Abstract
During bacterial degradation of methoxylated lignin monomers, such as vanillin and vanillic acid, formaldehyde is released through the reaction catalyzed by vanillic acid demethylase. When Burkholderia cepacia TM1 was grown on vanillin or vanillic acid as the sole carbon source, the enzymes 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI) were induced. These enzymes were also expressed during growth on Luria-Bertani medium containing formaldehyde. To understand the roles of these enzymes, the hps and phi genes from a methylotrophic bacterium, Methylomonas aminofaciens 77a, were introduced into B. cepacia TM1. The transformant strain constitutively expressed the genes for HPS and PHI, and these activities were two- or threefold higher than the activities in the wild strain. Incorporation of [14C]formaldehyde into the cell constituents was increased by overexpression of the genes. Furthermore, the degradation of vanillic acid and the growth yield were significantly improved at a high concentration of vanillic acid (60 mM) in the transformant strain. These results suggest that HPS and PHI play significant roles in the detoxification and assimilation of formaldehyde. This is the first report that enhancement of the HPS/PHI pathway could improve the degradation of vanillic acid in nonmethylotrophic bacteria.
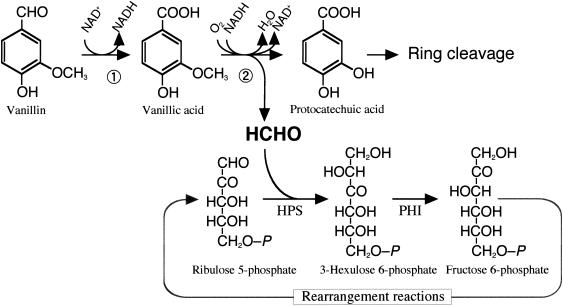
Lignin is the most abundant aromatic compound in the biosphere. Therefore, there have been various studies of its use. Chemical degradation of lignin produces a high yield of vanillin (8), which is an important starting material for the production of various flavors and fragrances (13). The bacterial pathway for vanillin degradation has been extensively investigated (12). In the early steps of the pathway, vanillin is oxidized to vanillic acid, whose aromatic methyl ether is demethylated through hydroxylation to yield protocatechuic acid and formaldehyde. In contrast to the degradation pathway for protocatechuic acid (11), the fate of formaldehyde in vanillin-utilizing microorganisms has not been studied. Nevertheless, lignin is recognized to be a source of formaldehyde present in nature (3, 20).
In methylotrophic microorganisms, formaldehyde is a key intermediate in the metabolism of several C1 compounds and enters into both dissimilation and assimilation pathways. On the other hand, formaldehyde is a highly reactive compound that has a toxic effect on all organisms through its nonspecific reactivity with proteins and nucleic acids (5, 6). In methylotrophic organisms, both dissimilatory and assimilatory pathways for formaldehyde could contribute to the detoxification of formaldehyde (16).
Recently, genetic studies on the ribulose monophosphate (RuMP) pathway for formaldehyde fixation have been performed with methylotrophic bacteria, Methylomonas aminofaciens 77a (15), Mycobacterium gastri MB19 (9), and Bacillus brevis S1 (25). A homology search of protein databases revealed that the 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI) enzymes in the RuMP pathway exhibit high levels of similarity to a variety of unidentified proteins of nonmethylotrophic prokaryotes, including members of the domains Bacteria and Archaea (22), as well as methylotrophs. In a nonmethylotroph, Bacillus subtilis, both HPS and PHI expression is induced by formaldehyde, and a HPS-negative strain exhibits a formaldehyde-sensitive phenotype (24).
