Abstract
Prochlorothrix hollandica is the only filamentous chlorophyll b (Chlb)-containing oxyphotobacterium that has been found in freshwater habitats to date. Chlb serves as a light-harvesting pigment which is bound to special binding proteins (Pcb). Even though Prochlorothrix was initially characterized as a highly salt-sensitive species, we detected it in a brackish water environment that is characterized by salinities of up to 12 practical salinity units. Using PCR and reverse transcription, we amplified pcb gene fragments of phytoplankton samples taken along a salinity gradient in the eutrophic Darss-Zingst estuary (southern Baltic Sea). After sequencing, high levels of homology to the pcbB and pcbC genes of P. hollandica were found. Furthermore, autofluorescence of Prochlorothrix-like filaments that indicated that Chlb was present was detected in enrichment cultures prepared from the estuarine phytoplankton. The detection of Chlb-containing filaments, as well as the pcb and 16S ribosomal DNA sequences, suggests that Prochlorothrix is an indigenous genus in the Darss-Zingst estuary and may also inhabit many other brackish water environments. The potential of using pcb gene detection to differentiate Prochlorothrix from morphologically indistinguishable species belonging to the genera Pseudanabaena and Planktothrix (Oscillatoria) in phytoplankton analyses is discussed.
The Prochlorophyta is an unusual group among the oxyphotobacteria. Like the chloroplasts in higher plants, the members of this group contain chlorophyll b (Chlb) as an accessory pigment. For many years, the symbiont Prochloron didemni (19) was the only known representative of the Prochlorophyta. The first filamentous strain (5) in the group of Chlb-synthesizing prokaryotes, the free-living species Prochlorothrix hollandica, was isolated in 1984 from a lake in The Netherlands (4). Prochlorothrix sp. strain NIVA-8/90, tentatively named Prochlorothrix scandica, has been proposed as a second species (28). In contrast to the salt-sensitive filamentous oxyphotobacteria, coccoid Chlb-containing species of the genus Prochlorococcus are very abundant in the central oceans (7, 26). However, phylogenetic analyses of 16S ribosomal DNA (rDNA) sequences have clearly indicated that the oxyphotobacteria do not form a separate bacterial lineage but are specially pigmented members of the old cyanobacterial evolutionary radiation (39).
Many investigations have concentrated on the evolutionary importance of the Chlb-containing oxyphotobacteria and the specificities of their photosynthetic machinery (3, 12, 19, 23, 37, 38). However, since most attention has been paid to the biology of the globally important genus Prochlorococcus in marine ecosystems (8, 11, 21, 22, 26, 33), our understanding of Prochlorothrix ecology is limited. The isolation of Prochlorothrix strains from the Loosdrecht lakes (6) and Lake Malaren (28) and the results of laboratory studies (5) imply that Prochlorothrix spp. are freshwater organisms with a preference for shallow eutrophic water. The highest levels of Prochlorothrix were found during the summer in shallow, phosphate-limited regions of the Loosdrecht lakes (5). The oxygenic photosynthetic activity was found to be highly resistant to inhibition by sulfide (30). Experiments focusing on phosphorus nutrition resulted in characterization of P. hollandica as a high-affinity storage strategist (9).
While Prochlorococcus cells dominate phytoplankton communities in wide areas of the oceans (27), Prochlorothrix occurs at most times of the year at rather low levels, while cyanobacteria that are very similar morphologically seem to dominate. It is particularly difficult to distinguish Prochlorothrix from members of the cyanobacterial genera Pseudanabaena and Planktothrix (Oscillatoria). In the past, epifluorescence microscopy was used to investigate phytoplankton from the Loosdrecht lakes (43), the original source of P. hollandica (5). This technique is based on the phycoblisome autofluorescence that is characteristic of filamentous cyanobacteria but is missing in Prochlorothrix because of its different pigmentation. Detecting Prochlorothrix based on the absence of fluorescent trichomes is not a very reliable method (5, 43). Modern molecular techniques may be much more suitable for this purpose. The currently used methods, such as 16S rDNA denaturing gradient gel electrophoresis and sequence analysis, are restricted to analysis of species that occur at rather high levels in the environment (42). Methods based on genes which are restricted to specific bacterial groups could provide greater sensitivity. Genes encoding Chla/b binding proteins (pcb) may be good candidates for specific detection of Chlb-containing oxyphotobacteria. These genes exhibit only low levels of similarity to members of the extended gene family encoding eukaryotic Chla/b and Chla/c light-harvesting proteins (18). The antenna polypeptides are encoded by three genes in P. hollandica (pcbABC). The pcbC gene is significantly different from pcbA and pcbB, which exhibit high levels of similarity to iron stress-induced isiA genes of cyanobacteria (41). The genes of the multicistronic pcbABC operon are transcribed largely independent of the light intensity applied (24). Seven different pcb-like genes were found to be expressed in a Prochlorococcus strain (12). In general, the known sequences of pcb genes from P. hollandica allow the design of PCR primers which are highly specific for this genus.
In this paper we present evidence that the genus Prochlorothrix is much more widespread than the previous occasional observations indicated. During our investigation of cyanobacterial diversity in an estuary in the Baltic Sea (unpublished data), DNA fragments with high levels of sequence similarity to pcb genes of P. hollandica were obtained. The search for Prochlorothrix-like filaments and their characteristic autofluorescence signatures in enrichment cultures was augmented with tests for the salt tolerance of P. hollandica cells to verify the genetic results.
