injection and varied by less than 0.2 units from pH 7.0 across the site. Decreases in U(VI) were coincident with the accumulation of Fe(II) (Fig. 4) and prior to the loss of sulfate (see Fig. 6). After 50 days of acetate injection, U(VI) began to increase within much of the well field (Fig. 3). Coincident with the rise in U(VI), Fe(II) generally decreased (Fig. 4) and acetate within the well field decreased to nondetectable levels (Fig. 5). Acetate concentrations in the tank remained stable and bromide remained detectable within the well field (Fig. 2), suggesting that the decrease in acetate was due to increased consumption at the point of injection. These changes after 50 days of injection were accompanied by a decrease in sulfate (Fig. 6) and the appearance of a black precipitate, presumably ferrous sulfide, in the groundwater. Sulfide was detected within the groundwater during this time (data not shown). The loss in sulfate (2 to 3 mM) was approximately stoichiometric with the loss of acetate (2 to 3 mM).
FIG. 3.
U(VI) in groundwater samples. The layout is described in the Fig. 2 legend.
FIG. 4.
Fe(II) in groundwater samples. The layout is described in the Fig. 2 legend.
FIG. 6.
Sulfate concentrations in groundwater samples. The layout is described in the Fig. 2 legend.
FIG. 5.
Acetate in groundwater samples. The layout is described in the Fig. 2 legend.
Impact of acetate injection on the microbial community.
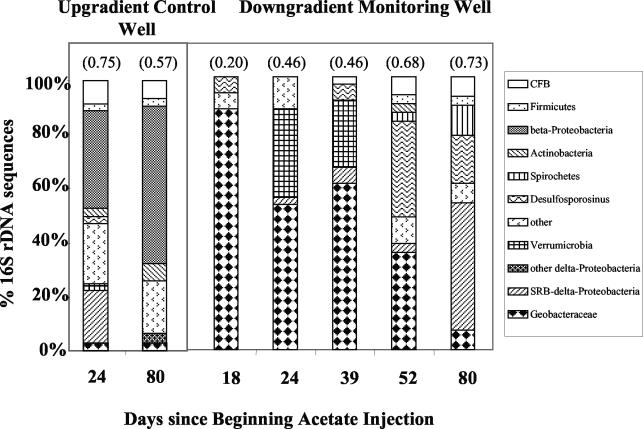
To monitor changes within the subsurface microbial community during the injection of acetate, 16S rDNA sequences detected in the groundwater from representative monitoring wells were analyzed from clone libraries. Coverage for all libraries was greater than 86% (14). In an upgradient control well, 16S rDNA sequences of β-proteobacteria predominated, with a low percentage of 16S rDNA sequences closely related to those of Geobacter or sulfate-reducing species (Fig. 7). Changes in microbial diversity within the control well over the course of the study were not significant (t test evaluation, 5% cutoff [21]). β-Proteobacteria tended to remain the dominant detected group throughout the experiment (Fig. 7).
FIG. 7.
Clone library analyses of changes in the groundwater microbial community from a representative downgradient well (M-07) within the second row of monitoring wells compared to data from an upgradient control well (B-02) during the experiment. The results are compiled from sequences of at least 30 clones analyzed for each time point. Calculated diversity values (Shannon-Weaver index) are provided in parentheses at the top of each clone library. Similar results were obtained from a nearby downgradient well.
In contrast, the injection of acetate resulted in a substantial enrichment of “Geobacteraceae” and decreased calculated diversity within the treatment zone (Fig. 7). At 17 days after the start of the acetate injection “Geobacteraceae” accounted for 89% of the groundwater microbial community (a second verification analysis resulted in 87% Geobacter enrichment, data not shown). Sequences that fell within the Geobacter genus accounted for 83% of the sequences of the “Geobacteraceae.” “Geobacteraceae” continued to comprise more than 50% of the groundwater microbial community through at least the first 39 days of the experiment. Other well-known U(VI)-reducing organisms, such as Shewanella or Desulfovibrio species, were not detected.
As acetate injection continued beyond 39 days the composition of the microbial community began to shift from a “Geobacteraceae”-dominatated community to a community dominated by organisms known for sulfate reduction. This shift was reflected in the microbial community at 52 days, where the abundance of “Geobacteraceae” had dropped to 35% of the groundwater microbial community and calculated species diversity tended to increase. Other prominent groups at this time point included organisms exhibiting similarity to gram-positive, sulfate-reducing Desulfosporosinus species (80 to 88% sequence identity), which comprised 33% of the microbial community. Beyond 52 days the relative abundance of “Geobacteraceae” detected within the microbial community continued to decrease, falling to below 7% by day 80, when the microbial community became dominated (45%) by members of the family “Desulfobacteraceae” (80 to 95% similarity), organisms more commonly known for acetate oxidation coupled to sulfate reduction. Desulfosporosinus-like organisms comprised 17% of the population at this point (day 80), and one clone exhibited 87% similarity to Desulfotomaculum acetoxidans, a gram-positive, acetate-oxidizing sulfate reducer.