Burkholderia cepacia TM1 grows on several lignin monomers, such as vanillin and vanillic acid, but uses no C1 compound as the sole carbon source (18). From the results of analyses of metabolic intermediates, demethylation of vanillic acid seems to be a rate-limiting step of vanillin degradation (19). In this work, we found that the activities of both HPS and PHI were induced by formaldehyde and by vanillin and vanillic acid in B. cepacia TM1. Furthermore, the vanillic acid degradation was significantly improved on overexpression of the genes for HPS and PHI originating from the methylotrophic bacterium M. aminofaciens 77a. This work suggests that the formaldehyde-fixing enzymes play important roles in the scavenging and fixation of formaldehyde during bacterial degradation of vanillic acid.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
B. cepacia TM1 was isolated from humus as described by Tanaka and Hirokane (18). The broad-host-range vector pBBR122 (4) and its derivatives were used to transform B. cepacia TM1. Each bacterial strain was cultured at 28°C. Luria-Bertani (LB) medium and a mineral salt medium were used to cultivate the bacteria. The basal composition of the mineral salt medium was as follows: 0.26% (wt/vol) (NH4)2SO4, 0.1% K2HPO4, 0.05% KH2PO4, 0.02% MgSO4 · 7H2O, 0.001% CaCl2 · 2H2O, 0.0001% FeSO4 · 7H2O, 0.005% yeast extract, 0.0002% CH3COONH4, and the sole carbon source (at the concentrations indicated in Table 1). The B. cepacia transformants were grown on medium containing kanamycin at a final concentration of 300 μg/ml. Escherichia coli DH5α and pT7-blue (Novagen, Madison, Wis.) were the host vector systems used for cloning of the PCR product. E. coli was grown at 37°C in LB or M9 mineral salt medium (17) in the presence of ampicillin (50 μg/ml) or kanamycin (25 μg/ml), when necessary.
TABLE 1.
HPS and PHI activities in cell extracts of B. cepacia TM1 grown on media with various carbon sourcesa
| Medium and carbon source (concn) | Enzyme activity (U/min/mg of protein)
|
|
|---|---|---|
| HPS | PHI | |
| LB medium | <0.01 | <0.01 |
| LB medium + formaldehyde (0.7 mM) | 0.37 | 0.21 |
| Mineral salt medium + gluconic acid (45 mM) | <0.01 | 0.01 |
| Mineral salt medium + vanillin (15 mM) | 0.30 | 0.20 |
| Mineral salt medium + vanillic acid (15 mM) | 0.28 | 0.15 |
| Mineral salt medium + protocatechuic acid (15 mM) | 0.05 | 0.04 |
The assays were done in duplicate, and each reading did not vary from the mean by more than 30%.
DNA manipulations.
DNA was manipulated as previously described (17). pBRM1 was constructed to express HPS and PHI from B. cepacia TM1. The region containing rmpA, rmpI, and rmpB was amplified by PCR using primers Frmp-A1 (5′-GGAATTCGTTTCAATCGCCTAGATGCC-3′) and Rrmp-B1 (5′-GGAATTCTGATAGAAAGGTAACGGCGA-3′). PCR was performed using EX Taq DNA polymerase (Takara Shuzo Co., Kyoto, Japan) with plasmid pUH1 (23) as the template, which contains the RuMP pathway gene cluster from M. aminofaciens 77a genomic DNA. The PCR product was purified, ligated with pT7-blue, and then introduced into E. coli DH5α. The resultant plasmid was prepared from the E. coli transformant and then digested with EcoRI. The resultant fragment was purified and introduced into the EcoRI site of pBBR122.
Transformation of B. cepacia TM1.
B. cepacia TM1 colonies were grown on 30 ml of SOB medium (17) containing 10 mM MgSO4 to an absorbance at 610 nm of 0.5. The temperature was 28°C. The cells were harvested by centrifugation (7,000 × g) at 4°C for 5 min and then washed twice with ice-cold water. The washed cell pellet was resuspended in an ice-cold 10% glycerol solution. The cells in the resultant suspension, 40 μl, displayed competence. pBBR122 (a control) or pBRM1 (0.5 μg) was transferred into the competent-cell suspension by electroporation using a Gene-Pulser II apparatus (Bio-Rad Laboratories, Hercules, Calif.) as follows: voltage, 2.5 kV; initial field strength, 12.5 kV/cm; resistance, 200 Ω; and capacitance, 25 μF. Transformants were selected by plating the cells on LB medium containing 300 μg of kanamycin per ml.
