Abstract
The identification of phylogenetic clusters of bacteria that are common in freshwater has provided a basis for probe design to target important freshwater groups. We present a set of 16S ribosomal RNA gene-based oligonucleotide probes specific for 15 of these freshwater clusters. The probes were applied in reverse line blot hybridization, a simple method that enables the rapid screening of PCR products from many samples against an array of probes. The optimized assay was made stringent to discriminate at approximately the single-mismatch level. This made 10 of the probes highly specific, with at least two mismatches to the closest noncluster member in the global database. Screening of PCR products from bacterioplankton of 81 diverse lakes from Belgium, The Netherlands, Denmark, Sweden, and Norway showed that the respective probes were reactive against 5 to 100% of the lake samples. Positive reactivity of six highly specific probes showed that bacteria from actinobacterial clusters ACK-M1 and Sta2-30 and from verrucomicrobial cluster CLO-14 occurred in at least 90% of the investigated lakes. Furthermore, bacteria from alpha-proteobacterial cluster LD12 (closely related to the marine SAR11 cluster), beta-proteobacterial cluster LD28 and cyanobacterial cluster Synechococcus 6b occurred in more than 70% of the lakes. Reverse line blot hybridization is a new tool in microbial ecology that will facilitate research on distribution and habitat specificity of target species at relatively low costs.
The introduction of molecular methods into microbial ecology and the selection of rRNA genes as tools for bacterial classification (28, 29, 35, 36) initiated a massive effort to describe bacterial diversity in the environment (15). Random cloning of PCR-amplified genes, the most common procedure for describing diversity, has added thousands of sequences of 16S rRNA genes to the nucleotide databases. These data, which could not have been retrieved through cultivation of bacteria (3, 15), are gradually generating a better and more complete view of composition and diversity in several habitats. For instance, it is now clear that both ocean and inland waters have limited bacterial diversity. That is, the evenness in these aquatic systems is relatively low because a few groups of bacteria dominate the systems (10, 12, 13, 23, 24, 39). Moreover, these dominating groups appear to be widespread over the globe. For freshwater, we recently analyzed all of the available 16S rRNA gene sequences from clone libraries and identified 34 phylogenetic groups that appeared in more than one data set from different lakes or rivers (39). The identification of relatively narrow groups such as these (91 to 99% sequence similarity) creates a starting point for exploration of the functions and ecology of bacteria and allows the design of probes for rapid screening and comparison of bacterial communities in many different freshwater ecosystems.
In the present study we describe molecular probes targeting 16S rRNA genes from 15 of the freshwater clusters and explain how these probes are used in reverse line blot hybridization (16), a method new to microbial ecology. We also present results from a study of 81 North and West European lakes in which we used reverse line blot hybridization with these probes to demonstrate the ubiquity of a number of freshwater bacterial groups.
MATERIALS AND METHODS
Lake samples.
Six Dutch, 28 Belgian, and 32 Danish shallow lakes (<5 m depth) were sampled in 2000 or 2001 within the framework of the EU project BIOMAN. Characteristics of these diverse lakes can be found at http://www.nioo.knaw.nl/cl/me/. Depth-integrated pelagic lake samples were separated into two size fractions through filtration (pore sizes larger and smaller than 20 μm). After prefiltration through a 125-μm-pore-size nylon net, the larger fraction was collected on a 20-μm-pore-size nylon net and flushed from this net (using 0.2-μm-pore-size filtered lake water) to be collected on a 5-μm-pore-size MF-Millipore MCE filter (Millipore, Bedford, Mass.). The smaller fraction was obtained by collecting the filtrate from the 20-μm-pore-size filtration onto a 0.2-μm-pore-size MF-Millipore MCE filter. The filters were stored at −80°C until further processing. The collection of depth-integrated epilimnetic samples from 10 lakes in Central-East and North Sweden and four Norwegian lakes from Svalbard (Spitsbergen) has been described previously (20-22). In addition, a depth-integrated epilimnetic sample was collected from Lake Ånnsjön, an oligotrophic clear water lake in Central-West Sweden. The Swedish and Norwegian lakes are diverse in many parameters, including trophic status (from oligotrophic to eutrophic) and depth (from 1- to 64-m maximum depth). The samples were collected either in July 1996 or in July 1997. Bacterioplankton was collected on a 0.2-μm-pore-size filter after prefiltration through a 1-μm-pore-size glass fiber filter.
DNA isolation.
For DNA isolation from BIOMAN samples, 0.5 g of zirconium beads (0.1 mm in diameter), 0.5 ml of TE buffer (10 mM Tris [pH 7.6], 1 mM EDTA), and 0.5 ml of buffer-saturated phenol (pH 7 to 8) were added to the tubes containing a quarter filter with collected cells. The tubes were vigorously shaken (5,000 rpm) on a Mini-Beadbeater (Biospec Products, Bartlesville, Okla.) for 90 s with intermittent cooling on ice and then centrifuged for 5 min at 10,000 × g. The upper, aqueous phase was collected and extracted once with phenol-chloroform-isoamyl alcohol (25:24:1). The DNA was precipitated by adding 1/10 volume of 3 M sodium acetate (pH 5) and 2 volumes of 96% (vol/vol) ethanol, followed by incubation overnight at −20°C and centrifugation for 30 min at 14,000 × g. Subsequently, the DNA was dissolved in 100 μl of water and purified on a Wizard column (Promega, Madison, Wis.). For the BIOMAN samples, 20-μl aliquots of DNA solution from six monthly samplings (May to October) were combined, and this 120 μl was purified on a Wizard column to obtain a summer season-integrated DNA sample for each lake.