PLFA analysis.
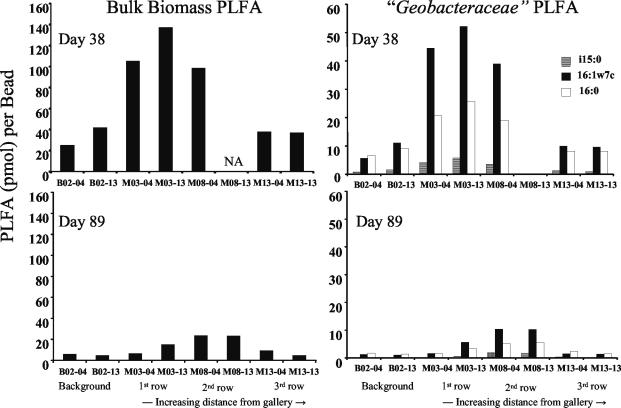
The microbial community in the subsurface was also evaluated by suspending Bio-Sep polymer beads in perforated containers at two depths within monitoring wells and then extracting and analyzing the PLFA content (36, 45). All capsules were retrieved and replaced with fresh capsules at two time points (day 38 and 89) during acetate injection. Beads from the first two rows of monitoring wells in the treatment zone that were sampled after 38 days of acetate injection had 2.5 to 7 times more bulk biomass PLFA than those from upgradient control wells or wells positioned further downgradient in the treatment zone. The PLFAs in the two rows of wells immediately downgradient of the injection gallery were enriched in three fatty acids, 16:1ω7c, i15:0, and 16:0, known to be associated with “Geobacteraceae” (24), compared to the upgradient control wells or the downgradient wells furthest from the injection gallery (Fig. 8). Later in the experiment (>50 days), overall biomass and the proportion of Geobacter signature lipids within the well field decreased (Fig. 8).
FIG. 8.
Changes observed in bulk biomass PLFA and signature lipids for Geobacter species detected on Bio-Sep beads in capsules suspended in four selected control and monitoring wells (B-02, M-03, M-08, and M-13) at two different depths for each well within the saturated subsurface (designated sampling points 4 and 13 for each well) from two different time points during the field test.
DISCUSSION
The results provide the first field evidence that it is possible to effectively remove uranium from contaminated groundwater in situ by stimulating the activity of Geobacter species in the subsurface. When acetate was added to enhance the growth of Geobacter species, U(VI) was actively removed. When conditions no longer favored the growth of Geobacter species and sulfate-reducing microorganisms flourished, uranium removal was less effective. These results suggest that implementation of a long-term in situ bioremediation strategy should optimize conditions for continued growth and/or survival of Geobacter species.
Removal of U(VI) from contaminated groundwater. (i) Fe(III) reduction phase.
Loss of soluble U(VI) began almost immediately following the start of acetate injection at the Old Rifle site, and levels dropped ca. 70% within 50 days, with concentrations in some wells falling below the UMTRA treatment goal of 0.18 μM. This demonstrates that U(VI)-reducing microorganisms have the potential to immobilize uranium in situ. Thus, the suggestion that microbial U(VI) reduction forms nanoparticles of the U(IV) mineral, uraninite, that will still be mobile in the subsurface (39) does not appear to be a significant concern. This should have been apparent from earlier laboratory studies (15, 30) which demonstrated that, although the initial U(IV) products of microbial U(VI) reduction are small and readily pass through microporous filters, within a matter of hours the initial U(IV) products aggregate into larger U(IV) precipitates which are highly insoluble and which cannot pass through the filters. In fact, the results of the study raising concerns about nanoparticles (39) are consistent with this finding, as the electron micrographs clearly show aggregation of uraninite nanoparticles into much-larger particles. This helps explain why, despite the potential formation of nanoparticles of U(IV), the most recent study found that over 98% of the dissolved uranium in a uranium-contaminated sediment was removed from solution when microbial U(VI) reduction was stimulated in laboratory incubations (39). Thus, even if the initial products of U(VI) reduction are small uraninite crystals, processes such as agglomeration and/or absorption onto sediments remove them from the groundwater.