MATERIALS AND METHODS
Sampling sites and samples.
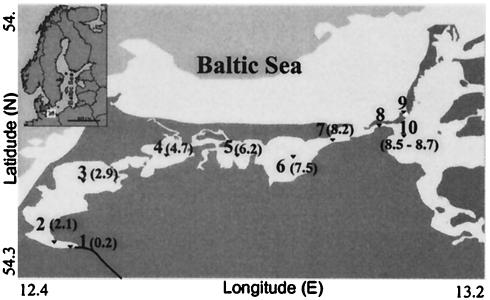
Plankton samples were taken in July 2001 from various locations (Fig. 1) in the Darss-Zingst estuary. The samples originated from 10 stations, which comprise a 55-km line along a salinity gradient from 0.2 to 8.7 practical salinity units (PSU). All salinity values were derived from measurements made with a conductivity-measuring cell (LT 197; WTW Weilheim) according to the definition of the conductivity ratio of standard seawater (salinity, 35 PSU) and a solution containing 32.4356 g of KCl per kg of solution at 15°C (International Association for the Physical Sciences of the Ocean). The water temperature ranged from 22 to 25°C. The sampling sites were designated by using a series of increasing numbers that followed the west-east axis of the estuary and increasing salinity (Fig. 1). Samples for RNA extraction were quickly frozen on board.
FIG. 1.
Map of sampling sites in the Darss-Zingst estuary. Sampling sites 1 to 10 correspond to buoys Recknitz, R2, R84, R37/44, R1/B65, B53, B37/46, B12/13, B18/19, and B35, respectively. The salinity at each sampling site (in PSU) is indicated in parentheses. The inset is a map of the Baltic Sea.
Cells of the isolate from Lake Malaren, Prochlorothrix sp. strain NIVA-8/90 (the organism proposed as P. scandica), were obtained as a frozen pellet.
Phytoplankton counting and differential epifluorescence microscopy.
Phytoplankton cells were counted by using Lugol-fixed samples and an inverted microscope (20, 40) at a magnification of ×256. For the dominant cyanobacteria and members of the Chlorophyceae, which form large spherical colonies, numbers of cells were determined by examining 10 randomly chosen colonies per sample with green light excitation at a magnification of ×1,250. From the mean cell number, the total number of cells per species was calculated. Cell diameters were measured to calculate cell volumes (10), and the amount of carbon biomass was estimated by using 11.25% carbon per fresh weight (15) at a specific density of 1.04 g cm−3. Additionally, unicellular rod-shaped cyanobacteria in the pico size class (diameter, <2 μm) were quantified by using glutaraldehyde-fixed samples (which were filtered through a 0.2-μm-pore-size Millipore filter prior to analysis) and epifluorescence illumination (green light; BP 545 nm; magnification, ×1,250), and a mean cell volume of 0.86 μm3 (unpublished data) was used for biomass calculations. Differential epifluorescence microscopy and microphotography were performed with the equipment described by Schubert and Schumann (36), which allowed detection of Chla (photosystem II) autofluorescence of living cells after highly selective excitation in the blue absorption bands of either Chla or Chlb.
DNA and RNA techniques.
For extraction of nucleic acids, 70 mg (fresh weight) of phytoplankton cells was used. The cells were disrupted by bead beating (5,000 rpm; 10 s; 0.1-mm-diameter glass beads). For extraction of chromosomal DNA, a Qiagen plant extraction kit (Qiagen) was used. An additional preextraction step with hot phenol (Aqua-Roti-Phenol; Roth) for 10 min (pH 4.5 to 5.0; 65°C) was used for RNA extraction. Total RNA was isolated by using a High Pure RNA isolation kit (Roche Biochemicals). All PCR were performed by using Taq PCR Master Mix (Qiagen), and reverse transcription (RT) reactions were carried out with SuperScriptII Rnase H reverse transcriptase (Gibco BRL Life Technologies).
Cyanobacterial 16S rDNA fragments (424 bp) were amplified with degenerate standard primers CYA359F and CYA781R (25). The 16S rDNA primers that specifically targeted P. hollandica (16S-Pholl-fw [5′-ACA CAG CTT AAC TGT GGG AGA-3′] and 16S-Pholl-rev [5′-AGT TGG CTG CTC TTT GTC CCT-3′]) were based on the alignment obtained with the software BIOEDIT (14). Besides the exact match with P. hollandica, the primer sequences are identical to sequences of the unidentified cyanobacterial clones LD16 and LD22 (accession no. AJ007866 and AJ006285) from the Loosdrecht lakes. These clones exhibit more than 99% identity to P. hollandica (44). All primers were used at an annealing temperature of 55°C. The pcbB gene was obtained by first using primers optisi-fw (5′-AAD TAY GAH TGG TGG GC-3′) and hlwha-l-rev (5′-GCG TGC CAS AGR TGA CC-3′) and then using reverse primer pyfadt-rev (5′-CGT TTC GGC AAA RTA RGG-3′) in a second seminested PCR. Specific amplification of the pcbC gene was performed with primers pcbC-fw (5′-GTA ATA TCC GCC TCG TAG AC-3′) and pcbC-rev (5′-CTA ACC GTC AGA CCT TAA CC-3′). The PCR program and all other procedures have been described previously (13). Sequencing of cloned fragments was done at least in triplicate by using a capillary sequencer (Beckman-Coulter).