Cell extract.
Cells were harvested by centrifugation at 7,000 × g, and the cell pellet was washed twice with 50 mM potassium phosphate buffer (pH 7.5). The washed cell pellet was resuspended in 50 mM potassium phosphate buffer containing 5 mM MgCl2, 1 mM dithiothreitol, and 0.15 mM phenylmethylsulfonyl fluoride. Cells were disrupted by sonication (201 M sonicator; Kubota, Tokyo, Japan) at 9 KHz and 170 W for 10 min. The sonicate was clarified by centrifugation at 15,000 × g for 20 min at 4°C. The resultant supernatant was used as the cell extract.
Protein analyses.
Protein concentrations were determined by the Bradford method (2) using a Bio-Rad protein assay kit with bovine serum albumin as a standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7) was performed on a 12.5% polyacrylamide gel. Immunoblotting was performed by the method of Towbin et al. (21), and to detect the complex of HSP and anti-HSP antibody, an ECL detection kit (Amersham Biosciences, Uppsala, Sweden) was used. The activities of HPS and PHI were assayed by measuring the rate of ribulose-5-phosphate-dependent disappearance of formaldehyde as described previously (1).
Formaldehyde fixation into cell constituents.
14C-labeled formaldehyde was purchased from ICN Biomedicals, Inc. (Irvine, Calif.). The transformed strains of B. cepacia were grown on LB medium, which contained 3 mM formaldehyde and 15 μCi of [14C]formaldehyde (specific radioactivity, 57 mCi/mmol). Before preparation of the cell extract, the harvested cells were washed with 50% methanol in order to remove free formaldehyde. Cell extract was prepared by using the BugBuster HT protein extract reagent kit (Merck, Frankfurter, Germany). Each cell extract (50 μg of protein) was spotted on a filter membrane (3MMChr filter paper; Whatman, Maidstone, England) and then exposed to X-ray film (Hyperfilm MP; Amersham Bioscience Co., Piscataway, N.J.) for 2 days. The β-radiation of each cell extract in Clear-sol scintillation fluid (Nacalai Tesque, Kyoto, Japan) was measured with a Tri-Carb 1600CA liquid scintillation counter (Packard, Downers Grove, Ill.).
Formaldehyde and vanillic acid concentration.
The formaldehyde concentration was determined by the method of Nash as previously described (10). Vanillic acid was measured by high-performance liquid chromatography using an octyldecylsilyl-silica gel column (TSKgel ODS-80TM; TOSOH, Tokyo, Japan) as previously described (18).
RESULTS
Formaldehyde fixation activities in B. cepacia TM1.
Table 1 shows the HPS and PHI activities of cell extracts of B. cepacia TM1 grown in a variety of media. Significant activities of both HPS and PHI were detected in vanillin- and vanillic acid-grown cells, but no or only very weak activities of the two enzymes were found in cells grown on LB medium and the medium containing gluconate or protocatechuic acid as the sole carbon source. When the bacterium was grown on LB medium, the activities of HPS and PHI were induced by formaldehyde (0.7 mM), and the levels of the two enzyme activities were the same as those in the vanillin- and vanillic acid-grown cells. Taking into account the degradation pathway for vanillic acid (14), these results suggest that HPS and PHI are induced by formaldehyde derived from the growth substrate, i.e., vanillin or vanillic acid.
Expression of M. aminofaciens hps and phi in B. cepacia TM1.