DNA isolation from the Swedish and Norwegian samples was performed by lysing the cells in sodium dodecyl sulfate (SDS) and extracting with phenol-chloroform-isoamyl alcohol, followed by purification using Sephadex spin columns, as described previously (19).
PCR.
We tested combinations of two different 5′-biotinylated forward primers (biot-F27 [5′-biot-AGA GTT TGA TCM TGG CTC AG-3′] and biot-F357 [5′-biot-CCT ACG GGA GGC AGC AG-3′]) and four different reverse primers (R518 [5′-ATT ACC GCG GCT GCT GG-3′], R907 [5′-CCG TCA ATT CCT TTG AGT TT-3′], R1221 [5′-CAT TGT AGC ACG TGT GTA GCC-3′], and R1492 [5′-GRT ACC TTG TTA CGA CTT-3′]). The numbers in primer names refer to the Escherichia coli 16S rRNA gene position (rrnB operon) corresponding to the 3′ end of the primers. All six primers match most bacterial 16S rRNA gene sequences. PCR amplification was performed by using an MBS 0.5S thermocycler (ThermoHybaid, Ashford, United Kingdom) in a 50-μl reaction mixture containing ca. 100 ng of purified DNA, 20 μg of bovine serum albumin (New England Biolabs, Beverly, Mass.), 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 200 μM concentrations of each deoxynucleotide, 1.5 mM MgCl2, 2.5 U of recombinant Taq DNA polymerase (Invitrogen, Merelbeke, Belgium), and 0.5 μM concentrations of each primer. After preincubation at 94°C for 5 min, samples were subjected to 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. After the last cycle, the temperature was kept at 72°C for another 5 min and then brought to 4°C.
PCR products were analyzed through ethidium bromide staining in agarose gel. Quantification was done through analysis of digital images of a dilution series of PCR products in comparison to two dilutions of the Smartladder quantification standard (Eurogentec, Seraing, Belgium) by using the software package Phoretics 1D Advanced (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom).
Reverse line blot hybridization.
In reverse blot hybridization, probes are bound to a membrane and the target DNA is applied in solution (8, 26). Reverse line blot hybridization is a modification introduced by Kaufhold et al. (16) in which the probes are applied as lines instead of dots. Hybridization with labeled target DNA in lines perpendicular to the probe lines enables simultaneous testing of several samples to several probes. A detailed protocol is available at http://www.nioo.knaw.nl/cl/me/.
(i) Membrane preparation.
For covalent linkage of probes, a Biodyne C membrane (Pall Europe, Ltd., Portsmouth, United Kingdom) was activated by incubation for 10 min in a rolling bottle at room temperature in 16% 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide (Sigma-Aldrich, St. Louis, Mo.), rinsed with tap water, and placed on a PC200 support cushion (Immunetics, Cambridge, Mass.) in a Miniblotter 45 manifold (Immunetics), with screws handtightened. Slots were filled with 150 μl of probe solution consisting of 125 to 5,000 pmol of probe (Table 1) in 500 mM NaHCO3 (pH 8.4). Probes were allowed to bind at room temperature for 5 min, after which the probe solution was aspirated from the slots. The blot was removed from the manifold by using forceps and incubated for 8 min at room temperature in 100 mM NaOH in a rolling bottle to inactivate the membrane. Then, the membrane was shaken gently in 200 ml of 2× SSPE-0.1% SDS at 60°C for 5 min (SSPE is composed of 18 mM NaCl, 1 mM NaH2PO4, and 0.1 mM NaEDTA [pH 7.4]). For storage, the prepared membrane was washed in 200 ml of 20 mM sodium EDTA (pH 8) at room temperature for 15 min, sealed in seal foil (Audion Electro, Weesp, The Netherlands) to avoid dehydration, and kept at 4°C until use.
TABLE 1.