The initial removal of U(VI) from the groundwater appeared to be associated with Fe(III) reduction. There was an accumulation of Fe(II) and a substantial enrichment of Geobacter species in the groundwater. This is similar to results from laboratory incubations of uranium-contaminated subsurface sediments in which U(VI) was reduced concurrently with Fe(III) and “Geobacteraceae” became the predominant organisms in the sediments (12, 13, 18). It was possible to more quantitatively monitor Fe(III) reduction and the growth of “Geobacteraceae” in the laboratory incubations because it was possible to sample the sediments over time. Most of the Fe(II) produced from Fe(III) reduction is associated with the sediments (27). Furthermore, it is not certain how well the microbial community in the groundwater reflects that attached to the sediments. Although Geobacter species must attach to available Fe(III) oxides in order to reduce them (35), it is expected that at any time a proportion of the Geobacter organisms will be free swimming because, once they reduce Fe(III) oxides in the immediate area, they need to become motile in order to find new sources of Fe(III) (8). However, even though the monitoring of Fe(III) reduction and the microbial community was more qualitative in the field experiment, the trends in the initial phase of the field experiment are clearly similar to those observed in the laboratory incubations.
(ii) Sulfate reduction phase.
With continued injection of acetate, there was a loss of sulfate associated with an increase in 16S rDNA sequences most closely related to the sequences of sulfate-reducing microorganisms and an eventual depletion of acetate from the treatment zone. This suggested that longer-term injection of acetate promoted the growth of sulfate-reducing microorganisms near the point of injection. There is an equimolar stoichiometry of acetate and sulfate consumption associated with the oxidation of acetate coupled to sulfate reduction, and the loss of sulfate from the groundwater flowing into the treatment zone corresponded closely with the acetate input. Furthermore, the loss of sulfate was accompanied by an accumulation of sulfide and a decrease in soluble Fe(II), consistent with the precipitation of iron sulfides.
These results suggest that the terminal electron-accepting process responsible for acetate oxidation in the treatment zone switched from Fe(III) reduction to sulfate reduction. Previous studies have demonstrated that, when Fe(III) oxides are available, Fe(III)-reducing microorganisms are able to outcompete sulfate reducers for electron donors (7, 26). Furthermore, sulfate reducers may preferentially reduce Fe(III) over sulfate but may not conserve energy to support growth from this reaction (10, 32). Thus, initially Geobacter species may have been competitive for the acetate entering the treatment zone. However, as Fe(III) oxides nearest the point of injection were depleted, sulfate reducers became more competitive. If so, there was more than enough sulfate in the groundwater for complete consumption of the incoming acetate by sulfate reducers before it reached downgradient portions of the treatment zone that still contained Fe(III) oxides. The complete consumption of acetate under sulfate-reducing conditions near the injection gallery prevented further metal reduction in these downgradient sediments. This hypothesis, which is consistent with the available data, needs further verification, which would include detailed analysis of the distribution of Fe(III) oxides in the sediments.
As sulfate reduction became the predominant terminal electron accepting process in the treatment zone, the concentration of U(VI) in the groundwater increased somewhat. Some sulfate-reducing microorganisms have been shown to reduce U(VI) (25, 30, 32), but typically with hydrogen or lactate serving as the electron donor. Evaluation of several acetate-oxidizing sulfate reducers, including Desulfobacter and Desulfotomaculum species with 16S rDNA sequences closely related to those of organisms recovered from the treatment zone, suggested that they did not reduce U(VI) (32), but a Desulfotomaculum species isolated from marine sediments did reduce U(VI) with butyrate as the electron donor (40). Furthermore, 16S rDNA sequences closely related to those of Desulfosporosinus species were prevalent during the sulfate reduction phase, and it has been suggested that Desulfosporosinus species can reduce U(VI) (38, 39), although the ability of organisms in this genus to reduce U(VI) with acetate as the electron donor has not been demonstrated. The increase in U(VI) in the groundwater during the sulfate reduction phase suggests that the acetate-oxidizing sulfate reducers may not have been as effective as Geobacter species in reducing U(VI).
In summary, this study demonstrates that Geobacter species can be important agents for the in situ bioremediation of uranium and that sulfate-reducing microorganisms may be less effective in uranium removal. Thus, further study into strategies to promote the long-term maintenance of metal reduction and the activity of Geobacter species may yield a better in situ uranium bioremediation strategy.
Acknowledgments
This research was funded by the Natural and Accelerated Bioremediation Research program, Biological and Environmental Research, U.S. Department of Energy (grants DE-FG02-0ER62985 and DE-FG02-97ER62475).
We thank Guibo Xie (The Johns Hopkins University) for aiding in the construction of the injection gallery and storage shed and Darren Flood (Rain-for-Rent, Inc.) for procurement of the stainless steel tank. We thank Kathleen O'Neill for technical assistance and helpful discussion of 16S rDNA sequence information and Jason Proctor for insightful input with the statistical analyses.