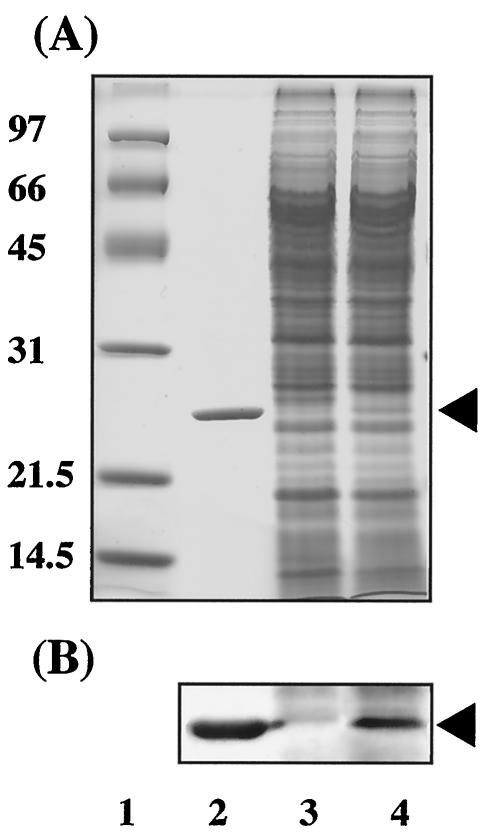
The HPS and PHI expression plasmid pBRM1, which carries rmpA, rmpI, and rmpB with the original promoter of M. aminofaciens 77a (15), was constructed and then introduced into B. cepacia TM1 by electroporation. The pBBR122-transformed strain was used as the control strain. A cell extract was prepared from each strain grown on LB medium. As shown by the SDS-PAGE profiles (Fig. 1A), a band corresponding to the molecular mass of HPS from M. aminofaciens 77a was detected in the lane containing B. cepacia TM1(pBRM1), and HPS antibodies recognized this band on immunoblot analysis (Fig. 1B). The specific activities of HPS and PHI in cell extracts of B. cepacia TM1(pBBR122) grown on LB medium lacking formaldehyde were both <0.01 U · min−1 · mg of protein−1. The specific activities of HPS and PHI in B. cepacia TM1(pBRM1) grown on LB medium lacking formaldehyde were 0.94 and 0.47 U · min−1 · mg of protein−1, respectively. These assays were done in duplicate, and each reading did not vary from the mean by more than 10%. The specific activities of HPS and PHI in cell extracts of B. cepacia TM1(pBRM1) were 2.5- and 2.2-fold higher, respectively, than those in the wild-type strain grown on LB medium containing 0.7 mM formaldehyde. There was a slight increase in enzyme activities in the presence of formaldehyde. This increase was comparable in the presence or absence of the plasmid-borne genes, which suggests that the increase is derived from low-level induction of the indigenous HPS and PHI of B. cepacia TM1. These results suggested that the promoter originating from the plasmid contributes to the expression of HPS and PHI; however, expression of HPS and PHI is not regulated by formaldehyde in the cells of B. cepacia TM1. Since HPS and PHI are expressed only in the presence of formaldehyde in B. cepacia TM1, both activities of the transformant cells grown on LB medium without formaldehyde are assumed to be the products of genes on plasmid pBRM1.
FIG. 1.
SDS-PAGE of a cell extract prepared from B. cepacia TM1 transformants grown on LB medium. (A) The bands on the SDS-polyacrylamide gel were stained with Coomassie brilliant blue R-250. (B) Immunoblot analysis with anti-HPS polyclonal antibodies. Lane 1, molecular mass standards; lane 2, purified HPS obtained from M. aminofaciens 77a; lane 3, B. cepacia TM1(pBBR122); lane 4, B. cepacia TM1(pBRM1). In panel A, the positions of molecular mass standards (in kilodaltons) are indicated to the left of the gel. The position of HPS is indicated by arrowheads to the right of the gels.
Growth of transformants on LB medium containing formaldehyde.
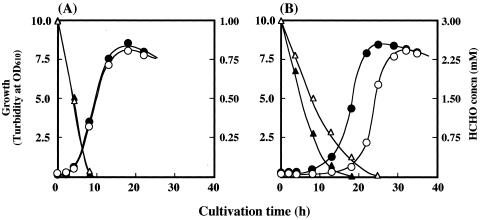
The effects of formaldehyde on the growth of B. cepacia TM1(pBRM1) and control strain B. cepacia TM1(pBBR122) were compared (Fig. 2). The growth profiles of the two strains in LB medium containing 1 mM formaldehyde were almost the same. In LB medium containing 3 mM formaldehyde, the lag period of B. cepacia TM1(pBRM1) was much shorter than that of the control strain, and the rate of formaldehyde consumption by the transformant was clearly faster than that by the control strain. Once both strains had begun to grow, however, the growth rates and final cell yields were almost the same. These findings indicate that formaldehyde inhibits the initial growth of the parent strain and that the transformant is capable of overcoming the toxicity of formaldehyde more efficiently. When the formaldehyde concentration in LB medium was increased to 5 mM, neither strain grew during the experimental period (data not shown).