Sequences and characteristics of probes
| Probe | Cluster | (Sub)divisiona | Probe sequence | E. coli position | Tmb (°C) | Amt appliedc (pmol) | No. of nongroup sequences with single mismatchd |
|---|---|---|---|---|---|---|---|
| LD12-143 | LD12 | Alpha-P | TTT CCC AAT GTT GTC CTG AA | 143-162 | 59 | 312.5 | 0 |
| Rho-BAL47-396 | Rhodoferax sp. strain BAL47 | Beta-P | TCC TGC ACG CGG AAT G | 396-411 | 61 | 2,500 | 474 |
| Rpickettii-490 | Ralstonia pickettii | Beta-P | AGC CGG TCC TTA TTC TTC C | 490-508 | 58 | 1,250 | 131 |
| CR-FL23-464 | Polynucleobacter necessariuse | Beta-P | CCG TCA TTC ACA GCT GTG TTA | 464-485 | 60 | 2,500 | 0 |
| LD28-451 | LD28 | Beta-P | TCA TGA TTT ATT CGA TCA TAA GTT TT | 451-476 | 58 | 1,250 | 0 |
| GKS98-442 | GKS98 | Beta-P | TGT GCG ATT TCT TTC CTG C | 442-460 | 60 | 1,250 | 0 |
| Mpsychro-211 | Methylobacter psychrophilus | Gamma-P | TCA TCT CAT AGC GAA AGG TCC | 211-231 | 59 | 1,250 | 7 |
| ACK-M1-193 | ACK-M1 | Actinobacteria | TCT TCA ACC GAA AAA CTT TCC | 193-228 | 58 | 125 | 0 |
| Sta230-187 | Sta2-30 | Actinobacteria | AAT TCT TTC AAA ACT CAG TGA TGC | 187-210 | 58 | 625 | 0 |
| Urk014-126 | Urk-014 | Actinobacteria | GAC CGG GGT TGG TTT CTC | 126-142 | 60 | 2,500 | 0 |
| CL0-14-464 | CL0-14 | Verrucomicrobia | GTA TGA GCC TGG CCA TTA GTC | 464-485 | 58 | 2,500 | 0 |
| FukuN18-221 | FukuN18 | Verrucomicrobia | CGC GAG CTC ATC AGG AAG | 221-238 | 61 | 2,500 | 0 |
| Syn/Pro-415 | Synechococcus cluster 6b | Cyanobacteria | GTT TAC AGC CCA GAG GCC T | 415-433 | 59 | 5,000 | 0 |
| Oagardhii-425 | Oscillatoria agardhii | Cyanobacteria | CTG AGA AAA GGG GTT GAC AAT | 425-445 | 58 | 2,500 | 1 |
| Aflosaquae-199 | Aphanizomenon flos-aquae | Cyanobacteria | TCT TCT TCA GGC AGC AAG C | 199-231 | 59 | 625 | 2 |
| Uni518 | Most organisms | GTA TTA CCG CGG CTG CTG G | 518-536 | 64 | 625 | NAf |
Alpha-P, Beta-P, and Gamma-P, alpha-, beta-, and gamma-proteobacteria, respectively.
Tm, melting temperature according to Breslauer et al. (6) at a 50 nM probe concentration and with 50 mM salt.
That is, the amounts of probes applied in slots, for chemical linkage to membrane, in 150 μl of 500 mM NaHCO3 (pH 8.4).
None of the probes has a perfect match to sequences that are not in the defined clusters (based on the GenBank, EMBL, and DDBJ databases).
Probe CR-FL23-464 matches to a subcluster of the P. necessarius cluster (see Fig. 2).
NA, not applicable.
(ii) Hybridization.
After incubation in 2× SSPE-0.1% SDS at room temperature for 5 min, the probe-carrying membrane was placed on the support cushion in the manifold, rotated 90° from the orientation of probe application. Each PCR product was denatured in 150 μl of 2× SSPE-0.1% SDS in a 0.5-ml microcentrifuge tube at 99°C for 10 min and immediately thereafter chilled on ice. All PCR products were then applied to the slots, and slots without PCR product were filled with 2× SSPE-0.1% SDS. In the final assay, we used 0.6 pmol of PCR product of natural sample per slot. Hybridization was performed in the manifold at 42°C for 60 min on a horizontal surface without agitation. The membrane was washed by two subsequent incubations in a rolling bottle (with mesh) in 200 ml of 2× SSPE-0.5% SDS at 52°C for 10 min (stringent wash).
(iii) Detection.
In the next step, the washed membrane was incubated in 10 ml of a 1:4,000 dilution of peroxidase-labeled streptavidin conjugate (Roche, Mannheim, Germany) in 2× SSPE-0.5% SDS, in a rolling bottle at 42°C, for 45 min. The membrane was then washed twice in 200 ml of 2× SSPE-0.5% SDS at 42°C for 10 min and rinsed twice in 200 ml of 2× SSPE at room temperature for 5 min. After incubation in 10 ml of BM chemiluminescence blotting substrate (Roche) for 1 min the membrane was covered with an overhead sheet and X-ray film for detection of chemiluminescence. The film was exposed 30 min for PCR products from control templates (16S rRNA gene clones) or overnight for natural samples. Signal strengths (pixel volumes) were recorded from the X-ray film by using a charge-coupled device camera and analyzed by using the software package Phoretics 1D Advanced (Nonlinear Dynamics).
Probe design.
The probes were designed by eye from the complete small subunit alignment downloaded from the European rRNA database (37), which we expanded with environmental 16S rRNA gene sequences from EMBL, GenBank, and DDBJ. Probes were chosen to match most members of the monophyletic freshwater clusters completely and have maximal mismatch to noncluster members with the help of the dedicated comparative sequence editor software (DCSE) (9). Selected probes (Table 1) were checked against all sequences in EMBL, GenBank, and DDBJ by using BLASTN 2.2.1 (2) included at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast/). All probes are antisense.
Control templates.