Computer analysis.
We searched for sequence similarities in databases with the assistance of the BLAST software (1). Sequence alignment was performed with the software BIOEDIT (14). A consensus tree was constructed by using the multiple-sequence alignment and the software program PAUP (Phylogenetic Analysis Using Parsimony, beta version 4.0; David Swofford, Laboratory of Molecular Systematics, Smithsonian Institution). Rooted cladograms were constructed after phylogenetic analyses of the 16S rDNA sequences (maximum-parsimony method) and of the pcbB and pcbC sequences (neighbor-joining method) with 1,000 replications. The cladograms were constructed with TreeView (version 1.5; R. D. M. Page, 1998).
Cultivation and high-performance liquid chromatography (HPLC) analysis.
Estuarine phytoplankton were enriched by cultivating 1 ml of the natural phytoplankton assemblage in 3 ml of BG11 medium (32) at 23°C. The enrichment cultures were exposed to daylight.
P. hollandica SAG 10.89 cells were grown at 23°C in plates containing 15 ml of BG11 medium (32) supplemented with different concentrations of NaCl with constant illumination of 9 μmol of photons m−2 s−1 (photosynthetically active radiation; fluorescent tubes; color code 25; Philips). The NaCl concentrations used were 34.2 mM (2 PSU), 68.4 mM (4 PSU), and 102.6 mM (6 PSU). Cells were harvested after 10 days by filtration onto membrane filters (pore size, 2 μm) for determination of the net dry weight and compatible solutes. For analysis of compatible solutes by HPLC, the cells on filters were extracted in 2 ml of 80% ethanol for 3 h at 65°C. After an internal standard (50 μg of sorbitol) was added, the particulate material was removed by centrifugation, and the supernatant was dried in a Speed-Vac. Prepurification of low-molecular-weight carbohydrates was performed by sequentially dissolving the dried supernatants in different solvents (0.5 ml of deionized water, 0.5 ml of 100% ethanol, 100 μl of HPLC grade water). The concentrations of low-molecular-weight carbohydrates were analyzed with an HPLC setup as described by Schoor et al. (35). All chromatographic experiments were performed with a chromatograph consisting of LC-9A pumps, SIL-9A autoinjector (1- to 50-μl sample loop), a CTO-6A column oven, a DGU-4A solvent degasser, and a RID-6A refractive index detector (Shimadzu Corp., Kyoto, Japan). A reverse-phase column filled with Hypersil 120 ODS and a sugar-alcohol column (Aminex HPX-87C; Bio-Rad) were the columns used.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rDNA of P. hollandica (accession no. AJ007907), Leptolyngbya sp. strain PCC 7104 (AB039012), Thermosynechococcus elongatus BP-1 (AP005376), Prochlorococcus marinus SSW5 (X63140), Prochloron sp. (X63141), uncultured cyanobacterial clones LD7 (AJ007864) and LD16 (AJ007866), Escherichia coli PK3 (X80731), Gloeobacter violaceus PCC 8105 (AF132791), Prochlorococcus marinus subsp. pastoris NATL2 (AF311219), Prochlorococcus marinus MIT 9303 (AF001469), Planktothrix agardhii NIVA CYA59 (AB045939), Synechocystis sp. strain PCC 6803 (AB041938), Synechococcus sp. strain PCC 7002 (AJ000716), Prochlorococcus marinus (X63140), Nostoc sp. strain PCC 7120 (AP003595), Synechococcus sp. strain WH 7805 (AF001478), and Trichodesmium erythreum IMS 101 (NZ_AAAU01000054) and the nucleotide sequences of the pcbBC genes of P. hollandica (X97043) and Prochlorococcus marinus CCMP 1375 (AF198526 and AF198528) were obtained from databases. Partial 16S rDNA and pcbC gene sequences of Prochlorothrix sp. strain NIVA-8/90 (accession no. AJ534944 and AJ534947) and of the uncultured estuarine Prochlorothrix (AJ534945 and AJ534946) were obtained in this study and were deposited in the databases.
RESULTS
Detection of pcb-like and 16S rDNA sequences in samples from the Darss-Zingst estuary.