FIG. 2.
Growth profiles of B. cepacia TM1 transformants grown on LB medium containing formaldehyde at a final concentration of 1 mM (A) or 3 mM (B). Growth of B. cepacia TM1(pBRM1) (•) and TM1(pBBR122) (○) and formaldehyde concentration (triangles) in the culture medium of B. cepacia TM1(pBRM1) (▴) and TM1(pBBR122) (▵) are shown. OD610, optical density at 610 nm.
Formaldehyde uptake into the biomass.
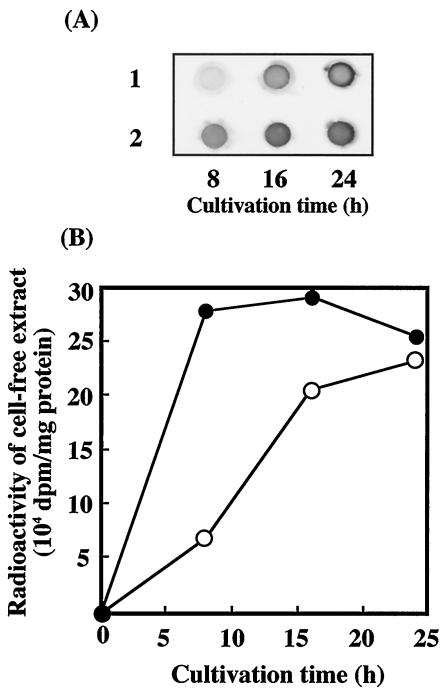
The incorporation of [14C]formaldehyde into cells grown on LB medium was examined. The radioactivity of the cell extracts of the B. cepacia transformants, TM1(pBRM1) and TM1(pBBR122), increased with growth (Fig. 3), and the incorporation rate of strain TM1(pBRM1), in which hps and phi were overexpressed, was much higher than that of the control strain, TM1(pBBR122). These results suggest that formaldehyde is incorporated into the cell constituents of both strains through a metabolic pathway involving HPS and PHI.
FIG. 3.
Formaldehyde incorporation of B. cepacia TM1 transformants grown on LB medium containing [14C]formaldehyde. (A) X-ray film image for 14C activities of the extracted proteins (50 μg for each dot). B. cepacia TM1(pBBR122) (row 1) and TM1(pBRM1) (row 2) grown for various times were tested. (B) β-Radiation of the extracted protein (150 μg). Symbols: •, B. cepacia TM1(pBRM1); ○, B. cepacia TM1(pBBR122).
Growth of the transformants on vanillic acid.
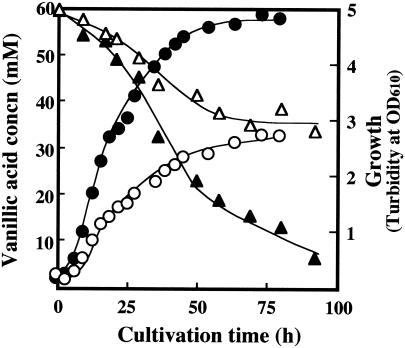
B. cepacia TM1(pBRM1) and the control strain, TM1(pBBR122), were grown on mineral salt medium containing vanillic acid as the sole source of carbon. Several concentrations of vanillic acid were tested. The growth profiles of the two strains in the medium containing vanillic acid at 10 to 40 mM were almost the same. As shown in Fig. 4, significant differences between the two strains were observed in the utilization of vanillic acid at a higher concentration (60 mM). B. cepacia TM1(pBRM1) exhibited a higher growth rate and yield and also a higher rate and degree of substrate consumption. In the control strain, when 60 mM vanillic acid was provided, no more than 25 mM was consumed. However, the control strain completely consumed vanillic acid up to 40 mM (data not shown). With a higher concentration of vanillic acid (60 mM), since the transformant strain constitutively expressed the formaldehyde-fixing enzyme system at a level higher than that of the control strain, the transformant strain could detoxify and assimilate formaldehyde more efficiently than the control strain. However, with a lower concentration of vanillic acid (<40 mM), it is speculated that the endogenous expression of HPS and PHI tolerates formaldehyde toxicity.