As positive controls, we used 16S rRNA gene plasmid clones, retrieved in earlier studies, that matched the probes exactly. Clones LD8 (accession number AJ006281), LD12 (Z99997), LD18 (AJ006284), and LD28 (Z99999) were described elsewhere (40). SY1-18 (AF107499) and Urk0-14 (AJ416173) were described by Zwart et al. (39). Columbia River clones CRE-FL38 (AF141459), CR-FL25 (AF141406), CR-PA50 (AF141431), CR-FL23 (AF141405), CR-PA40 (AF141428), CR-PA24 (AF141422), CRE-FL18 (AF141445), and CR-FL3 (AF141389) were described by Crump et al. (7). For probe Syn/Pro-415 we had no matching clone but instead used PCR product amplified from Synechococcus sp. strain Y0011 (AY183114), which had an exact match. To investigate hybridization to mismatching templates, we used 16S rRNA gene clones LD1 (AJ007870), LD2 (AJ007871), LD3 (AJ007872), LD17 (Z99998), LD21 (AJ007873) (40), Urk0-03 (AJ416170), Urk0-13 (AJ416172), Urk0-17 (AJ416174), Sta2-06 (AJ416202), Sta5-10 (AJ416258), Sta5-26 (AJ416268), Med0-02 (AJ416157), and Med0-06 (AJ416158) (39).
RESULTS
Selection of amplification product and probe design.
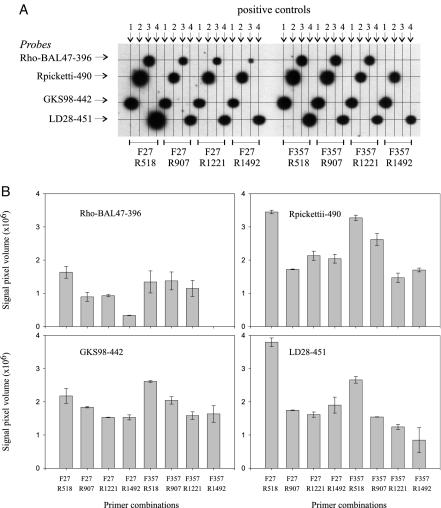
In order to seek an optimal 16S rRNA gene PCR product for hybridization reactions, we tested PCR products generated with eight different combinations of primer pairs on four oligonucleotide probes. For each probe, a different perfectly matching 16S rRNA gene template was used to generate PCR products of different lengths. All hybridizations were done in duplicate with 0.4 pmol of PCR product. Figure 1 shows that PCR products generated with primer combinations F27-R518, and F357-R518 generally had greater reactivity than PCR products generated with F27 or F357 in combination with R907, R1221, or R1492. The reactivity was highest for primer set F27-R518 for the probes Rho-BAL47-396, Rpickettii-490, and LD28-451, whereas primer set F357-R518 produced the strongest signal to probe GKS98-442. A possible explanation for the differences in reactivity may be sought in the lengths and positions of the PCR products. The relatively short PCR products from F357-R518 and F27-R518 (ca. 193 and 526 nucleotides, respectively) may have less opportunity for internal folding, when single stranded, than products generated with the reverse primers R907, R1221, or R1492. Such internal folding can interfere with probe binding. However, the product from primer set F357-R907 (ca. 585 nucleotides) is not much longer than the product from F27-R518 but reacts more weakly in all cases. Therefore, the position in the 16S rRNA gene may also be important for determining the stability of secondary structures of single-stranded DNA.
FIG. 1.
Reactivity of four probes against homologous PCR products of different lengths. (A) Digitized image of X-ray film. Membrane-bound probes horizontally, hybridized PCR products vertically. The positive control templates (perfect match) used were clone CR-PA24 (control 1) for GKS98-442, clone CR-PA50 (control 2) for Rpickettii-490, clone CRE-FL38 (control 3) for probe Rho-BAL47-396, and clone LD28 (control 4) for LD28-451. PCR products were generated from these templates by using eight different combinations of primers (indicated at the bottom). A total of 0.4 pmol of PCR product was used per slot. Grid lines were added to the image to indicate position of probes and PCR products. (B) Hybridization signals for homologous reactions. Signal strengths were measured by using Phoretics 1D Advanced and are expressed as pixel volume, which is the sum of all pixel values, corrected for background, within the signal area. Bars indicate average of two replicate reactions. Error bars indicate high and low values of the two replicates.
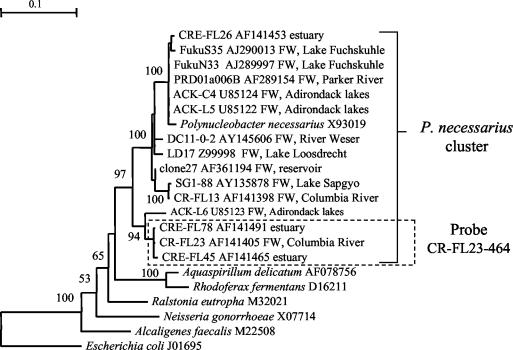
For the final assay format we chose the PCR product defined by primers F27 and R518 since it offers more opportunity to develop probes than the short sequence stretch defined by F357 and R518. Within the region from positions 27 to 518 (E. coli numbering), we designed probes to cover 15 of the 34 described freshwater clusters (39), aiming at maximal coverage within each cluster and maximal difference to taxa outside of the cluster (Table 1). The cluster targeted by probe CR-FL23-464 is a very narrow subcluster of the Polynucleobacter necessarius cluster as shown in Fig. 2. Phylogeny of the clusters not included in this tree can be inspected in the earlier study (39). Whereas 10 of the probes have at least two mismatches to all noncluster members (RDP probecheck and BLASTN at GenBank), 5 probes have single mismatches with a number of taxa outside of the cluster (Table 1).
FIG. 2.