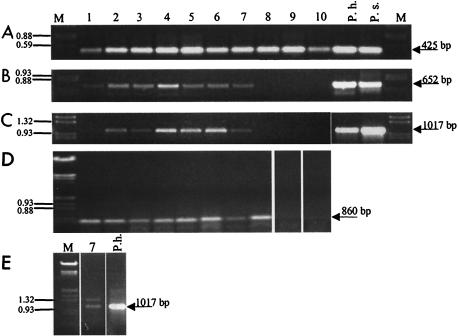
Gene fragments that exhibited high levels of sequence similarity (99.9%) to pcbB from P. hollandica were amplified by RT-PCR from sampling sites 1 to 9 (Fig. 2D). Until now, this gene has been described only for P. hollandica and Prochlorococcus marinus. In order to verify that Prochlorothrix-like organisms are present in the Darss-Zingst estuary, specific primers for amplification of the pcbC gene of P. hollandica were used in RT-PCR, as well as in PCR. Use of these pcbC-specific primers resulted in amplification of the expected 1-kb fragment with DNA from sampling sites 2 to 7 (Fig. 2C). Fragments of the same size were also obtained with cDNA, which were synthesized from total RNA from sampling sites 1, 3, 6, 7, and 9 (Fig. 2E). The similarities of the DNA fragments obtained to pcbC of P. hollandica were verified by Southern hybridization experiments, in which the fragments were recognized by a specific pcbC probe obtained from P. hollandica (data not shown). Restriction analyses with NcoI resulted in a fragment size pattern which was identical to that expected based on the P. hollandica pcbC sequence (data not shown). Several PCR fragments from all sampling sites were cloned and sequenced. All of the sequences were 99% identical to pcbC from P. hollandica regardless of the sampling site; i.e., just one genotype was found.
FIG. 2.
Separation of PCR fragments obtained with DNA (A to C) or cDNA (D and E) from 10 sampling sites. The lane numbers indicate the sampling sites (Fig. 1). The following different primer pairs were used on the DNA level: degenerate cyanobacterial 16S rDNA primers CYA359F and CYA781R (A), Prochlorothrix-specific 16S rDNA primers 16S-Pholl-fw and 16S-Pholl-rev (B), and Prochlorothrix-specific pcbC primers pcbCfw and pcbCrev (C). On the cDNA level, degenerate primers optisi-fw, hlwha-l-rev, and pyfadt-rev for pcbB amplification in nested PCR (D) and Prochlorothrix-specific pcbC primers pcbCfw and pcbCrev (E) were used. DNA from cultures of P. hollandica (P.h.) and P. scandica (P.s.) served as positive controls. Lanes M contained a marker (EcoRI/HindIII-digested λ DNA).
Finally, we searched for P. hollandica-like 16S rDNA sequences in environmental samples (Fig. 2A). This was done initially with cyanobacterium-specific primers (25), which amplify a 425-bp internal 16S rDNA fragment. Restriction analyses and sequencing of at least 10 randomly obtained 16S rDNA clones never resulted in a sequence similar to that of P. hollandica. In all cases, the sequences were similar to those of phycobilisome-containing cyanobacteria (data not shown). Prochlorothrix-type organisms could not be detected by this approach. However, the use of P. hollandica-specific 16S rDNA primers allowed amplification of the expected 650-bp fragments. The sequences of these fragments exhibited about 99% identity to sequences of P. hollandica. In addition, significant amounts of 16S rDNA fragments characteristic of P. hollandica were detected in pcbC-positive samples (Fig. 2B). Significant amounts of P. hollandica-like fragments were not detected in DNA from sampling sites 9 and 10.
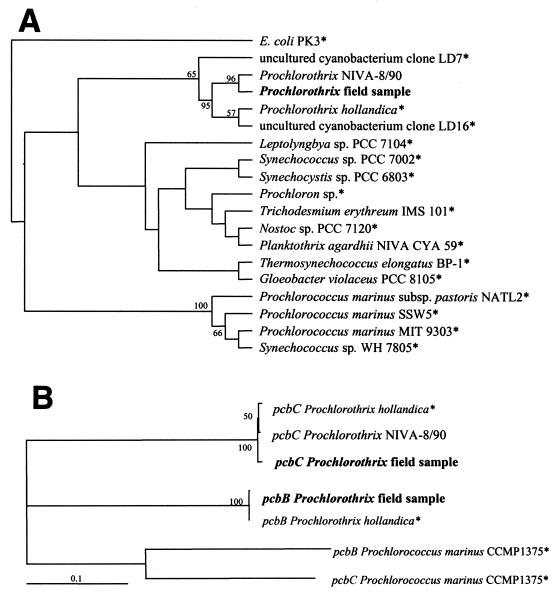
The partial sequences of the pcbC, pcbB, and 16S rDNA genes were used for phylogenetic comparisons with similar sequences from the databases (Fig. 3A). Corresponding sequences were also obtained from the proposed new Prochlorothrix species, P. scandica (strain NIVA-9/80) (29). The pcb gene sequences from the estuarine phytoplankton samples clustered closely with those of P. hollandica, while the sequences of Prochlorococcus marinus were clearly not closely related (Fig. 3B). The pcbC sequence of P. scandica NIVA-8/90 was similar to the sequence of P. hollandica. In spite of minor differences, the resulting amino acid sequences were 99% identical (Table 1). Thus, the pcbC sequences of the Darss-Zingst estuary clones, P. scandica, and P. hollandica are almost identical. Analyzing the partial 16S rDNA sequences again led to close grouping of the 16S rDNA sequences in the genus Prochlorothrix (Fig. 3A). According to this alignment, the 16S rDNA sequence of the environmental samples exhibited a slightly higher level of similarity to the sequence of P. scandica NIVA-8/90 than to the sequence of P. hollandica.
FIG. 3.