FIG. 4.
Growth profiles of B. cepacia TM1 transformants grown on mineral salt medium containing vanillic acid as the sole carbon source at a final concentration of 60 mM. Growth of B. cepacia TM1(pBRM1) (•) and TM1 [pBBR122] (○) and vanillic acid concentration (triangles) in the culture medium of B. cepacia TM1(pBRM1) (▴) and TM1(pBBR122) (▵) are shown. OD610, optical density at 610 nm.
DISCUSSION
In this work, we focused on the fate of formaldehyde during the degradation of a lignin monomer, vanillin or vanillic acid, by B. cepacia TM1 and revealed that formaldehyde-fixing reactions in the RuMP pathway could play an important role in formaldehyde detoxification during growth on these lignin monomers. Furthermore, the utilization of vanillic acid by B. cepacia TM1 was significantly improved on overexpression of the HPS and PHI genes from M. aminofaciens 77a. B. cepacia TM1 incorporated formaldehyde into the cell constituents, and the incorporation rate was much improved by overexpression of hps and phi. These results suggest that the formaldehyde released in the vanillic acid demethylase reaction is fixed into the cell mass through the RuMP pathway, as shown in Fig. 5. The original strain exhibited lower HPS and PHI activities than the methylotrophic bacterium M. aminofaciens 77a. Therefore, the formaldehyde utilization reaction might be rate-limiting in the utilization of vanillic acid by B. cepacia TM1, especially at high concentrations of vanillic acid. Apparently, the overexpressed enzymes contribute to the suppression of formaldehyde toxicity. Also, the formaldehyde fixation could contribute to the increase in cell yield on vanillic acid, as shown in Fig. 4. This transformant will provide a more effective means for the biological conversion of lignin monomers into useful compounds.
FIG. 5.
Proposed pathway of vanillic acid degradation linked with the RuMP formaldehyde fixation pathway in B. cepacia TM1. Formaldehyde derived from vanillic acid is incorporated into the RuMP pathway. Ribulose-5-phosphate, the acceptor of formaldehyde, may be regenerated from fructose-6-phosphate as in the RuMP pathway of methylotrophs. Enzyme 1, vanillin dehydrogenase; enzyme 2, vanillic acid demethylase.
It is evident that genes for the formaldehyde-fixing system are widely distributed in nonmethylotrophic Bacteria and Archaea, as well as methylotrophic bacteria. The physiological roles of HPS and PHI in the nonmethylotrophic organisms have been confirmed using B. subtilis. The organism produces the enzymes only when the growth medium contains formaldehyde and detoxifies the formaldehyde through enzyme reactions (24). In the case of B. cepacia, the detoxification of formaldehyde is indispensable for growth on lignin monomers, which are the main carbon sources in the original habitat of this strain. Many methoxyl groups of lignin are believed to be released through microbial activity and to be an important source of C1 compounds in nature. This work suggests that nonmethylotrophic bacteria that use lignin monomers can incorporate formaldehyde into cell constituents through formaldehyde-fixing reactions. It is speculated that such a microbial metabolism of C1 compounds considerably contributes to the carbon flow in nature.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to R.M.