REFERENCES
- 1.Abdelouas, A., W. Lutze, and H. E. Nuttall. 1999. Uranium contamination in the subsurface: characterization and remediation, p. 433-473. In P. C. Burns and R. Finch (ed.), Uranium: mineralogy, geochemistry and the environment, vol. 38. Mineralogical Society of America, Washington, D.C.
- 2.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. Binder, S. W. Chisholm, R. Olsen, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, R. T., and D. R. Lovley. 2000. Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34:2261-2266. [Google Scholar]
- 6.Anderson, R. T., and D. R. Lovley. 2002. Microbial redox interactions with uranium: an environmental perspective, p. 205-223. In M. Keith-Roach and F. Livens (ed.), Interactions of microorganisms with radionuclides. Elsevier Science Limited, Amsterdam, The Netherlands.
- 7.Chapelle, F. H., and D. R. Lovley. 1992. Competitive exclusion of sulfate reduction by Fe(III)-reducing bacteria: a mechanism for producing discrete zones of high-iron ground water. Ground Water 30:29-36. [Google Scholar]
- 8.Childers, S. E., S. Ciufo, and D. R. Lovley. 2002. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416:767-769. [DOI] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 10.Coleman, M. L., D. B. Hedrick, D. R. Lovley, D. C. White, and K. Pye. 1993. Reduction of Fe(III) in sediments by sulphate-reducing bacteria. Nature 361:436-438. [Google Scholar]
- 11.Eden, P. E., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 12.Finneran, K. T., R. T. Anderson, K. P. Nevin, and D. R. Lovley. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 11:339-357. [Google Scholar]
- 13.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 14.Good, I. 1953. The population frequency of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 15.Gorby, Y. A., and D. R. Lovley. 1992. Enzymatic uranium precipitation. Environ. Sci. Technol. 26:205-207. [Google Scholar]
- 16.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure in estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. L., B. Pace, G. J. Olsen, D. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmuir, D. 1978. Uranium solution-mineral equilibria at low temperatures with applications to sedimentary ore deposits. Geochim. Cosmochim. Acta 42:547-569. [Google Scholar]
- 21.Lloyd, M., J. Zar, and J. Karr. 1968. On the calculation of information-theoretical measures of diversity. Am. Midland Naturalist 79:257-272. [Google Scholar]
- 22.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Industr. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 23.Lovley, D. R. 2001. Reduction of iron and humics in subsurface environments, p. 193-217. In J. K. Frederickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss, Inc., New York, N.Y.
- 24.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R., and E. J. P. Phillips. 1992. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26:2228-2234. [Google Scholar]
- 26.Lovley, D. R., and E. J. P. Phillips. 1987. Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl. Environ. Microbiol. 53:2636-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovley, D. R., and E. J. P. Phillips. 1988. Manganese inhibition of microbial iron reduction in anaerobic sediments. Geomicrobiol. J. 6:145-155. [Google Scholar]
- 28.Lovley, D. R., and E. J. P. Phillips. 1994. Novel processes for anoxic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl. Environ. Microbiol. 60:2394-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 32.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 33.Messing, J., and J. Vieira. 1982. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene 19:269-276. [DOI] [PubMed] [Google Scholar]
- 34.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Wu, and E. F. DeLong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antartica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 36.Pinkart, H. C., D. B. Ringelberg, Y. M. Piceno, S. J. Macnaughton, and D. C. White. 2002. Biochemical approaches to biomass measurements and community structure analysis, p. 101-113. In D. E. Stahl, C. H. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 37.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2002. Nanometre-size products of uranium bioremediation. Nature 419:134. [DOI] [PubMed] [Google Scholar]
- 40.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(VI), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 41.U.S. Department of Energy. 1999. Final site observational work plan for the UMTRA project Old Rifle site GJO-99-88-TAR. U.S. Department of Energy, Grand Junction, Colo.
- 42.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 43.White, D. C., R. J. Bobbie, J. S. Heron, J. D. King, and S. J. Morrison. 1979. Biochemical measurements of microbial mass and activity from environmental samples, p. 69-81. In J. W. Costerton and R. R. Colwell (ed.), Native aquatic bacteria: enumeration, activity and ecology, ASTM STP 695. American Society for Testing and Materials, Philadelphia, Pa.
- 44.White, D. C., and D. B. Ringelberg. 1998. Signature lipid biomarker analysis, p. 255-272. In R. S. Burlage, R. Atlas, D. A. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, N.Y.
- 45.White, D. C., J. O. Stair, and D. B. Ringelberg. 1996. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Ind. Microbiol. 17:185-196. [Google Scholar]