Rooted cladograms obtained after phylogenetic analyses by the maximum-parsimony method (performed with PAUP, beta version 4.0; Laboratory of Molecular Systematics, Smithsonian Institution) of a 527-bp fragment of the 16S rDNA sequences (maximum-parsimony method) (A) and of a 828-bp fragment of the pcbB and pcbC sequences (neighbor-joining method) (B). Organisms whose sequences were obtained from databases are indicated by asterisks. Selected bootstrap values based on 1,000 replications are shown at the nodes; only values greater than 50% are shown. The cladograms were constructed with TreeView (version 1.5; R. D. M. Page).
TABLE 1.
Levels of similarity of the DNA sequences from Prochlorothrix field samples to sequences obtained from databases and from analyses in this studya
| Organism | % Similarity
|
||
|---|---|---|---|
| 16S rDNA | pcbB | pcbC | |
| Prochlorothrix sp. strain NIVA-8/90 | 99.6 | 99.0 | |
| Prochlorothrix hollandicab | 98.9 | 99.9 | 99.3 |
| Prochloron sp.b | 90.5 | NDc | ND |
| Prochlorococcus marinus SSW5b | 91.3 | ND | ND |
| Prochlorococcus marinus CCMP 1375b | ND | 45.3 | 44.0 |
| Cyanobacterium clone LD7b | 94.9 | ND | ND |
| Cyanobacterium clone LD16b | 98.7 | ND | ND |
| Planktothrix agardhii NIVA CYA53b | 90.5 | ND | ND |
16S rDNA sequences (527 bp) and sequences of chlorophyll binding protein genes (pcbB and pcbC; 826 bp) were compared.
Sequence obtained from database.
ND, not determined.
Detection of Prochlorothrix-like trichomes in estuarine phytoplankton.
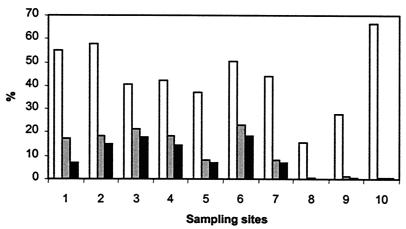
Phytoplankton counts from the 10 sampling sites (Fig. 4) revealed that about one-half of the members of the Oscillatoriales could be morphologically attributed to the microscopically indistinguishable taxa P. hollandica and Pseudoanabaena. This morphotype represented an important fraction of the phytoplankton biomass (mean, 12%) at stations 1 to 7, but for stations 8 to 10 the percentage was less than 1% (mean, 0.4%), which basically corresponds to the results of the PCR analyses.
FIG. 4.
Relative biomasses of Planktothrix-like filaments in different fractions of the phytoplankton in samples from the Darss-Zingst estuary. The percentages of biomass were calculated for Oscillatoriales (open bars), total cyanobacteria (grey bars), and total phytoplankton (solid bars).
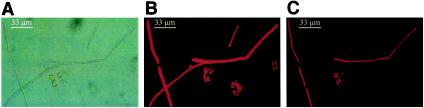
The molecular data strongly suggest that P. hollandica-like organisms are present in the Darss-Zingst estuary. This encouraged us to search for P. hollandica-like trichomes. Since the Darss-Zingst estuary is a highly eutrophic aquatic system, phytoplankton are abundant, and many filamentous cyanobacteria of the Planktothrix-Pseudoanabaena type are present (Fig. 5A). This fact made it impossible to detect P. hollandica trichomes in water samples reliably. Culture experiments to enrich mixtures of filamentous cyanobacteria were started. The cultures were examined after 2 weeks by light microscopy and differential epifluorescence microscopy (Fig. 5B and C). Some trichomes produced clear autofluorescence signals after selective Chlb-exciting illumination, in contrast to other trichomes that had the same morphology and to colony-forming chroococcal cyanobacteria. Some green algae proved that the Chlb excitation was specific. This finding supported the assumption that parts of the filamentous cyanoplankton contain Chlb, like Prochlorothrix filaments. Furthermore, PCR fragments from DNA extracted from the enrichment culture contained the same Prochlorothrix-like sequences as the environmental samples contained.
FIG. 5.
Micrographs of cyanobacteria and eukaryotic algae in enrichment cultures from the Darss-Zingst estuary after 14 days of incubation. (A) Bright-field microscopy. (B and C) Epifluorescence microscopy of autofluorescence with illumination that preferentially excited Chla (B) and Chlb (C).
Our data suggest that P. hollandica-like organisms are able to live throughout the estuary under low-salinity conditions, although such organisms have been isolated previously only from freshwater. P. hollandica SAG 10.89 was cultivated in NaCl-supplemented BG11 medium to mimic estuarine conditions. Growth was observed in media containing NaCl at salinities up to 6 PSU. In salt-treated cells, accumulation of sucrose was detected, while this sugar did not accumulate in P. hollandica cells under the low-salt conditions of standard BG11 medium (data not shown). These data indicate that sucrose might be used for osmoregulation, which can support acclimation to the salinities present in the Darss-Zingst estuary.