Minimum evolution tree showing the phylogenetic position of the 16S rRNA sequences targeted by probe CR-FL23-464 within the P. necessarius cluster. The tree was constructed from 100 bootstrapped trees obtained through neighbor-joining analysis by using the program Treecon (33). The alignment used stretches from positions 59 to 470 (E. coli numbering). Not all bootstrap values are shown. FW, clone obtained from freshwater water column.
In order to use multiple probes in a single hybridization assay, we selected sequences with similar theoretical melting temperatures (58°C < Tm < 61°C; Table 1). As a control for sample reactivity, we used a universal probe Uni518, which matches most 16S rRNA gene sequences and has a higher Tm (64°C).
Testing the probes.
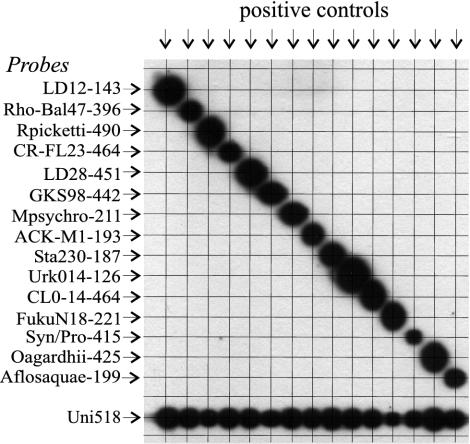
To test the efficiency of binding, we used PCR products from perfectly matching control DNA for each of the probes. Although theoretical melting temperatures were similar and equal amounts of control PCR products were used, homologous hybridization signals differed markedly between probes. To obtain similar reactivities, we adjusted the applied probe concentrations such that the respective positive controls yielded similar signals for the homologous reactions at the final conditions (Table 1 and Fig. 3). In all cases higher concentrations of probe yielded greater signal strengths (not shown). Low or high reactivity of the different probes could not be explained from their theoretical melting temperatures, illustrating that the behavior of probes bound to solid phase can deviate from the predicted behavior of probes in solution.
FIG. 3.
Reactivity of 15 probes against respective positive controls (see Materials and Methods). Arrows indicate the direction of application of probes (horizontally) and PCR products (vertically). A total of 0.2 pmol of PCR product (F27-R518) was used per slot.
Specificity.
Figure 3 also shows that there was no cross-reactivity between the probes. To further test the specificity of the assay, we used PCR products from cloned 16S rRNA genes that had one, two, or three mismatches with the probes (Table 2). No reactivity was observed to the tested templates with a single base insertion or deletion or two or three mismatches. A single mismatch prevented detection in two cases but not in the case of clone Sta5-26 that cross-reacted to probe Sta230-187. The fact that this G-T mismatch (G in the template sense strand and T in the probe) apparently does not prevent binding to the probe is in line with the findings of Allawi and SantaLucia (1). These authors concluded from nuclear magnetic resonance analyses of oligonucleotide duplexes that the effect of the G-T mismatch is context dependent. In the triplet AGA/TTT, the G-T mismatch contributes +1.05 kcal/mol to duplex free energy at 37°C and is therefore destabilizing to probe template binding. In the triplet CGC/GTG, as is found in the clone Sta5-26/probe Sta230-187 duplex, the G-T pair contributes −1.05 kcal/mol and thus is the most stable G-T mismatch.
TABLE 2.
Cross-reactivity in reverse line blot hybridization of probes to control templates with few mismatches
| Probe and clone | Target sequence | Cross- reactivityb |
|---|---|---|
| Probe Rho-BAL47-396 | CATTCCGCGTGCAGGA | |
| Clone URK0-13 | ---G------------ | − |
| Clone Sta5-10 | ---G------------ | − |
| Clone LD2 | ---C------------ | − |
| Clone Med0-02 | ---C------------ | − |
| Clone LD3 | ---G--A--------- | − |
| Clone LD21 | ---C-------A---- | − |
| Clone LD1 | ---C-------G---- | − |
| Clone URK0-17 | ---G--------G--- | − |
| Probe Rpickettii-490 | GGAAGAATAAGGACCGGCT | |
| Clone LD17 | T----------C------- | − |
| Clone LD3 | T------------T----- | − |
| Clone URK0-13 | -T-----------T----- | − |
| Probe CR-FL23-464a | TAACACAG•CTGTGAAUGACGG | |
| Clone LD17 | --------T------------- | − |
| Probe Sta230-187 | GCATCACTGAGTTTTGAAAGAATT | |
| Clone Sta5-26 | -----G------------------ | + |
| Probe Urk014-126 | GAGAAACCAACCCCGGTC | |
| Clone Sta2-06 | --------TG---T---- | − |
| Probe Syn/Pro-415 | AGGCCTCTGGGCTGTAAAC | |
| Clone LD6 | ------G----T------- | − |
| Clone URK0-03 | ------TC----------- | − |
| Clone MED0-06 | ------TC----------- | − |
Probe CR-FL23-464 and clone LD17 differ by an insertion or deletion as indicated by the bullet.
That is, the presence (+) or absence (−) of reactivity of probes to the mismatching templates (0.2 pmol of PCR product [F27-R518] per slot).
Analysis of lake samples.