REFERENCES
- 1.Arfman, N., K. J. de Vries, H. R. Moezelaar, M. M. Attwood, G. K. Robinson, M. van Geel, and L. Dijkhuizen. 1992. Environmental regulation of alcohol metabolism in thermotolerant methylotrophic Bacillus strains. Arch. Microbiol. 157:272-278. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Codd, G. A., L. Dijkhuzin, and F. R. E. Tabita. 1990. Advances in autotrophic microbiology and one-carbon metabolism, vol. 1, p. 163-191. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Elzer, P. H., M. E. Kovach, R. W. Phillips, G. T. Robertson, K. M. Peterson, and R. M. Roop II. 1995. In vivo and in vitro stability of the broad-host-range cloning vector pBBR1MCS in six Brucella species. Plasmid 33:51-57. [DOI] [PubMed] [Google Scholar]
- 5.Ferdman, M. Y. 1973. Reaction of nucleic acids and nucleoproteins with formaldehyde. Prog. Nucleic Acid Res. Mol. Biol. 13:1-49. [DOI] [PubMed] [Google Scholar]
- 6.Grafstrom, R. C., A. J. Fornace, Jr., H. Autrup, J. F. Lechner, and C. C. Harris. 1983. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science 220:216-218. [DOI] [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Mathias, A., and A. E. Rodrigues. 1995. Production of vanillin by oxidation of pine kraft lignins with oxygen. Holzforschung 49:273-278. [Google Scholar]
- 9.Mitsui, R., Y. Sakai, H. Yasueda, and N. Kato. 2000. A novel operon encoding formaldehyde fixation: the ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19. J. Bacteriol. 182:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nash, T. 1953. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem. J. 55:416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overhage, J., A. U. Kresse, H. Priefert, H. Sommer, G. Krammer, J. Rabenhorst, and A. Steinbuchel. 1999. Molecular characterization of the genes pcaG and pcaH, encoding protocatechuate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overhage, J., H. Priefert, and A. Steinbuchel. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Priefert, H., J. Rabenhorst, and A. Steinbuchel. 2001. Biotechnological production of vanillin. Appl. Microbiol. Biotechnol. 56:296-314. [DOI] [PubMed] [Google Scholar]
- 14.Priefert, H., J. Rabenhorst, and A. Steinbuchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai, Y., R. Mitsui, Y. Katayama, H. Yanase, and N. Kato. 1999. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol. Lett. 176:125-130. [DOI] [PubMed] [Google Scholar]
- 16.Sakai, Y., T. Nakagawa, M. Shimase, and N. Kato. 1998. Regulation and physiological role of the DAS1 gene, encoding dihydroxyacetone synthase, in the methylotrophic yeast Candida boidinii. J. Bacteriol. 180:5885-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Tanaka, M., and H. Hirokane. 2000. Oxidation of aromatic aldehyde to aromatic carboxylic acid by Burkholderia cepacia TM1 isolated from humus. J. Biosci. Bioeng. 90:341-343. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, M., Y. Hirokane, R. Mitsui, and T. Tsuno. 2001. Continuous oxidation of aromatic aldehyde to aromatic carboxylic acid by Burkholderia cepacia TM1 in a cell-holding reactor. J. Biosci. Bioeng. 91:267-271. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, B. F. 1983. Aerobic and anaerobic catabolism of vanillic acid and some other methoxy-aromatic compounds by Pseudomonas sp. strain PN-1. Appl. Environ. Microbiol. 46:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towbin, H., T. Staehelin, and J. Gordon. 1992. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Bio/Technology 24:145-149. [PubMed] [Google Scholar]
- 22.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanase, H., K. Ikeyama, R. Mitsui, S. Ra, K. Kita, Y. Sakai, and N. Kato. 1996. Cloning and sequence analysis of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microb. Lett. 135:201-205. [DOI] [PubMed] [Google Scholar]
- 24.Yasueda, H., Y. Kawahara, and S. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yurimoto, H., R. Hirai, H. Yasueda, R. Mitsui, Y. Sakai, and N. Kato. 2002. The ribulose monophosphate pathway operon encoding formaldehyde fixation in a thermotolerant methylotroph, Bacillus brevis S1. FEMS Microb. Lett. 214:189-193. [DOI] [PubMed] [Google Scholar]