DISCUSSION
We corroborated initial indications that Prochlorothrix-like organisms are present in the natural phytoplankton community of coastal waters in the southern Baltic Sea. DNA fragments exhibiting high levels of sequence identity to pcbB, pcbC, or 16S rDNA of P. hollandica were amplified from DNA isolated from different sampling sites along a gradient of salinities ranging from 0.2 to 8.2 PSU. In particular, the pcbC sequence can be used as a specific marker for P. hollandica-like strains, since this gene is different from other pcb genes in oxyphotobacteria and from similar genes in eukaryotic algae (41). Furthermore, P. hollandica-like trichomes were enriched from the estuary and produced autofluorescence signatures that indicated that Chlb was present. Assuming that mRNA is a useful marker of cell viability because of its short half-life compared to that of DNA, Prochlorothrix-like cells are able to propagate at least in the enclosed parts of the Darss-Zingst estuary. Horizontal drifting of buoyant phytoplankton is caused by net horizontal water currents with velocities that are usually in the range from 0.1 to 0.5 m s−1 (34) (broad and narrow parts). Therefore, 30 to 150 h is the minimum drifting time from sampling site 1 to site 10 in the estuary. These time estimates were obtained by excluding mixing in the larger basins and periods of salt water inflow with inverted drift direction. Addition of Prochlorothrix originating from freshwater input is unlikely, since pcb mRNA half-lives were shown to be in the range from minutes (in light) to hours (in darkness) (24).
The occurrence of viable Prochlorothrix at salinities up to 8 PSU was initially surprising, because Burger-Wiersma et al. (5) characterized P. hollandica as a freshwater organism with a very low salt tolerance. Growth of P. hollandica ceased completely in the presence of 100 mM NaCl (17% of the seawater concentration or 6 PSU) and was inhibited in the presence of >25 mM NaCl (ca. 1.5 PSU). In contrast, our growth experiments with P. hollandica SAG 10.89 did not show growth inhibition at such salinities, and optimum growth was detected in the presence of about 85 mM NaCl (14% of the seawater concentration or 5 PSU) under the conditions used in this study. We assume that our cultivation method, which decreased shearing forces and other parameters that are not known, led to these differences in salt tolerance. Mechanical destruction of cell surfaces in particular might be critical for regulation of osmotic pressure by accumulation of compatible solutes and ion export. Cells of P. hollandica SAG 10.89 accumulated sucrose upon exposure to increased NaCl concentrations. Sucrose accumulation was found to be characteristic of the cyanobacterial strains with the lowest halotolerance (31). The optimal growth conditions for P. hollandica in cultures at salinities of about 4 PSU parallel those expected for Prochlorothrix-like cells in the estuary. Even though pcbB and pcbC DNA fragments were found in the western to central part of the Darss-Zingst estuary at all sampling sites, undetectable amounts of these fragments in the northeastern transition zone to the open Baltic Sea (station 10) support the suggestion that the organisms are restricted to salinities below 10 PSU.
The high levels of similarity of the 16S rDNA and functional pcb sequences (more than 99%) put the Prochlorothrix field sample sequences into the Prochlorothrix cluster with the type strain of P. hollandica. Only a few additional isolates, like Prochlorothrix strain NIVA-8/90, provisionally named P. scandica (28), and some uncultured clones from Lake Loosdrecht, exhibited high levels of similarity to P. hollandica (44). Based on the pcbB and pcbC sequence comparisons, the levels of similarity are even higher. Therefore, we believe that the Prochlorothrix-like gene fragments found in the Darss-Zingst estuary in fact indicate that Prochlorothrix species are present. On the 16S rDNA level the Prochlorothrix representative from the Darss-Zingst estuary showed a slightly higher level similarity to P. scandica NIVA-8/90 from Lake Malaren. The pcbB-pcbC sequence comparison implied that there is a closer relationship to P. hollandica. Nevertheless, it should be considered how significant differences are when levels of 16S rDNA sequence identity in the range from 98.8 to 99.6% are used to distinguish different species. At this time we are reluctant to assign the sequences to different species, and generally we refer to P. hollandica. In the case of Prochlorococcus species, all comparisons of 16S rDNA sequence homologies showed high degrees of identity, but there are stable ecotypes with completely different genome sizes (16). Therefore, further investigations of the genus Prochlorothrix could reveal more detailed information.
Monthly phytoplankton monitoring in the estuary failed to detect Prochlorothrix because of its inconspicuous and somewhat unclear morphological features. It is necessary to distinguish this taxon from the frequently occurring, morphologically similar cyanobacteria (i.e., Pseudanabaena limnetica [Oscillatoria limnetica]). These organisms can be distinguished by some ultrastructural features (29) or by different pigment-dependent autofluorescence signatures (42). However, electron microscopy is too costly to be used as a tool to search for phytoplankton species. In addition, acclimation to nutrient depletion and irradiance could lead to a significantly changed pigment content of Pseudoanabaena or Planktothrix (Oscillatoria) species (26). Phycobilisome fluorescence is not a reliable tool for distinguishing filamentous cyanobacterium-like organisms in the phytoplankton (5, 43). We used differential Chla/b excitation of long-wavelength autofluorescence to differentiate Prochlorothrix from Chlb-free cyanobacteria. This positive indication allowed us to prove that Prochlorothrix was present. However, a heavily reduced Chlb antenna per cell also could lead to incorrect identification. Therefore, prechecking phytoplankton samples for the presence of Prochlorothrix-like DNA sequences with PCR can be a useful technique for reducing the risk of misinterpreting microscopic investigations of natural phytoplankton samples. Even in samples in which Prochlorothrix-like trichomes accounted for less than 1% of the total phytoplankton biomass (Fig. 4), Prochlorothrix could be detected by PCR. In addition, potential errors of microscopic investigation can be excluded completely when a set of Prochlorothrix-like filaments can be verified to be Prochlorothrix by using a PCR approach at the level of single trichomes. The latter method has already been successfully applied to cyanobacterial filaments (2, 17). Possibly, Prochlorothrix is much more widely distributed than currently expected. In fact, Prochlorothrix-like filaments were suspected to be components of cyanobacterial blooms in the Baltic Sea (Gulf of Finland, Baltic Proper) during monitoring cruises (http://meri.fimr.fi/Algaline/eng/EnAlgaline.nsf). Methods for the unambiguous identification of these organisms are necessary. Further investigations should lead to estimates of the ecological importance of Prochlorothrix in brackish phytoplankton.