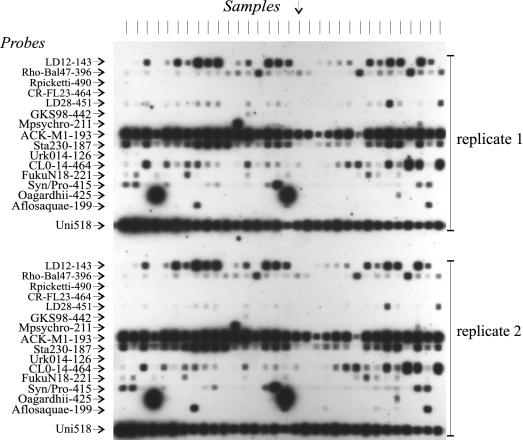
In order to test applicability of the assay on natural samples and to investigate the occurrence of bacteria represented in the probe panel, we tested samples from 81 European freshwater lakes. Figure 4 shows a typical blot in which we tested 32 Danish lakes (size fraction < 20 μm). The blot shows considerable differences in reactivity patterns between lakes but also shows widespread reactivity to some of the probes. Two lakes had such strong signals with the Oscillatoria agardhii probe (Oagardhii-425) that they overlapped neighboring reaction sites on the blotting membrane. These lakes were reported to have summer blooms of this cyanobacterial species (E. Jeppesen, unpublished data). The probe set was applied twice on the same blot: once on the upper part and once on the lower part. In this way, in a single assay, each tested PCR product is hybridized in duplicate to each particular probe. Among 512 reactions (32 samples on 16 probes), 231 were positive in both replicates, whereas 11 weak signals were detectable in only one of the replicates (98% similarity).
FIG. 4.
Reactivity of probes to PCR products from 32 Danish lake samples. Probes were applied in duplicate. Arrows indicate the direction of application of probes (horizontally) and PCR products (vertically; 0.6 pmol per slot). PCR products (F27-R518) were obtained from pooled lake samples (fractions smaller than 20 μm).
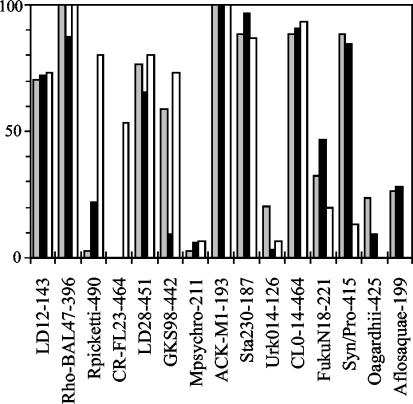
Figure 5 summarizes the results for all 81 lake samples. Five probes (LD12-143, Rho-BAL47-396, LD28-451, ACK-M1-193, Sta230-187, and Syn/Pro-415) represent bacterial groups present in >50% of the lakes tested in all regions. Of these, ACK-M1-193 generated signals in all of the 81 lakes. The probes that were reactive in only a few lakes were Mpsychro-211, CR-FL23-464, Urk014-126, and Oagardhii-425 with 4, 8, 9, and 11 positive lakes, respectively. Remarkably, all eight lakes that were positive for probe CR-FL23-464 were in Sweden.
FIG. 5.
Percentage of lakes positive to the probes in Denmark (n = 32, gray bars), Belgium and The Netherlands combined (n = 34, solid bars), and Sweden and Norway combined (n = 15, open bars). The Danish and Belgian-Dutch samples were scored positive if the cell fraction smaller than 20 μm and/or the fraction greater than 20 μm was positive.
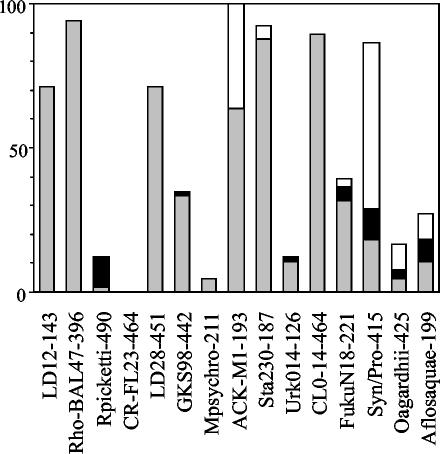
For the Danish, Belgian, and Dutch samples the numbers of positive lakes (Fig. 5) were calculated by combining results from the small and the large size fraction. Figure 6 shows differences between these size fractions. For nine probes, positive reactivity was only, or predominantly, found in the small size fraction. When a lake tested positive for the cyanobacterial probes Syn/Pro-415, Oagardhii-425, and Aflosaquae-199, it was frequently positive for these probes in both the small and the large size fractions. Lakes positive for probe Rpickettii-490 predominantly reacted to this probe in the large size fraction and not in the small size fraction.
FIG. 6.
Percentage of positive results in small- and large-size fractions. Danish, Belgian, and Dutch samples were evaluated together (n = 66). Bars: ░⃞, percentage of samples positive for small-size fraction (<20 μm); □, percentage of samples positive for large-size fraction (>20 μm); ▪, percentage of samples positive for both the small- and large-size fractions.
DISCUSSION
The number of deposited 16S rRNA gene sequences from different habitats is steadily growing, and it is becoming easier to design molecular probes for ecologically relevant organisms. Reverse line blot hybridization is a simple and rapid method in which such probes can be used to screen a large number of samples for known groups of bacteria. With the described blotting manifold, a single blot allows simultaneous hybridization of 45 samples to a maximum of 43 different probes. Membrane preparation, hybridization, and detection can be completed within a day. In addition, the blot with attached probes can be stored for months and reused at least 10 times without significant loss of signal by stripping templates between experiments. Taken together, the potential for simultaneous testing of multiple samples, the reusability of blots, and the use of equipment available to most molecular laboratories make this method compare favorably in cost and time efficiency to current microarray methods. The main advantage of microarrays over reverse line blot hybridization is the much larger number of probes that can be used. However, for many research questions in which the target organisms are known, an array of 43 different probes may be sufficient.