Acknowledgments
We thank H. Schubert (Department of Aquatic Ecology, University of Rostock) for the setup used for differential epifluorescence microscopy. B. Brzezinka is acknowledged for her technical support. We are grateful to the crew of the Zingst Biological Station (University of Rostock) for their kind support of the field work. Cells of P. scandica were kindly provided by H. C. P. Matthijs (Department of Microbiology, University of Amsterdam). Cells of P. hollandica were kindly provided by M. Lorenz (Sammlung von Algenkulturen, University of Göttingen).
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, G. L., B. A. Handley, P. Vacharapiyasophon, J. R. Stevens, and P. K. Hayes. 2000. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (cyanobacteria) filaments in the Baltic Sea. Microbiology 146:2865-2875. [DOI] [PubMed] [Google Scholar]
- 3.Bullerjahn, G. S., and A. F. Post. 1993. The prochlorophytes: are they more than just chlorophyll a/b-containing cyanobacteria? Crit. Rev. Microbiol. 19:43-59. [DOI] [PubMed] [Google Scholar]
- 4.Burger-Wiersma, T., M. Veenhuis, H. J. Korthals, C. C. M. Van de Wiel, and L. R. Mur. 1986. A new prokaryote containing chlorophylls a and b. Nature 320:262-264. [Google Scholar]
- 5.Burger-Wiersma, T., L. J. Stal, and L. M. Mur. 1989. Prochlorothrix hollandica gen. nov., sp. nov., a filamentous oxygenic photoautotrophic prokaryote containing chlorophylls a and b: assignment to Prochlorotrichaceae fam. nov. and order Prochlorales Florenzano, Balloni, and Materassi 1986, with emendation of the ordinal description. Int. J. Syst. Bacteriol. 39:250-257. [Google Scholar]
- 6.Burger-Wiersma, T. 1991. Prochlorothrix hollandica—a filamentous prokaryotic species containing chlorophylls-a and chlorophylls-b. Arch. Hydrobiol. 92:555-558. [Google Scholar]
- 7.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. Waterbury, and N. Welschmeyer. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340-343. [Google Scholar]
- 8.DiTullio, G. R., D. A. Hutchins, and K. W. Bruland. 1993. Interaction of iron and major nutrients controls phytoplankton growth and species composition in the tropical North Pacific Ocean. Limnol. Oceanogr. 38:495-508. [Google Scholar]
- 9.Ducobu, H., J. Huisman, R. R. Jonker, and L. R. Mur. 1998. Competition between a prochlorophyte and a cyanobacterium under various phosphorus regimes: comparison with the Droop model. J. Phycol. 34:467-476. [Google Scholar]
- 10.Edler, L. 1979. Recommendations on methods for marine biological studies in the Baltic Sea. Phytoplankton and chlorophyll. The Baltic Marine Biologists Publication No. 5. Opulus Press, Uppsala, Sweden.
- 11.Garczarek, L., W. R. Hess, J. Holtzendorff, G. W. van der Staay, and F. Partensky. 2000. Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. USA 97:4098-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garczarek, L., G. W. M. van der Staay, W. R. Hess, F. Le Gall, and F. Partensky. 2001. Expression and phylogeny of the multiple antenna genes of the low-light-adapted strain Prochlorococcus marinus SS120 (oxyphotobacteria). Plant Mol. Biol. 46:683-693. [DOI] [PubMed] [Google Scholar]
- 13.Geiß, U., J. Vinnemeier, A. Kunert, I. Lindner, B. Gemmer, M. Lorenz, M. Hagemann, and A. Schoor. 2001. Detection of the isiA gene across cyanobacterial strains: potential for probing iron deficiency. Appl. Environ. Microbiol. 67:5247-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Heerkloß, R., and U. Vietinghoff. 1981. Biomasseäquivalente planktischer und benthischer Organismen in den Darß-Zingster Boddengewässern. Wiss. Z. Univ. Rostock 30:31-36. [Google Scholar]
- 16.Hess, W. R., G. Rocap, C. S. Ting, J. Larimer, S. Stilwagen, J. Lamerdin, and S. W. Chisholm. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth. Res. 70:53-71. [DOI] [PubMed] [Google Scholar]
- 17.Laamanen, M. J., M. F. Gugger, J. M. Lehtimaki, K. Haukka, and K. Sivonen. 2001. Diversity of toxic and nontoxic Nodularia isolates (cyanobacteria) and filaments from the Baltic Sea. Appl. Environ. Microbiol. 67:4638-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Roche, J., G. W. van der Staay, F. Partensky, A. Ducret, R. Aebersold, R. Li, S. S. Golden, R. G. Hiller, and B. R. Green. 1996. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. USA 93:15244-15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewin, R. A. 1977. Prochloron, type genus of the Prochlorophyta. Phycologia 16:217. [Google Scholar]
- 20.Lund, W. S., C. Kipling, and E. D. LeCren. 1958. The inverted microscope method of estimating algal numbers and the statistical basis of estimations by counting. Hydrobiologia 11:144-170. [Google Scholar]
- 21.Mann, E. L., and S. W. Chisholm. 2000. Iron limits the cell division rate of Prochlorococcus in the eastern equatorial Pacific. Limnol. Oceanogr. 45:1067-1076. [Google Scholar]
- 22.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 23.Morden, C. W., and S. S. Golden. 1989. PsbA genes indicate common ancestry of prochlorophytes and chloroplasts. Nature 337:382-385. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaitchik, O. A., and G. S. Bullerjahn. 1998. Transcript analysis of the pcbABC genes encoding the antenna apoproteins in the photosynthetic prokaryote, Prochlorothrix hollandica. FEMS Microbiol. Lett. 168:187-194. [DOI] [PubMed] [Google Scholar]
- 25.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojala, A. 1993. The influence of light quality on growth and phycobiliprotein/chlorophyll a fluorescence quotients of some species of freshwater algae in culture. Phycologia 32:22-28. [Google Scholar]
- 27.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinevich, A. V., O. M. Skulberg, H. C. P. Matthijs, H. Schubert, E. Willen, O. V. Gavrilova, and N. V. Velichko. 1999. Characterization of a novel chlorophyll b-containing Prochlorothrix species (Prochlorophyta) and its photosynthetic apparatus. Microbios 100:159-174. [Google Scholar]
- 29.Pinevich, A. V., H. C. P. Matthijs, O. V. Gavrilova, S. G. Averina, and N. V. Velichko. 1996. New ultrastructural aspects of membranes and cell inclusions in Prochlorothrix hollandica (Prochlorales, Cyanobacteria). Microbios 87:217-225. [Google Scholar]
- 30.Post, A. F., and B. Arieli. 1997. Photosynthesis of Prochlorothrix hollandica under sulfide-rich anoxic conditions. Appl. Environ. Microbiol. 63:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, R. H., L. J. Borowitzka, M. A. Mackay, J. A. Chudek, R. Foster, S. R. C. Warr, D. J. Moore, and W. D. P. Stewart. 1986. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol. Rev. 39:51-56. [Google Scholar]
- 32.Rippka, R., J. Deruelles J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-16. [Google Scholar]
- 33.Scanlan, D. J., and N. J. West. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 34.Schlungbaum, G., H. Baudler, and C. Neumann. 1995. Langzeitreihen zur Beschaffenheitsentwicklung in den Gewässern der Darß-Zingster Boddenkette. Dtsch. Hydrogr. Z. Suppl. 5:183-196. [Google Scholar]
- 35.Schoor, A., N. Erdmann, U. Effmert, and S. Mikkat. 1995. Determination of the cyanobacterial osmolyte glucosylglycerol by high-performance liquid chromatography. J. Chromatogr. A 704:89-97. [Google Scholar]
- 36.Schubert, H., and R. Schumann. 2002. Epifluorescence microscopy: identification of higher taxa of phytoplankton, p. 95-108. In S. Rao (ed.), Pelagic ecology methodology. Balkema Publishers, Tokyo, Japan.
- 37.Tomitani, A., K. Okada, H. Miyashita, H. C. Matthijs, T. Ohno, and A. Tanaka. 1999. Chlorophyll b and phycobilins in the common ancestor of cyanobacteria and chloroplasts. Nature 400:159-162. [DOI] [PubMed] [Google Scholar]
- 38.Turner, S., T. Burger-Wiersma, S. J. Giovannoni, L. R. Mur, and N. R. Pace. 1989. The relationship of a prochlorophyte, Prochlorothrix hollandica, to green chloroplasts. Nature 337:380-382. [DOI] [PubMed] [Google Scholar]
- 39.Urbach, E., D. J. Scanlan, D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 1998. Rapid diversification of marine picoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46:188-201. [DOI] [PubMed] [Google Scholar]
- 40.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Int. Ver. Limnol. 9:1-38. [Google Scholar]
- 41.van der Staay, G. W., N. Yurkova, and B. R. Green. 1998. The 38 kDa chlorophyll a/b protein of the prokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene. Plant Mol. Biol. 36:709-716. [DOI] [PubMed] [Google Scholar]
- 42.van Hannen, E. J., G. Zwart, M. P. van Agterveld, H. J. Gons, J. Ebert, and H. J. Laanbroek. 1999. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl. Environ. Microbiol. 65:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Liere, L., L. Breebaart, and Y. J. Dullemont. 1989. Determining the relative number of prochlorophytes in lake phytoplankton using epifluorescence microscopy. Br. J. Phycol. 24:391-394. [Google Scholar]
- 44.Zwart, G., W. D. Hiorns, B. A. Methé, M. P. van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]