For the study of aquatic ecosystems, reverse line blot hybridization can be particularly useful because a relatively small number of bacterial groups dominate both the freshwater and marine water column (31). The identification of 34 phylogenetically narrow bacterial groups found in many freshwater ecosystems (39) provided a basis for the design of probes for 15 of these groups. The theoretical specificity of 10 probes is high, with at least two mismatches to any noncluster member. The specificity of the other five probes is lower because they have single mismatches to the closest noncluster members in the database. The stringency of the assay, achieved through the washing step at 52°C, makes it unlikely that sequences with two or more mismatches or sequences with insertion/deletions will hybridize to the probes, as apparent from the cross-reactivity tests (Table 2). Sequences with a single mismatch will be detected in some cases and not in others, depending on the probe sequence.
The assay in the present study was developed for characterization of bacterial communities in natural freshwater bodies. We therefore used the assay to survey the bacterial communities of 81 diverse lakes in North and West Europe. All of the probes were reactive to PCR products from a number of lakes, showing that all probes can detect bacteria in natural samples. Seven probes (LD12-143, RhoBAL47-396, LD28-451, ACK-M1-193, STA230-187, CL0-14-464, and Syn/Pro-415) were reactive to >70% of the 81 lakes. The results for one of these probes, RhoBal47-396, should be interpreted cautiously because many database sequences that are not related to the RhoBal47 cluster have a single mismatch to this probe. The other six probes have at least two mismatches to the nearest noncluster sequence, providing strong evidence that bacteria from the represented clusters are very common in North and West European lakes.
These six freshwater clusters were also commonly identified in random cloning studies of lakes and rivers in North America, Europe, and Asia (4, 5, 7, 12, 14, 27, 32, 39, 40). The actinobacterial cluster ACK-M1, detected in all of the 81 lakes with probe ACK-M1-193, was also present in 9 of 12 freshwater ecosystems from which a 16S ribosomal DNA sequence data set has been obtained (39). The cluster Sta2-30, which is a sister cluster of ACK-M1, was detected in 74 of the 81 lakes (91%) and was represented in 7 of the 12 freshwater sequence sets. The alpha-proteobacterial cluster LD12 was detected in 58 of 81 lakes (72%) and was represented in 8 of 12 sequence sets. The beta-proteobacterial cluster LD28, detected in 59 of the 81 lakes (73%), was represented in 6 of 12 sequence sets. The verrucomicrobial cluster CL0-14 and the cyanobacterial cluster Synechococcus 6b, detected in 90 and 73% of lakes, respectively, were each represented in 3 of 12 sequence sets. Clearly, both approaches, screening with probes and clone library construction, have detection limits and biases. Therefore, absence from a sequence library or the absence of reactivity to the probes does not evidence the absence of the targeted groups in the inspected ecosystems. However, for bacteria from these six phylogenetic clusters the two approaches independently support the wide geographic distribution and ubiquity in freshwater ecosystems.
The detection in two different size fractions for 66 BIOMAN samples allows evaluation of the microscale distribution of the investigated bacterial populations. Most targeted bacteria, predominantly occurred in the <20-μm size fraction (clusters LD12, Rhodoferax sp. strain BAL47, LD28, GKS98, Methylobacter psychrophilus, Sta2-30, Urk0-14, CL0-14, and FukuN18), indicating that bacteria from these clusters are either free-living or attached to particles smaller than 20 μm in diameter. In contrast, the three targeted cyanobacterial groups frequently occurred in the >20-μm fraction, as well as in the smaller fraction, probably due to their filamentous morphology and/or colony formation.
The probe Rpickettii-490 has a single mismatch to a number of noncluster members, allowing for possible misidentification. Still, the distribution in the size fractions is interesting, since in most positive samples, only the larger fraction was positive. One of the cultivated species targeted by the probe, Ralstonia pickettii, is occasionally found in clinical specimens (18, 38) and is known to survive in aqueous solutions, both distilled water and saline (17, 18). Other species from this cluster, such as Ralstonia solanacearum, are associated with plant disease (11, 30). It is possible that bacteria from this cluster were attached to plant or animal material in the positive lake samples, thus explaining their occurrence in the larger size fraction.
Assays such as the one presented here and microarray methods based on PCR are not suitable for obtaining accurate quantitative data. In addition to the known biases of PCR (25, 34), there are differences in the efficiency of probe binding that cannot yet be predicted reliably. In the development of the current assay, we found that different probes with similar theoretical melting temperatures, applied in equal concentrations, yielded considerable differences in the strengths of the homologous hybridization signals. Still, comparison of relative reactivities between samples obtained by using the same probe can provide indications of relative abundance. For the O. agardhii probe, very strong signals were measured in samples from lakes that had blooms of this cyanobacterial species. For more accurate quantification, other methods, such as in situ hybridization, should be used. Glöckner et al. performed fluorescence in situ hybridization with a probe that encompassed both of the actinobacterial clusters ACK-M1 and Sta2-30 (12). These authors showed that the supergroup (designated HCGI) containing both clusters was a dominant group (between 10 and 49% of DAPI [4′,6′-diamidino-2-phenylindole]-stainable cells) in Lake Baikal and Lake Gossenkölle (Austria) and Lake Fuchskuhle (Germany). Apparently, bacteria from this actinobacterial group, from which no cultivated representatives are known, are not only ubiquitous but also numerically important in diverse freshwater ecosystems.
Samples from the set of lakes analyzed here produced a variety of hybridization patterns with our panel of 15 probes. Therefore, although a few taxa are almost omnipresent, differentiation on the basis of probe reactivity was possible. Future research will have to point out which environmental factors differing between the lakes steer the occurrence of different bacterial groups. Microbial ecology in general faces the challenge of relating environmental conditions to community structure. With the increasing knowledge of relevant organisms in freshwater, oceans, soils, and other habitat types, reverse line blot hybridization can be a very useful tool for tackling this challenge.
Acknowledgments
We thank all water managers and lake owners involved in this project for their willing cooperation. We are indebted to Anneliese Ernst for providing DNA from cyanobacterial strain Y0011.
This work was supported by EC project BIOMAN EVK2-1999-00046.
Footnotes
Publication no. 3194 of NIOO-KNAW (Netherlands Institute of Ecology).
REFERENCES
- 1.Allawi, H. T., and J. SantaLucia. 1997. Thermodynamics and NMR of internal GT mismatches in DNA. Biochemistry 36:10581-10594. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1996. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 5.Bosshard, P. P., Y. Santini, D. Gruter, R. Stettler, and R. Bachofen. 2000. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol. Ecol. 31:173-182. [DOI] [PubMed] [Google Scholar]
- 6.Breslauer, K. J., R. Frank, H. Blöcker, and L. A. Marky. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83:3746-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dattagupta, N., P. M. M. Rae, E. D. Huguenel, E. Carlson, A. Lyga, J. A. Shapiro, and J. P. Albarella. 1989. Rapid identification of microorganisms by nucleic-acid hybridization after labeling the test sample. Anal. Biochem. 177:85-89. [DOI] [PubMed] [Google Scholar]
- 9.De Rijk, P., and R. De Wachter. 1993. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 9:735-740. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genin, S., and C. Boucher. 2002. Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol. Plant Pathol. 3:111-118. [DOI] [PubMed] [Google Scholar]
- 12.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagström, A., T. Pommier, F. Rohwer, K. Simu, W. Stolte, D. Svensson, and U. L. Zweifel. 2002. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiorns, W. D., B. A. Methé, S. A. NierzwickiBauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and nonradioactive allele-specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-25. [DOI] [PubMed] [Google Scholar]
- 17.Kulakov, L. A., M. B. McAlister, K. L. Ogden, M. J. Larkin, and J. F. O'Hanlon. 2002. Analysis of bacteria contaminating ultrapure water in industrial systems. Appl. Environ. Microbiol. 68:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labarca, J. A., W. E. Trick, C. L. Peterson, L. A. Carson, S. C. Holt, M. J. Arduino, M. Meylan, L. Mascola, and W. R. Jarvis. 1999. A multistate nosocomial outbreak of Ralstonia pickettii colonization associated with an intrinsically contaminated respiratory care solution. Clin. Infect. Dis. 29:1281-1286. [DOI] [PubMed] [Google Scholar]
- 19.Lindström, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 20.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 21.Lindström, E. S. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598-605. [DOI] [PubMed] [Google Scholar]
- 22.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 23.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 24.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 25.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saiki, R. K., P. S. Walsh, C. H. Levenson, and H. A. Erlich. 1989. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 86:6230-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenova, E. A., and K. D. Kuznedelov. 1998. A study of the biodiversity of Baikal picoplankton by comparative analysis of 16S rRNA gene 5′-terminal regions. Mol. Biol. 32:754-760. [PubMed] [Google Scholar]
- 28.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1984. The analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science 224:409-411. [DOI] [PubMed] [Google Scholar]
- 29.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S ribosomal RNA sequences. Appl. Environ. Microbiol. 49:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumithra, K. U., M. Krishnappa, T. K. Vasanth, H. S. Shetty, C. N. Mortensen, and S. B. Mathur. 2000. Seed-borne nature of Ralstonia solanacearum in eggplant (Solanum melongena L.) cultivars in India. Seed Sci. Technol. 28:291-299. [Google Scholar]
- 31.Torsvik, V., L. Øvreås, and T. F. Thingstad. 2002. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 32.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 33.Van de Peer, Y., and R. De Wachter. 1994. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 34.Walsh, P. S., H. A. Erlich, and R. Higuchi. 1992. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1:241-250. [DOI] [PubMed] [Google Scholar]
- 35.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuyts, J., Y. Van de Peer, T. Winkelmans, and R. De Wachter. 2002. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 30:183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoneyama, A., H. Yano, S. Hitomi, K. Okuzumi, R. Suzuki, and S. Kimura. 2000. Ralstonia pickettii colonization of patients in an obstetric ward caused by a contaminated irrigation system. J. Hosp. Infect. 46:79-80. [DOI] [PubMed] [Google Scholar]
- 39.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 40.Zwart, G., W. D. Hiorns, B. A. Methé, M. P. Van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]